SUMMARY
Expansin proteins, which loosen plant cell walls, play critical roles in normal plant growth and development. The horizontal acquisition of functional plant-like expansin genes in numerous xylem-colonizing phytopathogenic bacteria suggests that bacterial expansins may also contribute to virulence. To investigate the role of bacterial expansins in plant diseases, we mutated the non-chimeric expansin genes (CmEXLX2 and RsEXLX) of two xylem-inhabiting bacterial pathogens, the Actinobacterium Clavibacter michiganensis subsp. michiganensis (Cmm) and the β-proteobacterium Ralstonia solanacearum (Rs), respectively. The Cmm ΔCmEXLX2 mutant caused increased symptom development on tomato, which was characterized by more rapid wilting, greater vascular necrosis and abundant atypical lesions on distant petioles. This increased disease severity correlated with larger in planta populations of the ΔCmEXLX2 mutant even though the strains grew equally well as wildtype in vitro. Similarly, when inoculated onto tomato fruit, ΔCmEXLX2 caused significantly larger lesions with larger necrotic centers. In contrast, the Rs ΔRsEXLX mutant had reduced virulence on tomato following root inoculation, but not following direct petiole inoculation, suggesting that the RsEXLX expansin contributes to early virulence at the root infection stage. Consistent with this finding, ΔRsEXLX attached to tomato seedling roots better than wildtype Rs, which may prevent mutants from invading the plant’s vasculature. These contrasting results demonstrate the diverse roles of non-chimeric bacterial expansins and highlight their importance in plant-bacterial interactions.
Keywords: horizontal gene transfer, expansin, bacterial plant pathogenesis, Clavibacter michiganensis, Ralstonia solanacearum
INTRODUCTION
Plant primary cell walls are highly dynamic structures that fluctuate between rigid and relaxed states, enabling basic biological processes such as growth, enlargement, and cell division (Cosgrove, 1993; Cosgrove, 2005). Regulating cell wall elasticity and plasticity is an intricate process that requires many cell wall loosening enzymes, including expansins (Cosgrove, 1993; Sampedro and Cosgrove, 2005). Plant expansins loosen the rigid carbohydrate matrix of the cell wall through an uncharacterized non-lytic slippage mechanism (Cosgrove, 2000). This key biological function may explain why expansins are conserved in all vascular plants, with isoforms regulating cell wall changes required for cellular growth, vascular differentiation, fruit ripening, seed germination, abscission, and leaf development (Cho and Cosgrove, 2000; Im et al., 2000; Kende et al., 2004; Rose et al., 1997). Expansins are pH-dependent and are activated when plant growth hormones stimulate H+-ATPases, which lower the pH of the cell wall matrix by producing a proton differential across the plasma membrane (Cosgrove, 2000).
Recent evidence suggests that diverse microbes independently acquired and exploited this plant-derived enzyme for unknown reasons (Nikolaidis et al., 2014). It is hypothesized that multiple independent horizontal gene transfers have led to the microbial acquisition of plant expansins, with subsequent horizontal gene transfer events within bacterial and fungal phyla (Nikolaidis et al., 2014). Expansins are rare in bacteria; only 3% of sequenced bacteria have expansin xenologs, and these bacterial genera represent diverse ecological niches from free-living saprophytes to plant pathogens (Nikolaidis et al., 2014). Interestingly, systemic xylem pathogens are present in all plant pathogenic genera with an expansin gene: Xanthomonas, Xylella, Ralstonia, Dickeya, Pectobacterium, Acidovorax, and Clavibacter (Georgelis et al., 2015; Nikolaidis et al., 2014). The only exception is the genus Streptomyces (Nikolaidis et al., 2014). However, no expansin xenologs are present among the non-vascular phytopathogenic bacterial genera Pseudomonas or Agrobacterium (Nikolaidis et al., 2014). Two forms of expansins have been described in bacteria: a chimeric version, which has an endoglucanase domain fused to the expansin domain, and a non-chimeric version (Jahr et al., 2000; Nikolaidis et al., 2014). Most bacteria contain only a single expansin (either chimeric or non-chimeric), with the notable exception of the tomato pathogen Clavibacter michiganensis subsp. michiganensis (Cmm), which has both a chimeric (CmEXLX1) and non-chimeric version (CmEXLX2) (Georgelis et al., 2015; Nikolaidis et al., 2014). Another vascular tomato pathogen, Ralstonia solanacearum (Rs) only has a non-chimeric expansin, RsEXLX (Nikolaidis et al., 2014).
Cmm, the internationally quarantined causal agent of tomato bacterial canker, is an economically devastating seed-disseminated pathogen found in all major tomato-producing regions (Bryan, 1930; de León et al., 2011). Cmm enters plants epiphytically through natural openings, wounds, or infected seed (Bryan, 1930; Carlton et al., 1998; Tancos et al., 2013). Once inside its host, this bacterium systemically spreads throughout the vasculature, plugging and degrading xylem vessels, resulting in significant tissue maceration and impaired water transport, which in turn leads to characteristic wilting, marginal necrosis of leaflets, stem cankers, and fruit lesions (Bryan, 1930; Tancos et al., 2013; Wallis, 1977). Bacterial canker of tomato is difficult to control due to the lack of resistant cultivars and improper sanitation (Sen et al., 2013; Werner et al., 2002).
Rs is a soil-borne β-proteobacteria vascular pathogen that infects a wide host range of economically important crops, including monocots and dicots (Elphinstone, 2005). Root attachment is a critical early step in Rs pathogenesis (Yao and Allen, 2006). Rs infects roots near the elongation zone, secondary root emergence sites, and wounds before the bacterium penetrates the vascular cylinder and colonizes the xylem (Vasse et al., 1995). Rs spreads systemically throughout the vasculature, reaching population sizes >109 CFU/g of stem tissue (Tans-Kersten et al., 2004). These large bacterial populations, which produce abundant extracellular polymeric substances and macerate tissue, reduce xylem sap flow and cause wilting (Vasse et al., 1995). Like Cmm, Rs is difficult to manage due to the lack of resistant plant cultivars or other effective control strategies (Bae et al., 2015).
Microbial expansins are structurally similar to plant expansins, and the unique cellulose binding domains are highly conserved (Kerff et al., 2008; Pastor et al., 2015). Purified bacterial expansins from Bacillus subtilis, Xanthomonas campestris, Rs, and Cmm loosen plant cell walls without lytic activity in vitro, but their activity is modest relative to that of plant expansins (Bunterngsook et al., 2015; Georgelis et al., 2014; Kerff et al., 2008). Recent studies have shown that a non-chimeric expansin mutant of the saprophytic biocontrol agent B. subtilis had reduced attachment to maize roots (Kerff et al., 2008), but, in contrast, Xylella fastidiosa and Cmm mutants lacking the endoglucanase-expansin chimeras caused reduced disease symptoms (Ingel et al., 2015; Jahr et al., 2000). However, the function(s) of non-chimeric expansins remain unknown, especially in phytopathogenic species.
Here we investigate the effect of non-chimeric bacterial expansins on pathogenesis and colonization of tomato by two vascular pathogens: the Gram-positive Cmm and the Gram-negative Rs. Because of the pathogens’ niche overlap, we hypothesized that non-chimeric expansins from Cmm and Rs would contribute similarly to vascular colonization. However, we found contrasting roles for expansins in these two systems. While the Cmm ΔCmEXLX2 mutant was hypervirulent and grew faster in planta, an Rs ΔRsEXLX mutant hyper-attached to tomato roots and was reduced in virulence after root inoculation.
RESULTS
We used a genetic approach to investigate the roles of non-chimeric plant-like bacterial expansins in the pathogenesis of two vascular plant pathogenic bacteria. Gene nomenclature for bacterial expansins was based on the current standard for microbial expansins (Georgelis et al., 2015; Kende et al., 2004). CmEXLX2 was disrupted in a virulent Cmm New York strain (Cmm0317) that naturally lacked the CmEXLX1 plasmid-borne chimeric expansin (Tancos et al., 2015). As expected, the Cmm ΔCmEXLX2 and Rs ΔRsEXLX mutants did not express CmEXLX2 or RsEXLX, while the wildtype and complemented mutants both did (Figure S1, Figure S2). In vitro growth of the Cmm strains was not significantly different from wildtype in nutrient-rich LB medium (P=0.994) and nutrient-poor tomato xylem sap (P=0.729) (Figure S3). Similarly, ΔRsEXLX grew at the same rate as wildtype in both rich (P=0.493) and minimal medium (P=0.301) (Figure S3). The CmEXLX2 mutation did not influence host recognition in Mirabilis jalapa (four o’clock plants). ΔCmEXLX2 induced a strong hypersensitive response (HR) indistinguishable from the HR induced by the wildtype and complemented mutant strain (Table 1).
TABLE 1.
