Abstract
Large-scale epidemiological studies show that low exercise capacity is the highest risk factor for all-cause morbidity and mortality relative to other conditions including diabetes, hypertension, and obesity. This led us to formulate the energy transfer hypothesis (ETH): Variation in capacity for energy transfer is the central mechanistic determinant of the divide between disease and health. As a test of this hypothesis, we predicted that two-way selective breeding of genetically heterogeneous rats for low and high intrinsic treadmill running capacity (a surrogate for energy transfer) would also produce rats that differ for disease risks. The lines are termed low-capacity runners (LCRs) and high-capacity runners (HCRs) and, after 36 generations of selection, they differ by more than eightfold in running capacity. Consistent with the ETH, the LCRs score high for developing disease risks, including metabolic syndrome, neurodegeneration, cognitive impairment, fatty liver disease, susceptibility to cancer, and reduced longevity. The HCRs are resistant to the development of these disease risks. Here we synthe-size ideas on nonequilibrium thermodynamics and evolution from Ilya Prigogine, Hans Krebs, and Peter Mitchell to formulate theoretic explanations for the ETH. First, at every moment in time, the atoms and molecules of organisms are reorganizing to pursue avenues for energy transfer. Second, this continuous organization is navigating in a constantly changing environment such that “strategies” are perpetually in flux and do not leave a simple footprint (evolution). Third, as a consequence, human populations demonstrate a wide variation in capacity for energy transfer that mirrors mechanistically the divide between disease and health.
NOTHING IN BIOLOGY MAKES SENSE EXCEPT IN THE LIGHT OF NONEQUILIBRIUM THERMODYNAMICS
Three decades ago we started discussions on how to develop valid animal models to understand complex diseases mechanistically. Our view was that most commonly used animal models of disease were too simplistic and did not emulate the polygenic nature of complex disease and that alternative pathways would be of value. As a minimum, we thought that a preclinical animal model of complex disease should:
emulate important and strongly predictive clinical phenotypes,
be polygenic and hence realistic for human translation,
respond accordingly to positive and negative health environments, and
be explained by fundamental scientific principles (Koch and Britton 2008).
Along our path of thinking about how to derive a more accurate animal model, we had noted the emergence in the 1980s of a strong statistical association between low capacity for energy transfer and high risk for disease and all-cause mortality (Cooksey et al. 1978; Bonow et al. 1980). This linkage of complex disease risks with low aerobic capacity, which has been extensively confirmed in large-scale contemporary studies (Myers et al. 2002; Kokkinos et al. 2008), was the information that led us to formulate the “energy transfer hypothesis” (ETH) (Koch et al. 2012). We devised the rat equivalent (Koch and Britton 2001) of the Bruce protocol (Bruce et al. 1963) to estimate maximal treadmill running capacity that served as a surrogate for maximal capacity for energy transfer.
AN EXPERIMENTAL TEST OF THE ENERGY TRANSFER HYPOTHESIS
Our long-term relationship with the pioneering quantitative geneticist John P. Rapp (2000) led us to consider that artificial selective breeding could be used as an unbiased test of the ETH. For this test, we developed a contrasting animal model system via two-way artificial selection on the variation in maximal treadmill running capacity within a large founder population of genetically heterogeneous rats (N:NIH) (Hansen and Spuhler 1984). That is, we selected for rats that differ “intrinsically” at the extremes for running capacity as low-capacity runners (LCRs) and high-capacity runners (HCRs). We found that two-way artificial selective breeding of rats for low and high innate endurance running capacity (Koch and Britton 2001) also yields rats that differ for numerous disease risks, including the metabolic syndrome, cardiovascular complications, neurodegeneration, cognitive decline, premature aging, and reduced longevity (Koch et al. 2012).
Selection for Intrinsic Capacity (LCR/HCR Lines)
Since starting selection in 1996, progress continues for both lines. It is critical that at each generation, a within-family rotational breeding scheme using 13 mating pairs for each line is practiced (Koch and Britton 2001). This approach slows the rate of inbreeding to yield retention of background genetic variation, thus increasing the overall response to selection (Kimura and Crow 1963). By generation 7 of selection, the HCRs, relative to the LCRs, had a 12% greater maximal O2 uptake (VO2max) that was due exclusively to a greater O2 uptake and usage by skeletal muscle, without differences between the lines in convective O2 delivery to muscle by the cardiopulmonary system. At generation 15, the HCR rats had a 50% greater VO2max compared with the LCR rats that was partially accounted for by a 41% greater cardiac output in the HCR rats relative to the LCR (Gonzalez et al. 2006). The LCR rats also display a lower economy of running (i.e., higher oxygen cost of running) compared with the HCR rats (Wisloff et al. 2005).