Virulence of Clavibacter michiganensis subsp. michiganensis strains on tomato plants.
| Strain | HRa | AUDPCb | Disease incidencec | Petiole lesion length (cm)d | CFU/g in plantae
|
|
|---|---|---|---|---|---|---|
| 9 DPI | 21 DPI | |||||
| Cmm0317 | + | 139 (± 36) B | 15/15 | 0 | 2.78 × 108 B | 5.36 × 108 C |
| ΔCmEXLX2 | + | 430 (± 64) A | 15/15 | 2.16 (±0.70) | 3.72 × 109 A | 4.82 × 109 A |
| CΔCmEXLX2+ | + | 157 (± 21) B | 15/15 | 0.59 (±0.21) | 2.25 × 108 B | 2.30 × 109 B |
| Water control | − | 0.0 | 0/15 | 0 | 0.0 | 0.0 |
Induction of a hypersensitive response (HR) in Mirabilis jalapa. (+) positive for HR reaction; (−) negative for HR reaction.
The mean area under the disease progress curve (AUDPC) for disease severity for three independent experiments is given. Differences among strains were determined with PROC GLIMMIX (P<0.05) followed by Tukey-Kramer posttest (P<0.05). AUDPC values followed by the same superscript are not significantly different. Numbers in parentheses indicate the standard error associated with the AUDPC values.
Expressed as the number of wilting plants/number of inoculated plants characterized at 21 days post inoculation.
The mean length of lesions located on the inoculated petiole at 21 days post inoculation. Numbers in parentheses indicate the standard error associated with the lesion lengths.
The mean in planta population recovered from a 0.5 cm section of tomato stem tissue located 1-cm above the inoculation site at both 9 and 21 days post inoculation. Significant differences among strains were tested by PROC GLIMMIX (P<0.05) followed by Tukey-Kramer posttest (P<0.05). In planta populations followed by the same superscript are not different.
Plant expansins facilitate cell wall loosening as plant cells develop and grow. While the bacterial BsEXLX1 expansin contributed to the autolysis of B. subtilis, BsEXLX1 lacked peptidoglycan-lytic activity (Kerff et al., 2008). Because BsEXLX1 influenced B. subtilis cell wall integrity, we measured the autolysis rates of ΔCmEXLX2 and ΔRsEXLX in Cmm and Rs, respectively. Autolysis of ΔCmEXLX2 and ΔRsEXLX did not differ from the corresponding wildtype or complemented mutant strains (P=0.141 for Cmm and P=0.086 for Rs) (Figure S4).
The Cmm ΔCmEXLX2 mutant causes increased symptoms on tomato seedlings and fruit
Tomato seedlings inoculated with ΔCmEXLX2 consistently developed earlier and more severe wilt symptoms than the wildtype or complemented strains (P<0.0001) (Table 1). While 60% of plants inoculated with the wildtype and complement developed symptoms by 15 and 13 days post inoculation (DPI), respectively, ΔCmEXLX2 reached this disease incidence at 10 DPI. All three Cmm strains produced typical stem cankers at the site of inoculation, but ΔCmEXLX2 also produced multiple atypical cankers in the vascular bundles of distant petioles that appeared approximately 11–14 DPI (Table 1, Figure 1). These atypical petiole lesions extended along the tracks of the vascular bundles; necrosis was confined to the vasculature, with healthy tissue surrounding the lesions until later stages of disease development (Figure 1). Lesions on petioles were never observed on plants inoculated with the wildtype strain and were rarely formed by the complemented mutant strain.
FIGURE 1.
Atypical tomato petiole lesions associated with the Clavibacter michiganensis subsp. michiganensis expansin mutant ΔCmEXLX2. (A) An individual petiole lesion rupturing a vascular bundle. Necrosis appeared confined to the vasculature with healthy plant tissue surrounding the petiole lesions. (B) Multiple lesions localized to the vasculature of an individual petiole. Black arrows highlight the large canker lesions present along multiple vascular tracks.
The ΔCmEXLX2 mutant reached higher populations in planta at 1 cm above the inoculation site at both 9 DPI (P=0.0048) and 21 DPI (P<0.0001) (Table 1). To determine if systemic movement influenced symptom development and in planta population sizes, we also assessed Cmm populations 5 and 10 cm above the inoculation site at 21 DPI. All three Cmm strains were detectable 5 and 10 cm above the site of inoculation (Table S1A). Consistent with our findings at 1 cm above the inoculation site, the ΔCmEXLX2 population (2.41×109 CFU/g) at 5 cm was larger (P=0.024) than wildtype (6.85×108 CFU/g). However, population sizes of the two strains were not different 10 cm above the inoculation site (P=0.46) (Table S1A).
Plant vasculature enables water and nutrient transport; it is composed of a network of vascular bundles that contain aggregated xylem vessels and phloem tissue. To determine if CmEXLX2 influences movement of Cmm between xylem vessels or vascular bundles, we inoculated tomato plants with EGFP-expressing Cmm strains and visualized intra- or intervascular colonization. EGFP-expressing Cmm wildtype (CmEXLX2+) and ΔCmEXLX2 strains were both visible in xylem vessels at 3 cm above and below the inoculation site at 5, 7, and 9 DPI (Table S2A–B). This qualitative approach revealed no detectable differences in patterns of xylem colonization, lateral movement, or parenchyma cell colonization between the strains at any time point or distance from the inoculation site. By 9 DPI, all Cmm strains were present at high concentrations, and bacteria were visible in macerated vessel elements and in the surrounding parenchyma cells (Table S2B, Figure S5).
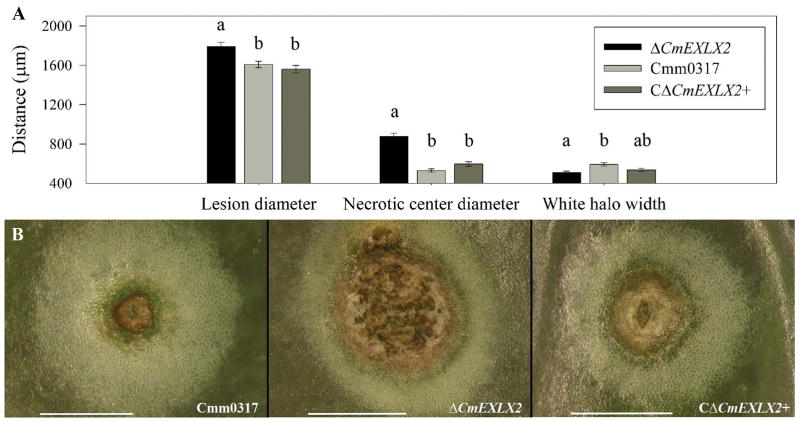
When inoculated onto immature green fruit, all three Cmm strains produced characteristic bacterial canker lesions, which have a necrotic center surrounded by a white halo. Fruit lesions appeared 3–4 DPI, but phenotypic differences between strains became evident at 7–10 DPI. The ΔCmEXLX2 lesions were larger and had more necrosis than lesions caused by either the wildtype or complemented mutant strain (P<0.0001), but ΔCmEXLX2 lesions were surrounded by thinner white halos compared to the wildtype lesions (P=0.0008) (Figure 2).
FIGURE 2.
Deleting Clavibacter michiganensis subsp. michiganensis (Cmm) expansin gene CmEXLX2 accelerates tomato fruit lesion development (A) Lesion and halo sizes on tomato fruit (n=100) inoculated with the three Cmm strains. Fruit lesion distances followed by the same letter are not significantly different. Differences among treatments for mean lesion size were determined with PROC GLIMMIX (P<0.05) followed by Tukey-Kramer posttest (P<0.01). (B) Representative images of tomato fruit lesions are shown below: Cmm0317 (wildtype), ΔCmEXLX2 (expansin mutant), and CΔCmEXLX2+ (complement). Scale bar: 1 mm. Error bars correspond to the standard error.
Symptom development on tomato seedlings infected with the Rs ΔRsEXLX mutant
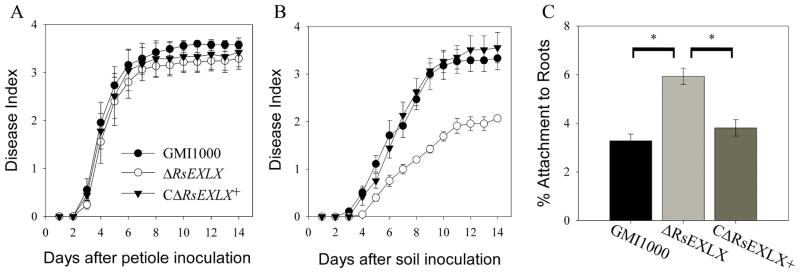
Since plant-like expansins have been acquired by many xylem-dwelling plant pathogenic bacteria, we hypothesized that expansins would also affect the virulence of another xylem pathogen, Rs. Because Rs infects host plants via the roots before systemically colonizing the xylem, two inoculation methods were used to distinguish possible roles of the Rs expansin. A soil-drench inoculation of unwounded plants mimics the natural route of Rs infection, while the cut-petiole technique bypasses root infection by introducing Rs directly into the xylem. In contrast to Cmm, ΔRsEXLX had wildtype virulence when directly introduced into tomato stems (P=0.106), but was reduced in virulence following soil-drench inoculation (P=0.0465) (Figure 3A–B, Figure S6). By 14 DPI, only 56% of plants inoculated with ΔRsEXLX displayed symptoms, compared to 87% and 89% of plants inoculated with the wildtype and complemented mutant strains, respectively.
FIGURE 3.
Virulence and root attachment of Ralstonia solanacearum (Rs) expansin mutant ΔRsEXLX. (A) Bacteria were directly inoculated into tomato stem vasculature by placing 500 CFU onto the surface of a freshly-cut leaf petiole (P=0.1056, repeated measures ANOVA), or (B) plants were naturalistically inoculated by drenching the soil around plants with ~5×108 CFU bacteria /g soil (P=0.0465, repeated measures ANOVA). Experiments were repeated 3 times with total N=45 plants per strain. (C) 104 CFU Rs were incubated for 2h with sterile tomato seedling roots. Percent of bacteria attached to roots after washing was measured by serial dilution plating ground roots. * indicates P<0.0001 by ANOVA. Error bars correspond to the standard error.
Expansins influence Rs, but not Cmm, attachment to host roots
The virulence defect of ΔRsEXLX following soil-drenching, but not direct xylem inoculation, suggested that RsEXLX contributes to early infection. To investigate the initial step of the infection process, bacterial attachment of Cmm and Rs to tomato seedling roots was measured. The ΔRsEXLX mutant hyper-attached to roots in comparison to wildtype (P<0.0001). Following a 2-hour incubation of tomato seedlings in a bacterial suspension, 6% of the ΔRsEXLX population attached to roots compared to only 3.3% and 3.8% of the wildtype and complemented strain populations, respectively (Figure 3C). In contrast, there were no differences in root attachment between the three Cmm strains (P=0.4647) (Table S1B).
To determine if the attachment phenotype was root-specific, we measured attachment of Rs and Cmm strains with an in vitro biofilm formation assay (Kwasny and Opperman, 2010). No differences in attachment were detected between the Rs wildtype and expansin mutant strains (P=0.111), or Cmm strains (P=0.117), but the Cmm complemented strain produced less biofilm than wildtype (Figure S7).
Heterologous expression of Rs expansin in Cmm
Because the ΔCmEXLX2 and ΔRsEXLX mutants had contrasting virulence phenotypes, we tested whether the heterologous expression of RsEXLX could complement the hypervirulence of ΔCmEXLX2. RsEXLX was fused with the Cmm expansin secretion sequence and promoter so that the Rs gene was expressed and secreted like the Cmm expansin; this hybrid fusion strain was named CΔCmP:RsEXLX + (Figure S8). Similar to previous Cmm experiments, no differences in root attachment were observed between strains (P=0.4647). The expansin-hybrid also behaved similarly to ΔCmEXLX2 and the complemented mutant, producing less biofilm than the wildtype strain (P=0.0094) when grown in pure tomato xylem sap. The expansin-hybrid did not restore the Cmm-wildtype-level virulence and canker severity when inoculated into tomato, but had a trend of less disease than ΔCmEXLX2 (P=0.5230) (Figure S9). Although both CmEXLX2 and RsEXLX are non-chimeric expansins, they only share 27% amino acid identity, which could explain why RsEXLX did not restore wildtype-level virulence (Table 2, Figure S10).
TABLE 2.
Overview of bacterial phenotypes associated with the mutation of bacterial expansins.
| Bacteria | Lifestyle | Primary route of colonization | Protein structure | Gene | % identitya | Expansin mutant phenotype
|
Reference | |
|---|---|---|---|---|---|---|---|---|
| Disease symptoms | Root attachment | |||||||
| Clavibacter michiganensis subsp. michiganensis | Vascular phytopathogen | Foliar | Non-chimeric | CmEXLX2 | 100 (100) | Increase | No difference | This study |
| Clavibacter michiganensis subsp. michiganensis | Vascular phytopathogen | Foliar | Chimeric | CmEXLX1 | 17 (54) | Decrease | n/a | (Jahr et al., 2000) |
| Ralstonia solanacearum | Vascular phytopathogen | Root | Non-chimeric | RsEXLX | 27 (34) | Decrease | Hyper-attach | This study |
| Bacillus subtilis | Saprophyte | Root | Non-chimeric | BsEXLX1 | 28 (36) | n/a | Decrease | (Kerff et al., 2008) |
Percent amino acid identify of the full-length protein, relative to the non-chimeric Clavibacter michiganensis subsp. michiganensis expansin CmEXLX2. The number in parenthesis denotes the amino acid identity of the most similar regions via the pairwise sequence alignment tool EMBOSS Matcher (Goujon et al., 2010).
DISCUSSION
Functional plant-like bacterial expansins were first studied in B. subtilis, and then identified in a wide array of plant-associated bacteria (Kerff et al., 2008; Nikolaidis et al., 2014). However, the biological function of non-chimeric bacterial expansins, which lack endoglucanase domains, has not been investigated in phytopathogenic bacteria. We found the loss of non-chimeric bacterial expansins divergently affected virulence of two vascular pathogens of tomato, Cmm and Rs (Nikolaidis et al., 2014).
The ΔCmEXLX2 mutant caused more severe disease on tomato, characterized by a faster onset of unilateral wilting, increased necrosis, and larger pathogen population sizes. Although all three Cmm strains produced typical stem lesions at the site of inoculation, only ΔCmEXLX2 caused atypical necrotic lesions on the vasculature of distant petioles. The quick onset of symptoms by ΔCmEXLX2 was not correlated with increased intra- or intervascular spread, because EGFP-expressing strains had similar rates of vascular infection both above and below the site of inoculation. In parallel with the increased stem tissue necrosis, ΔCmEXLX2 caused larger necrotic tomato fruit lesions. Fruit infected with ΔCmEXLX2 had larger, more blistered lesions with more necrosis and less noticeable ‘white halos’. Unfortunately, fruit lesions are a relatively unexplored disease symptom associated with bacterial canker, and the significance of the ‘white halo’ that surrounds the lesion remains unknown.
The ΔCmEXLX2 mutant strain reached larger in planta populations at 1 and 5 cm above the site of inoculation. However, the differences in population size between Cmm strains diminished as acropetal regions of the stem were colonized; at 10 cm above the inoculation site, population sizes of the three strains were not significantly different. Therefore, acropetal in planta populations and the vascular GFP-movement study both demonstrate that systemic movement alone does not appear to have caused the increased in planta populations. Because ΔCmEXLX2 growth was not significantly different from wildtype in vitro, the mutant’s increased in planta population size could have been due to the increased necrosis induced by the mutant. The older, more established ΔCmEXLX2 populations near the inoculation site may degrade vascular tissue more quickly, which likely released plant-derived nutrients into the nutrient-poor xylem (Fatima and Senthil-Kumar, 2015). The greater rates of necrosis observed on the stems, and fruit, may be directly associated with the increased availability of plant-derived nutrients and subsequent influx of bacterial growth as established populations macerate plant tissue.
The molecular mechanism underlying the increased necrosis of ΔCmEXLX2 is unknown. We favor the hypothesis that CmEXLX2 competes with the numerous Cmm cell-wall degrading enzymes (CWDEs) for unique binding sites on the plant cell wall. In support of this idea, Georgelis et al. (2012) demonstrated that the B. subtilis BsEXLX1 expansin competes with type-A cellulose-binding modules for binding sites in crystalline cellulose, which is likely due to the similarity between the D2 domains of BsEXLX1 and other type-A cellulose-binding modules (Georgelis et al., 2012). While canonical CWDEs like pectinases, cellulases, and hemicellulases cleave plant cell wall polysaccharides, plant α-expansins induce rapid cell wall extension with no lasting structural changes (McQueen-Mason et al., 1992; Yuan et al., 2001). Without substrate competition from the Cmm expansin, Cmm CWDEs, which include a polygalacturonase, and several pectate lyases, xylanases, cellulases, and other endoglucanases (Gartemann et al., 2008), may have increased efficiency, resulting in major structural changes and the breakdown of the plant cell wall (i.e. necrosis). Under infection-mimicking conditions, CmEXLX2 and the aforementioned Cmm CWDE proteins were measured at similar levels (Savidor et al., 2012).
The increased necrosis caused by ΔCmEXLX2 likely poses a fitness cost at another life cycle stage, such as during seed or leaf infections. For example, X. fastidiosa rpfF quorum sensing mutants are hypervirulent in grape, but are poorly transmitted by their insect vectors because they attach too strongly to host surfaces (Newman et al., 2004). We investigated the role of the expansin in root attachment by Cmm, but we did not find any differences between strains. The non-chimeric CmEXLX2 investigated in this study shares only 54% identity (amongst expansin domain regions) with the chimeric CmEXLX1 present in other Cmm strains (Table 2, Figure 4, and Figure S10). Previous studies have shown that disrupting endoglucanase-expansin chimeras in Cmm and X. fastidiosa reduces disease symptoms (Ingel et al., 2015; Jahr et al., 2000; Laine et al., 2000). This reduction of disease symptoms might be attributed to the adjacent endoglucanase domain present in chimeric expansins (Ingel et al., 2015; Jahr et al., 2000; Nikolaidis et al., 2014).
FIGURE 4.
Maximum-likelihood phylogenetic tree for bacterial expansins represented by Clavibacter michiganensis, Ralstonia solanacearum, and Bacillus subtilis. Alignment gaps were excluded, and the total number of sites used was 188 with 1000 repetitions. Bootstrap values are shown at the nodes if greater than 50%. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (Tamura et al., 2011).
We hypothesized that non-chimeric bacterial expansins of other vascular plant pathogens may attenuate virulence similar to CmEXLX2. The Gram-negative vascular pathogen Rs was selected for comparative studies because it shares tomato as a host, and purified RsEXLX has validated expansin activity in vitro (Georgelis et al., 2014). Unlike Cmm, the ΔRsEXLX mutant had wildtype virulence when directly introduced into the tomato vasculature. However when inoculated via the soil to the natural root infection court, ΔRsEXLX caused significantly less disease than its wildtype parent. In contrast, Rs mutants lacking cellulases or pectinases typically have reduced virulence following both soil-drench and cut-petiole inoculations (Huang and Allen, 1997; Huang and Allen, 2000; Liu et al., 2005). We infer that the ΔRsEXLX mutant’s hyper-attachment to tomato roots delays symptom development in the soil-drench assay because the attached ΔRsEXLX cannot efficiently enter and colonize the root vasculature.
Plant and bacterial expansins have diverse functions. Within the plant kingdom, distinct cell-specific expansins modulate different aspects of plant cellular growth (Cosgrove, 2000; Cosgrove, 2015). Arabidopsis thaliana maintains at least 24 α-expansins with several putative roles in cell wall modification, and Zinnia elegans has a subset of three xylem-specific expansins that vary in temporal and spatial expression (Cosgrove, 2000; Im et al., 2000). Similarly, bacterial non-chimeric expansins appear to have undergone intense selective pressures to adapt to their hosts. Cmm is primarily a foliar pathogen that accesses xylem vessels through wounds or natural openings, consistent with our finding that CmEXLX2 acts in the xylem and plays no detectable role in root attachment. In contrast, RsEXLX reduced Rs attachment to tomato roots, while the B. subtilis non-chimeric expansin increased B. subtilis attachment to maize roots (Kerff et al., 2008). Moreover, we demonstrated that RsEXLX is functionally different from CmEXLX2; unlike CmEXLX2, RsEXLX could not complement the hypervirulence phenotype of the Cmm ΔCmEXLX2 mutant. These results indicate that non-chimeric expansins can play at least three distinct and contrasting functions adapted to the biology of the microbe that wields them (Table 2). The structural targets of plant and microbial expansins remain unknown, but differences in expansin structure and isoelectric points likely influence substrate specificity and activity (Pastor et al., 2015).
Only a subset of plant-associated bacteria possess expansin genes. Although some saprophytes and plant pathogens have expansins, other notorious plant-associated microbes such as Pseudomonas or Rhizobium lack expansins (Nikolaidis et al., 2014). Expansins are pH-dependent, with an optimal pH range of 4.5 – 6.0, which overlaps the pH range of xylem fluid (pH~5.5) and tomato fruit tissue (pH~4.4) (Bollard, 1960; Cosgrove, 2005; Jones, 1999; Urrestarazu et al., 1996). Perhaps an acidic microenvironment, like that of xylem or fruit tissue, provides the optimal pH environment for expansin activity, unlike the relatively neutral pH of apoplastic fluid. We hypothesize that vascular-inhabiting bacterial pathogens horizontally acquired and maintained the expansin gene to exploit this acidic microenvironment. However, putative endoglucanase-expansin chimeras are present in several non-vascular plant-associated microbes, such as the non-vascular Xanthomonas species X. oryzae pv. oryzicola, X. translucens, and X. campestris pv. raphani (Nikolaidis et al., 2014; Ryan et al., 2011). Streptomyces also contains non-vascular phytopathogenic bacteria possessing bacterial expansins; however, some phytopathogenic Streptomyces sp., such as S. acidiscabies, can tolerate acidic soils with pH values ranging between 4 and 5.5, depending on the strain (Lambert and Loria, 1989a; Lambert and Loria, 1989b). Furthermore, S. acidiscabies is likely to be the most recent microbe to acquire a plant-like expansin because the sequence is more similar to plant expansins than any known microbial expansin (Nikolaidis et al., 2014). Regardless of the bacterial-colonized microenvironment, whether xylem or soil, acidity appears to be important.
The independent horizontal acquisition of a plant gene and its subsequent prokaryotic-specific adaptations highlights the dynamic nature of plant-bacterial interactions. Appropriation of plant expansins may have allowed microbes to manipulate or possibly mimic the biological processes of their hosts. In Cmm, CmEXLX2 influences vascular and fruit necrosis leading to larger in planta populations and increased symptom progression. In contrast, RsEXLX appears to contribute to root infection, possibly by modulating root attachment. It would be of interest to further characterize the role of RsEXLX in Rs root infections, and elucidate the targets and molecular mechanisms of these diverse bacterial plant-like expansins.
EXPERIMENTAL PROCEDURES
Bacterial strains/plasmids and primers used in this study are listed in Table 3 and S3, respectively. For further experimental procedural details, see Supporting Information Text S1.
TABLE 3.
Bacterial strains and plasmids used in this study.
| Strain or Plasmid | Descriptiona | Reference/Source |
|---|---|---|
| Strains | ||
| C. michiganensis subsp. michiganensis | ||
| Cmm0317 | Wildtype virulent New York strain | (Tancos et al., 2015) |
| ΔCmEXLX2 | Cmm0317 mutant strain with disrupted CmEXLX2 | This study |
| CΔCmEXLX2+ | ΔCmEXLX2 transformed with pHNExpA (complement) | This study |
| CΔCmP:RsEXLX + | ΔCmEXLX2 transformed with pHNRsExpA (C.m. subsp. michiganensis: R. solanacearum expansin hybrid) | This study |
| Escherichia coli | ||
| Zymo 5α (DH5α) | Cloning strain | Zymo Research |
| E.coli ER2925 | dam and dcm methylation-negative strain | New England Biolabs |
| R. solanacearum | ||
| GMI1000 | Wildtype strain isolated from tomato in French Guyana ; phylotype I sequevar 18 | (Salanoubat et al., 2002) |
| ΔRsEXLX | GMI1000 mutant with unmarked RsEXLX deletion | This study |
| CΔRsEXLX + | ΔRsEXLX with a wildtype copy of RSc0818comp in the chromosome | This study |
|
| ||
| Plasmids | ||
| C. michiganensis subsp. michiganensis | ||
| pGEM-T Easy | Cloning vector; Ampr; 3 kb | Promega |
| pHN216 | E. coli–Clavibacter shuttle vector; Gmr Nmr; 13.8 kb | (Laine et al., 1996) |
| pGnR-BsiWI | pGEM-T Easy:Gmr cassette (from pHN216); Ampr Gmr; 3.8 kb | This study |
| pGCMEβ | pGEM-T Easy:CmEXLX2; 3.6 kb | This study |
| pGCMEβGM | pGEM-T Easy:CmEXLX2::Gmr cassette; Ampr Gmr; 4.3 kb | This study |
| pIDT-FLExpA | IDT vector:Full length CmEXLX2 and promoter; Ampr; 3 kb | IDT |
| pHNExpA | Full-length CmEXLX2 cloned into pHN216; Nmr; 13.1 kb | This study |
| pHNRsExpA | CmEXLX2 promoter and signal peptide fused to RsEXLX cloned into pHN216; Nmr; 13.1 kb | This study |
| pK2-22 | eGFP-expressing plasmid; Nmr; 13.5 kb | (Chalupowicz et al., 2012) |
| R. solanacearum | ||
| pUFR80 | Cloning vector; Kanr Sucs; 7.8 kb | (Castañeda et al., 2005) |
| pUFR80-RSc0818KO | pUFR80:Unmarked ΔRsEXLX cassette; Kanr Sucs; 9.4 kb | This study |
| pUC18t-MiniTn7t(Gm) | Wide host range complementation vector; Gmr; 4.6 kb | (Choi et al., 2005) |
| pMiniTn7-RSc0818comp | pUC18t-MiniTn7t(Gm):full length RsEXLX ; Gmr; 6.1 kb | This study |
Gmr, Gentamicin acetyltransferase; Nmr, neomycin phosphotransferase; Ampr, β-lactamase; Kanr, aminoglycoside 3′-phosphotransferase; Sucs, levansucrase
Bacterial strains and growth conditions
The Cmm strain (Cmm0317) used in the present study was a virulent New York field strain, which naturally lacked the celA gene (chimeric expansin CmEXLX1) (Tancos et al., 2015). The CmEXLX2 mutant was transformed by insertional mutagenesis of Cmm (strain Cmm0317) with the pGCMEβGM plasmid as described (Stork et al., 2008; Tancos et al., 2013). Depending on the assay, Cmm isolates were incubated for 3–4 days at 27°C in Luria-Bertani (LB) (Miller, 1972), SB (Kirchner et al., 2001; Stork et al., 2008), or D2ANX media (Hadas et al., 2005). When required, LB medium was supplemented with the antibiotics gentamicin (40 μg/ml), neomycin (100 μg/ml), or ampicillin (100 μg/ml) (Fisher Scientific; Pittsburgh, PA).
The phylotype I sequevar 18 Rs strain GMI1000 was used in this study. Rs was routinely grown in casamino acid, peptone, glucose media (CPG) or Boucher’s minimal media (BMM) with 20 mM glucose at 28°C (Boucher et al., 1985). When required, media were supplemented with the antibiotic gentamicin (25 μg/ml) (Fisher Scientific; Pittsburgh, PA). The unmarked Rs ΔRsEXLX mutant was generated by natural transformation with pUFR80-RSc0818KO as described in (Boucher et al., 1985; Lowe et al., 2015). Briefly, transformants were plated on kanamycin to select for vector integration at the RsEXLX locus. Then kanamycin-resistant clones were counter-selected on 5% w/v sucrose to select for loss of the sacB-containing pUFR80 vector backbone by homologous recombination. Sucrose resistant clones were restruck on unamended media. PCR screening with RSc0818intF/R primers and GoTaq Green PCR mastermix (Promega, Madison, WI) was used to confirm the loss of RsEXLX.
PCR analysis and RNA expression
Plasmid constructs and chromosome integrations were confirmed with gene-specific PCR and sequencing (Table S3). Genomic Cmm DNA was extracted with the MasterPure Gram-Positive DNA purification kit (Epicentre, Madison, WI) according to the manufacturer’s instructions. Amplification was performed in a C1000 Touch Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA) using EmeraldAmp GT PCR Master Mix (Takara Bio Inc., Otsu, Shiga, Japan) or Advantage-GC 2 Polymerase Mix with a final GC-Melt concentration of 1.0 M for high GC sequences (Clontech Lab. Inc., Mountain View, CA), according to the manufacturer’s instructions. PCR products were purified with a DNA Clean & Concentrator – 25 kit (Zymo Research Inc., Irvine, CA) according to the manufacturer’s instructions.
RNA was extracted from 6 ml of Cmm suspension grown in LB medium to an OD600nm= 1.4. Total RNA was extracted using a ZR Fungal/Bacterial RNA MiniPrep kit (Zymo Research Corporation, Irvine, CA). Extraction was followed by an additional DNase treatment using TURBO DNA-free DNase (Fisher Scientific; Pittsburgh, PA). Quantity and quality (260/280 ratio) of DNase-treated RNA was determined with a NanoDrop ND 1000 (NanoDrop Technologies, Wilmington, DE) and a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA). Reverse transcription was performed with RNA to cDNA EcoDry Premix with random hexamers (Clontech Laboratories, Inc. Mountain View, CA) using 300 ng of RNA per reaction. To ensure no DNA remained in the RNA, controls were performed with pure RNA (100 ng/reaction).
Assessing in vitro growth and hypersensitive response assays
In vitro bacterial growth rates were compared in LB medium and pure tomato xylem sap over 48 h. Xylem sap was harvested from tomato seedlings (Solanum lycopersicum) cultivar Mountain Fresh Plus as described (Chalupowicz et al., 2012; Hiery et al., 2013). Pooled collected sap was filter-sterilized through a 0.22 μm PES membrane 500-ml filter system (Corning Inc. Corning NY) and frozen in aliquots at −20°C.
Bacteria for in vitro growth assays were initially grown in LB medium for 28 hours and then adjusted to OD600nm= 0.6 in water and inoculated (10 μl) into individual wells (n=9/strain) of a 96-well Falcon tissue culture plate containing LB medium or tomato sap (190 μl) (Corning Inc., Corning, NY). The plate was incubated at 28°C with shaking in a BioTek microplate reader (Winooski, VT). Bacterial density was measured at 590 nm every 2 h for 48 h. The mean area under the growth curve was calculated for both LB medium and pure tomato xylem sap. The experiment was performed three times.
Cmm strains were tested for ability to induce a hypersensitive response (HR) in Mirabilis jalapa (four o’clock plants). Cmm strains were grown for 24–32 h in LB medium (gentamicin and/or neomycin were added for ΔCmEXLX2 or the complemented strain) while shaking at 160 rpm in 125 ml flasks at 27°C, and the OD600nm was adjusted to 0.8 (108 CFU/ml) with sterile water. Bacterial suspensions were syringe-injected into the abaxial surface of the expanded M. jalapa leaf using a needleless 10-ml syringe.
In vitro growth of Rs strains was measured in CPG and BMM broth. Strains were grown overnight in CPG broth, pelleted, and washed twice in water. Bacterial suspensions were adjusted to an OD600 nm=0.1 in CPG or BMM and 200 μl of the cell suspensions were added to individual wells (n=3/strain) of a 96-well Falcon tissue culture plate (Corning Inc., Corning, NY). The plate was incubated at 28°C with shaking in a BioTek microplate reader (Winooski, VT). Bacterial density was measured at 600 nm every 15 min for 24 h. The experiment was performed three times.
Cell autolysis assays
Cmm and Rs strains were tested for differences in autolytic rates. Bacterial strains were suspended in 1× PBS with 0.02% SDS (pH=7.0), and 200 μl suspensions were seeded into a 96-well microtiter plate. Absorbance at 590 nm (Cmm) or 600 nm (Rs) of bacterial suspensions was monitored every 30 min for 10–12 h at 28°C without agitation, using a BioTek microplate reader (BioTek, Winooski, VT). The experiment was performed three times.
Pathogenicity and fruit infection assays
Cultures for Cmm strains were prepared and adjusted to an OD600nm= 0.8 as described above, and inoculated into 3 week old tomato seedlings by the cotyledon clipping method (n=5/treatment) (Xu et al., 2010). Tomato seedlings (cv. Mountain Fresh Plus) were grown in a Fafard professional formula growing mix (Sun Gro Horticulture, Agawam, MA) with a 14-hr light/10-hr dark photoperiod in the greenhouse. Tomato plants were screened daily for characteristic wilting and chlorosis associated with bacterial canker as previously described (Balaji et al., 2008; Tancos et al., 2015). Disease severity was quantified by counting the number of individual leaflets wilting relative to the total number of individual leaflets present on the three oldest leaves. Observations continued until all plants were wilting or until 21 days post inoculation (DPI). The mean area under the disease progress curve (AUDPC) was calculated from disease severity (Madden et al., 2007). Each of the three treatments had five plants per replicate, and the entire experiment was performed three times.
To quantify Cmm populations in planta, tomato seedlings were harvested at 9 DPI (n=3/treatment) and at 21 DPI (n=5/treatment). A 0.5-cm section of tomato stem tissue was harvested 1 cm above the inoculation site and homogenized with a sterile 5 mm stainless steel grinding bead (Qiagen, Valencia, CA) using a TissueLyser (Retsch, Newtown, PA) as described (Balaji and Smart, 2012; Tancos et al., 2013). Subsequent dilutions and population counts were performed as described (Tancos et al., 2013). The experiment was performed twice.
Immature green tomato fruit (8–12 mm in diameter) were inoculated with wildtype Cmm, ΔCmEXLX2, and the complemented mutant strain at a density of 108 cells/ml using a #2 horse-hair paintbrush as previously described (Medina-Mora et al., 2001; Tancos et al., 2013). Tomato fruit were brushed with sterile distilled water as a negative control. Fruit were harvested (n=4/strain) approximately 16 days post inoculation and divided into five or six equal vertical cross-sections. All of the lesions represented in two randomly selected sections were visualized using an Olympus SZX2 dissecting microscope (Olympus Corp., Shinjuku, Tokyo, Japan) connected to a Nikon Digital Sight-Qi1Mc camera (Nikon Corp., Chiyoda, Tokyo, Japan). Nikon’s NIS-Elements V. 4.1 software (Nikon Corp., Chiyoda, Tokyo, Japan) was used to measure the total lesion diameter, necrotic center diameter, and the width of the lesion’s ‘white halo’, at the widest point of the respective variables, for 100 individual lesions/strain. The experiment was performed twice.
Virulence of Rs isolates was determined on tomato plants following soil-soaking and cut-petiole inoculations. For Rs assays, tomato plants (cv. Bonny Best) were grown in ReadyMix Potting Soil in a 28 °C climate controlled chamber with a 12 hr day/12 hr night cycle. At 14 days post sowing, seedlings were transplanted into individual 5-inch pots. For soil-soaking inoculations, strains were grown overnight in 100 ml CPG in 250 ml flasks. Cultures were washed once in water and resuspended in water to OD600 nm=0.200. Bacterial suspensions (50 ml per plant) were poured into the soil of 17-day old unwounded plants (n=15 plants/strain), which corresponds to an inoculum of ~5×108 CFU/g soil. For cut-petiole inoculations, strains were grown overnight in 5 ml CPG in test tubes. Cultures were washed once, and cell density was adjusted to ~250 CFU/μl. The oldest petiole of 21-day old plants was delicately removed with a sharp razor blade. A 2 μl drop containing 500 CFU was placed on the cut petiole. For both Rs virulence assays, wilting symptoms of each plant were rated daily on a 0–4 disease index scale: 0, no symptoms; 1, ≤ 25% leaves wilted; 2, ≤ 50% leaves wilted; ≤ 75% leaves wilted; ≤ 100% leaves wilted.
Cmm movement and its influence on in planta growth
To assess differences in bacterial movement in planta, OD600nm= 0.8 suspensions of Cmm wildtype, ΔCmEXLX2, and the complemented mutant strain were prepared as described above. To measure acropetal movement, three week old tomato seedlings with 3 true leaves were inoculated by the cotyledon clipping method (n= 3/treatment). To quantify in planta bacterial populations at disparate distances, tomato seedlings were harvested at 21 DPI. A 0.5 cm section of tomato stem tissue was harvested 5 cm and 10 cm above the inoculation site and processed to determine bacterial populations as described above. Negative control plants were cotyledon clipped with sterile water. The experiment was performed three times with a total of 9 plants/strain.
Assessing differences in the lateral movement of Cmm strains
To visualize in planta movement of bacteria, wildtype and ΔCmEXLX2 strains were transformed with the eGFP transient expression vector pK2-22 (Chalupowicz et al., 2012). EGFP-expressing strains adjusted to an OD600nm= 0.8 were inoculated into 3 week old tomato seedlings by the cotyledon clipping method and plants (n=6/treatment/time point) were harvested and screened at 5 DPI, 7 DPI, and 9 DPI. At each time point, microscopic analysis was performed on tomato stem cross-sections taken at 1, 2, and 3-cm above the inoculation site and -1, -2, and -3-cm below the inoculation site. We measured the number of protoxylem, vascular bundles, and xylem parenchyma cells infected with the eGFP-expressing strains. Plant sections were visualized using an Olympus BX61 microscope connected to a confocal laser scanning microscope (CLSM) system (Olympus Fluoview FV-300, Melville, NY). An argon laser (488 nm excitation) and a green helium neon laser (543 nm excitation) was used to excite the eGFP bacteria and induce plant autofluorescence, respectively (Tancos et al., 2013).
Crystal violet staining assay for Cmm attachment
Differences in bacterial attachment were assessed with pure tomato xylem sap. Bacterial isolates were grown in LB broth (gentamicin and/or neomycin were added when necessary) as described above. Bacterial suspensions (125 μl) were added to the individual wells of a 24-well Falcon tissue culture plate (Corning Inc., Corning, NY) containing 375 μl of pure tomato xylem sap. The plates were briefly agitated and incubated statically at 27°C for five days, stained with 0.1% crystal violet, and solubilized with 30% acetic acid as described in (Davey and O’Toole, 2000; Kwasny and Opperman, 2010). Attached bacteria were quantified with an absorbance of 590 nm using the BioTek micoplate reader (Davey and O’Toole, 2000; Kwasny and Opperman, 2010). The experiment was performed three times with independently derived media for a total of 27 absorbance readings/strain/medium.
Root attachment assays
To assess root attachment by Cmm and Rs strains, surface disinfested tomato seeds were germinated on wet sterile filter paper until roots were approximately 2 cm in length. Emergent seedlings were individually collected, placed on 1% water agar, inoculated along the root axis with 10 μl of bacterial suspension, and incubated at room temperature for 2 hours. Following incubation, the roots were aseptically removed and pooled in groups of four for each respective strain, and gently washed to remove any non-adherent bacteria. The pooled roots (n=4) were homogenized and dilution plated. The experiment was performed three times.
Supplementary Material
Protein alignment for the chimeric Clavibacter michiganensis subsp. michiganensis expansin (CmEXLX1), C. michiganensis subsp. michiganensis non-chimeric expansin (CmEXLX2), Ralstonia solanacearum non-chimeric expansin (RsEXLX), and the Bacillus subtilis non-chimeric expansin (BsEXLX1). Protein alignment was performed with the default settings of Clustal Omega (Goujon et al., 2010), and the image was generated with BoxShade in RTF_new format (http://www.ch.embnet.org). The shading correlates to amino acid similarity (gray boxes) and conservation (black boxes).
FIGURE S1. Ethidium bromide stained agarose gel showing analysis of expression of CmEXLX2 in vitro. Products of RT-PCR of CmEXLX2 with RNA obtained from wildtype (Cmm0317), CmEXLX2 mutant (ΔCmEXLX2), and complement (CΔCmEXLX2+) strains (top gel). As a constitutively expressed control, the housekeeping gene gyrB was amplified using the same RT-PCR conditions (bottom gel).
FIGURE S2. Deletion of RsEXLX in Ralstonia solanacearum. (A) Schematic of RsEXLX deletion. (B) Ethidium bromide stained agarose gel (1.5% w/v) showing the genotype of GMI1000, ΔRsEXLX, and CΔRsEXLX+. PCR was conducted on genomic DNA using RsEXLX specific primers (left) or Ralstonia solanacearum universal primers (right) as a control.
FIGURE S3. In vitro growth of Clavibacter michiganensis subsp. michiganensis (Cmm) and Ralstonia solanacearum (Rs) strains. (Top figures) Growth of Cmm0317 (wildtype), ΔCmEXLX2 (Cmm expansin mutant), CΔCmEXLX2+ (Cmm complement), and CΔCmP:RsEXLX + (R. solanacearum expansin-hybrid mutant) in nutrient-rich Luria-Bertani medium and in nutrient-poor tomato xylem sap medium. (Bottom figures) Growth of Rs GMI1000 (wildtype), ΔRsEXLX (Rs expansin mutant), and CΔRsEXLX+ (Rs complement) in Boucher’s minimal medium or CPG rich medium.
FIGURE S4. Autolysis of Ralstonia solanacearum (Rs) and Clavibacter michiganensis subsp. michiganensis (Cmm) strains. (A) Rs strains were incubated in PBS or in PBS with 0.02% w/v SDS. (B) Cmm strains incubated in PBS with 0.02% w/v SDS.
FIGURE S5. Inter-and intravascular colonization of tomato xylem vessels infected with eGFP-Clavibacter michiganensis subsp. michiganensis Cmm0317 (wildtype) and ΔCmEXLX2 (expansin mutant) at specified days post inoculation (DPI). All confocal microscopy images were generated by merging 2 channels (488 nm and transmitted light). Scale bar: 200 μm.
FIGURE S6. The mean area under the disease progress curve (AUDPC) for the disease severity of Ralstonia solanacearum GMI1000 (wildtype), ΔRsEXLX (Rs expansin mutant), and CΔRsEXLX+ (Rs complement) of tomato. Differences among strains were determined by one-way ANOVA (P<0.05) using Dunnett’s multiple comparison test (P<0.05). AUDPC values followed by the same superscript are not significantly different. Three independent experiments are presented. Error bars correspond to the standard error.
FIGURE S7. In vitro biofilm attachment of Clavibacter michiganensis subsp. michiganensis Cmm0317 (wildtype), ΔCmEXLX2 (expansin mutant), CΔCmEXLX2+ (complement), and CΔCmP:RsEXLX + (R. solanacearum expansin-hybrid mutant) in tomato sap. Error bars correspond to the standard error.
FIGURE S8. GelRed stained agarose gel showing analysis of expression of CΔCmP:RsEXLX + in vitro. Products of RT-PCR of RsEXLX with RNA obtained from wildtype (Cmm0317) and CΔCmP:RsEXLX + strains (top gel). As a constitutively expressed control, the housekeeping gene gyrB was amplified using the same RT-PCR conditions (bottom gel).
FIGURE S9. The mean area under the disease progress curve (AUDPC) for disease severity of Clavibacter michiganensis subsp. michiganensis Cmm0317 (wildtype), ΔCmEXLX2 (Cmm expansin mutant), CΔCmEXLX2+ (Cmm complement), and CΔCmP:RsEXLX + (R. solanacearum expansin-hybrid mutant) of tomato. Differences among strains were determined using the lme4 package in R v. 3.3.2 to create a linear mixed effects model (P<0.05) followed by Tukey-Kramer posttest (P<0.05). AUDPC values followed by the same superscript are not significantly different. Three independent experiments are presented. Error bars correspond to the standard error.
Phenotypic traits of Clavibacter michiganensis subsp. michiganensis (Cmm) strains on tomato tissue. (A) In planta populations of Cmm strains colonizing tomato stems 5 and 10-cm above the site of inoculation following 21 days post inoculation. (B) In planta attachment of Cmm strains colonizing tomato roots following a 2 hour incubation.
Clavibacter michiganensis subsp. michiganensis colonization and vascular movement within tomato stem tissue with eGFP-expressing wildtype and CmEXLX2 mutant strains. A) Amount of colonization and movement present at 5 and 7 days post inoculation. B) Amount of colonization and movement present at 9 days post inoculation.
Oligonucleotides used in this study.
Materials and Methods
Acknowledgments
M. Tancos was supported by grant no. DGE-1144153 from the NSF-Graduate Research Fellowships Program. Additional support was provided by federal formula funds administered through the New York State Agricultural Experiment Station and the University of Wisconsin-Madison College of Agricultural and Life Sciences. T. Lowe-Power was funded by NIH National Research Service Award T32 GM07215 and by Agriculture and Food Research Initiative Competitive Grant # 2015-67011-22799 from the USDA NIFA. T. Tran was supported by NSF Grant # IOS1456636. Special thanks to Shaun Stice, Chase Crowell, Greg Vogel, and members of the Smart lab for assisting with experiments. We would also like to thank Dr. Tom Burr for critical reading of the manuscript. The authors declare no conflicts of interest.
References
- Bae C, Han SW, Song Y-R, Kim B-Y, Lee H-J, Lee J-M, Yeam I, Heu S, Oh C-S. Infection processes of xylem-colonizing pathogenic bacteria: possible explanations for the scarcity of qualitative disease resistance genes against them in crops. Theor Appl Genet. 2015 doi: 10.1007/s00122-015-2521-1. [DOI] [PubMed] [Google Scholar]
- Balaji V, Mayrose M, Sherf O, et al. Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 2008;146:1797–1809. doi: 10.1104/pp.107.115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji V, Smart CD. Over-expression of snakin-2 and extensin-like protein genes restricts pathogen invasiveness and enhances tolerance to Clavibacter michiganensis subsp. michiganensis in transgenic tomato (Solanum lycopersicum) Transgenic Res. 2012;21:23–37. doi: 10.1007/s11248-011-9506-x. [DOI] [PubMed] [Google Scholar]
- Bollard EG. Transport in the xylem. Annu Rev Plant Physiol. 1960;11:141–166. [Google Scholar]
- Boucher C, Barberis P, Trigalet A, Demery D. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- Bryan MK. Studies on bacterial canker of tomato. J Agric Res. 1930;41:825–851. [Google Scholar]
- Bunterngsook B, Eurwilaichitr L, Thamchaipenet A, Champreda V. Binding characteristics and synergistic effects of bacterial expansins on cellulosic and hemicellulosic substrates. Bioresour Technol. 2015;176:129–135. doi: 10.1016/j.biortech.2014.11.042. [DOI] [PubMed] [Google Scholar]
- Carlton WM, Braun EJ, Gleason ML. Ingress of Clavibacter michiganensis subsp. michiganensis into tomato leaves through hydathodes. Phytopathology. 1998;88:525–529. doi: 10.1094/PHYTO.1998.88.6.525. [DOI] [PubMed] [Google Scholar]
- Chalupowicz L, Zellermann E-M, Fluegel M, et al. Colonization and movement of GFP-labeled Clavibacter michiganensis subsp. michiganensis during tomato infection. Phytopathology. 2012;102:23–31. doi: 10.1094/PHYTO-05-11-0135. [DOI] [PubMed] [Google Scholar]
- Cho H, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6:850–61. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015;25:162–172. doi: 10.1016/j.pbi.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol. 1993;124:1–23. doi: 10.1111/j.1469-8137.1993.tb03795.x. [DOI] [PubMed] [Google Scholar]
- Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphinstone J. The current bacterial wilt situation: a global overview. In: Allen C, Piror P, Hayward AC, editors. Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex. St. Paul, MN: APS Press; 2005. pp. 9–28. [Google Scholar]
- Fatima U, Senthil-Kumar M. Plant and pathogen nutrient acquisition strategies. Front Plant Sci. 2015;6:750. doi: 10.3389/fpls.2015.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartemann K-H, Abt B, Bekel T, et al. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J Bacteriol. 2008;190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgelis N, Nikolaidis N, Cosgrove DJ. Bacterial expansins and related proteins from the world of microbes. Appl Microbiol Biotechnol. 2015:3807–3823. doi: 10.1007/s00253-015-6534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgelis N, Nikolaidis N, Cosgrove DJ. Biochemical analysis of expansin-like proteins from microbes. Carbohydr Polym. 2014;100:17–23. doi: 10.1016/j.carbpol.2013.04.094. [DOI] [PubMed] [Google Scholar]
- Georgelis N, Yennawar NH, Cosgrove DJ. Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc Natl Acad Sci. 2012;109:14830–14835. doi: 10.1073/pnas.1213200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, Mcwilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL – EBI. Nucleic Acids Res. 2010;38:695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadas R, Kritzman G, Klietman F, Gefen T, Manulis S. Comparison of extraction procedures and determination of the detection threshold for Clavibacter michiganensis ssp. michiganensis in tomato seeds. Plant Pathol. 2005;54:643–649. [Google Scholar]
- Hiery E, Adam S, Reid S, Hofmann J, Sonnewald S, Burkovski A. Genome-wide transcriptome analysis of Clavibacter michiganensis subsp. michiganensis grown in xylem mimicking medium. J Biotechnol. 2013;168:348–354. doi: 10.1016/j.jbiotec.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Huang Q, Allen C. An exo-poly-alpha-D-galacturonosidase, PehB, is required for wild-type virulence of Ralstonia solanacearum. J Bacteriol. 1997;179:7369–7378. doi: 10.1128/jb.179.23.7369-7378.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Allen C. Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants. Physiol Mol Plant Pathol. 2000;57:77–83. [Google Scholar]
- Im KH, Cosgrove DJ, Jones A. Subcellular localization of expansin mRNA in xylem cells. Plant Physiol. 2000;123:463–470. doi: 10.1104/pp.123.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingel BM, Wang P, Labavitch J, Roper MC. An endoglucanase/expansin hybrid protein and the type II secretion system affect the virulence and systemic colonization of Xylella fastidiosa. Phytopathology. 2015;105:S4.62. [Google Scholar]
- Jahr H, Dreier J, Meletzus D, Bahro R, Eichenlaub R. The endo-beta-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol Plant-Microbe Interact. 2000;13:703–14. doi: 10.1094/MPMI.2000.13.7.703. [DOI] [PubMed] [Google Scholar]
- Jones JB. Tomato plant culture: In the field, greenhouse, and home garden. Boca Raton, Fl: CRC Press, LLC; 1999. [Google Scholar]
- Kende H, Bradford KJ, Brummell Da, et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol. 2004;55:311–314. doi: 10.1007/s11103-004-0158-6. [DOI] [PubMed] [Google Scholar]
- Kerff F, Amoroso A, Herman R, et al. Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization. Proc Natl Acad Sci U S A. 2008;105:16876–81. doi: 10.1073/pnas.0809382105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner O, Gartemann KH, Zellermann EM, Eichenlaub R, Burger A. A highly efficient transposon mutagenesis system for the tomato pathogen Clavibacter michiganensis subsp. michiganensis. Mol Plant-Microbe Interact. 2001;14:1312–8. doi: 10.1094/MPMI.2001.14.11.1312. [DOI] [PubMed] [Google Scholar]
- Kwasny S, Opperman T. Static biofilm cultures of Gram-positive pathogens grown in a microtiter format used for anti-biofilm drug discovery. Curr Protoc Pharmacol. 2010 doi: 10.1002/0471141755.ph13a08s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine MJ, Haapalainen M, Wahlroos T, Kankare K, Nissinen R, Kassuwi S, Metzler MC. The cellulase encoded by the native plasmid of Clavibacter michiganensis ssp. sepedonicus plays a role in virulence and contains an expansin-like domain. Physiol Mol Plant Pathol. 2000;57:221–233. [Google Scholar]
- Lambert D, Loria R. Streptomyces acidscabies sp. nov. Int J Syst Bacteriol. 1989a;39:393–396. [Google Scholar]
- Lambert D, Loria R. Streptomyces scabies sp. nov., nom. rev. Int J Syst Bacteriol. 1989b;39:387–392. [Google Scholar]
- León L, de Siverio F, López MM, Rodríguez A. Clavibacter michiganensis subsp. michiganensis, a seedborne tomato pathogen: healthy seeds are still the goal. Plant Dis. 2011;95:1328–1338. doi: 10.1094/PDIS-02-11-0091. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang S, Schell MA, Denny TP. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol Plant-Microbe Interact. 2005;18:1296–1305. doi: 10.1094/MPMI-18-1296. [DOI] [PubMed] [Google Scholar]
- Lowe T, Ailloud F, Allen C. Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Mol Plant-Microbe Interact. 2015;28:286–297. doi: 10.1094/MPMI-09-14-0292-FI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden L, Hughes G, van den Bosch F. The study of plant disease epidemics. St. Paul: The American Phytopathological Society; 2007. [Google Scholar]
- McQueen-Mason SJ, Durachko D, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Mora C, Hausbeck MK, Fulbright DW. Bird ‘ s eye lesions of tomato fruit produced by aerosol and direct application of Clavibacter michiganensis subsp. michiganensis. Plant Dis. 2001;85:88–91. doi: 10.1094/PDIS.2001.85.1.88. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Newman KL, Almeida RP, Purcell AH, Lindow SE. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Natl Acad Sci. 2004;101:1737–1742. doi: 10.1073/pnas.0308399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis N, Doran N, Cosgrove DJ. Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol Biol Evol. 2014;31:376–86. doi: 10.1093/molbev/mst206. [DOI] [PubMed] [Google Scholar]
- Pastor N, Dávila S, Pérez-Rueda E, Segovia L, Martínez-Anaya C. Electrostatic analysis of bacterial expansins. Proteins Struct Funct Bioinforma. 2015;83:215–223. doi: 10.1002/prot.24718. [DOI] [PubMed] [Google Scholar]
- Rose J, Lee H, Bennett A. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, Vorholter FJ, Potnis N, Jones JB, Van Sluys M-A, Bogdanove AJ, Dow JM. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidor A, Teper D, Gartemann K-H, et al. The Clavibacter michiganensis subsp. michiganensis-tomato interactome reveals the perception of pathogen by the host and suggests mechanisms of infection. J Proteome Res. 2012;11:736–750. doi: 10.1021/pr200646a. [DOI] [PubMed] [Google Scholar]
- Sen Y, Feng Z, Vandenbroucke H, Wolf J, van der Visser RGF, van Heusden AW. Screening for new sources of resistance to Clavibacter michiganensis subsp. michiganensis (Cmm) in tomato. Euphytica. 2013;190:309–317. [Google Scholar]
- Stork I, Gartemann K, Burger A, Eichenlaub R. A family of serine proteases of Clavibacter michiganensis subsp. michiganensis: chpC plays a role in colonization of the host plant tomato. Mol Plant Pathol. 2008;9:599–608. doi: 10.1111/j.1364-3703.2008.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancos MA, Chalupowicz L, Barash I, Manulis-Sasson S, Smart CD. Tomato fruit and seed colonization by Clavibacter michiganensis subsp. michiganensis through external and internal routes. Appl Environ Microbiol. 2013;79:6948–6957. doi: 10.1128/AEM.02495-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancos MA, Lange HW, Smart CD. Characterizing the genetic diversity of the New York Clavibacter michiganensis subsp. michiganensis population. Phytopathology. 2015;105:169–179. doi: 10.1094/PHYTO-06-14-0178-R. [DOI] [PubMed] [Google Scholar]
- Tans-Kersten J, Brown D, Allen C. Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol Plant-Microbe Interact. 2004;17:686–695. doi: 10.1094/MPMI.2004.17.6.686. [DOI] [PubMed] [Google Scholar]
- Urrestarazu M, Sanchez A, Lorente FA, Guzman M. A daily rhythmic model for pH and volume from xylem sap of tomato plants. Commun Soil Sci Plant Anal. 1996;27:1859–1874. [Google Scholar]
- Vasse J, Frey P, Trigalet A. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1995;8:241–251. [Google Scholar]
- Wallis FM. Ultrastructural histopathology of tomato plants infected with Corynebacterium michiganense. Physiol Plant Pathol. 1977;11:333–342. [Google Scholar]
- Werner N, Fulbright D, Podolsky R, Bell J, Hausbeck MK. Limiting populations and spread of Clavibacter michiganensis subsp. michiganensis on seedling tomatoes in the greenhouse. Plant Dis. 2002;86:535–542. doi: 10.1094/PDIS.2002.86.5.535. [DOI] [PubMed] [Google Scholar]
- Xu X, Miller SA, Baysal-Gurel F, Gartemann K-H, Eichenlaub R, Rajashekara G. Bioluminescence imaging of Clavibacter michiganensis subsp. michiganensis infection of tomato seeds and plants. Appl Environ Microbiol. 2010;76:3978–88. doi: 10.1128/AEM.00493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Allen C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J Bacteriol. 2006;188:3697–3708. doi: 10.1128/JB.188.10.3697-3708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Wu Y, Cosgrove DJ. A fungal endoglucanase with plant cell wall extension activity. Plant Physiol. 2001;127:324–333. doi: 10.1104/pp.127.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein alignment for the chimeric Clavibacter michiganensis subsp. michiganensis expansin (CmEXLX1), C. michiganensis subsp. michiganensis non-chimeric expansin (CmEXLX2), Ralstonia solanacearum non-chimeric expansin (RsEXLX), and the Bacillus subtilis non-chimeric expansin (BsEXLX1). Protein alignment was performed with the default settings of Clustal Omega (Goujon et al., 2010), and the image was generated with BoxShade in RTF_new format (http://www.ch.embnet.org). The shading correlates to amino acid similarity (gray boxes) and conservation (black boxes).
FIGURE S1. Ethidium bromide stained agarose gel showing analysis of expression of CmEXLX2 in vitro. Products of RT-PCR of CmEXLX2 with RNA obtained from wildtype (Cmm0317), CmEXLX2 mutant (ΔCmEXLX2), and complement (CΔCmEXLX2+) strains (top gel). As a constitutively expressed control, the housekeeping gene gyrB was amplified using the same RT-PCR conditions (bottom gel).
FIGURE S2. Deletion of RsEXLX in Ralstonia solanacearum. (A) Schematic of RsEXLX deletion. (B) Ethidium bromide stained agarose gel (1.5% w/v) showing the genotype of GMI1000, ΔRsEXLX, and CΔRsEXLX+. PCR was conducted on genomic DNA using RsEXLX specific primers (left) or Ralstonia solanacearum universal primers (right) as a control.
FIGURE S3. In vitro growth of Clavibacter michiganensis subsp. michiganensis (Cmm) and Ralstonia solanacearum (Rs) strains. (Top figures) Growth of Cmm0317 (wildtype), ΔCmEXLX2 (Cmm expansin mutant), CΔCmEXLX2+ (Cmm complement), and CΔCmP:RsEXLX + (R. solanacearum expansin-hybrid mutant) in nutrient-rich Luria-Bertani medium and in nutrient-poor tomato xylem sap medium. (Bottom figures) Growth of Rs GMI1000 (wildtype), ΔRsEXLX (Rs expansin mutant), and CΔRsEXLX+ (Rs complement) in Boucher’s minimal medium or CPG rich medium.
FIGURE S4. Autolysis of Ralstonia solanacearum (Rs) and Clavibacter michiganensis subsp. michiganensis (Cmm) strains. (A) Rs strains were incubated in PBS or in PBS with 0.02% w/v SDS. (B) Cmm strains incubated in PBS with 0.02% w/v SDS.
FIGURE S5. Inter-and intravascular colonization of tomato xylem vessels infected with eGFP-Clavibacter michiganensis subsp. michiganensis Cmm0317 (wildtype) and ΔCmEXLX2 (expansin mutant) at specified days post inoculation (DPI). All confocal microscopy images were generated by merging 2 channels (488 nm and transmitted light). Scale bar: 200 μm.
FIGURE S6. The mean area under the disease progress curve (AUDPC) for the disease severity of Ralstonia solanacearum GMI1000 (wildtype), ΔRsEXLX (Rs expansin mutant), and CΔRsEXLX+ (Rs complement) of tomato. Differences among strains were determined by one-way ANOVA (P<0.05) using Dunnett’s multiple comparison test (P<0.05). AUDPC values followed by the same superscript are not significantly different. Three independent experiments are presented. Error bars correspond to the standard error.
FIGURE S7. In vitro biofilm attachment of Clavibacter michiganensis subsp. michiganensis Cmm0317 (wildtype), ΔCmEXLX2 (expansin mutant), CΔCmEXLX2+ (complement), and CΔCmP:RsEXLX + (R. solanacearum expansin-hybrid mutant) in tomato sap. Error bars correspond to the standard error.
FIGURE S8. GelRed stained agarose gel showing analysis of expression of CΔCmP:RsEXLX + in vitro. Products of RT-PCR of RsEXLX with RNA obtained from wildtype (Cmm0317) and CΔCmP:RsEXLX + strains (top gel). As a constitutively expressed control, the housekeeping gene gyrB was amplified using the same RT-PCR conditions (bottom gel).
FIGURE S9. The mean area under the disease progress curve (AUDPC) for disease severity of Clavibacter michiganensis subsp. michiganensis Cmm0317 (wildtype), ΔCmEXLX2 (Cmm expansin mutant), CΔCmEXLX2+ (Cmm complement), and CΔCmP:RsEXLX + (R. solanacearum expansin-hybrid mutant) of tomato. Differences among strains were determined using the lme4 package in R v. 3.3.2 to create a linear mixed effects model (P<0.05) followed by Tukey-Kramer posttest (P<0.05). AUDPC values followed by the same superscript are not significantly different. Three independent experiments are presented. Error bars correspond to the standard error.
Phenotypic traits of Clavibacter michiganensis subsp. michiganensis (Cmm) strains on tomato tissue. (A) In planta populations of Cmm strains colonizing tomato stems 5 and 10-cm above the site of inoculation following 21 days post inoculation. (B) In planta attachment of Cmm strains colonizing tomato roots following a 2 hour incubation.
Clavibacter michiganensis subsp. michiganensis colonization and vascular movement within tomato stem tissue with eGFP-expressing wildtype and CmEXLX2 mutant strains. A) Amount of colonization and movement present at 5 and 7 days post inoculation. B) Amount of colonization and movement present at 9 days post inoculation.
Oligonucleotides used in this study.
Materials and Methods