At generation 28 of selection (2011), the low and high selected lines differed by greater than eightfold for intrinsic maximal endurance running performance (Fig. 1A). Formal regression analysis of distance over generations yielded a p < 2.2 × 10−16 (i.e., at the machine epsilon in language R [R-project.org]), in support of a nonzero slope for both lines. These results are similar to the sustained responsiveness seen in other selectively bred lines such as the long-term artificial selection for oil concentration in maize (Zea mays). That is, after a century (100 generations), maize oil concentration continues to increase in response to selection pressure (Dudley and Lambert 2010).
Figure 1.
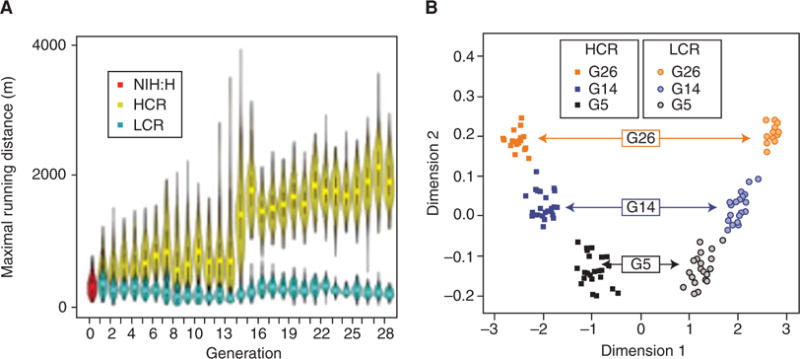
Running and genetic distance changes as a function of divergent selection. (A) Response to selection for 28 generations. Each symbol represents the distribution of running distance for each generation in each line. The symbols for each generation combine box plots and kernel density plots to depict the observed probability density. Males and females combined (n = 11,442 rats). NIH:H are the genetically heterogeneous founder stock rats. (B) Estimate of genetic “distance” between low-capacity runner (LCR) and high-capacity runner (HCR) lines. Multidimensional scaling showed that genetic distance between the lines increased across generations 5, 14, and 26. Dimension 1 = LCR-HCR lines, and dimension 2 generation. Dimension estimates for 142 rats shown. (A, From Ren et al. 2013; reprinted, with permission, from the authors.)
A long-term response to selection suggests that the trait for maximal running capacity is most likely influenced by many interacting quantitative trait loci (QTLs). As variants in some loci become fixed under selection, the previously hidden phenotypic effects of other variants can be “released” and come under selection, thereby powering prolonged responsiveness. This agrees with many previous observations that report long-term selection did not exhaust the genetic variation for the selected trait because of the existence of cryptic genetic variation (CGV) (Queitsch et al. 2002; Dworkin et al. 2003). CGV is defined as genetic variation that does not currently contribute to the phenotypes in a population but that can in the future as a result of changes in environment or new combinations of alleles. The strong continued response and operation of CGV suggest there is a future window in which more divergent phenotypes will be revealed.
The LCR and HCR rats were founded from the same base population (outcrossed N:NIH genetically heterogeneous stock [HS]) and have subsequently progressed as two separate strains. It is possible that genetic drift alone would have caused the two lines to diverge from each other and, as such, neutral differences would be distributed randomly over the entire genome. In contrast, as the two lines undergo different selective pressures (i.e., high or low running capacity), it is more likely that the positively selected loci that are functionally relevant for the phenotype would be driven apart even more quickly than by genetic drift and result in a reduction or elimination of genetic variabilities, as the frequency of linked loci in adjacent genomic regions also increases, creating a “selective sweep.” We genotyped 142 rats from three nonadjacent generations using the Affymetrix Rat Mapping 10K Genechips (Ren et al. 2013). The data reveal (Fig. 1B) that the genetic “distance” between the HCR and LCR lines increased from generation (G) 5 to 14, and to 26 as estimated using multidimensional scaling (MDS). MDS is a means of visualizing the level of similarity (or difference) between individual samples in a dataset. Dimension 1 reflects the HCR-LCR differences and dimension 2 reflects the differences between G5-14-26. The LCR-HCR lines formed separate genetic clusters at G5 and diverged further in G14 and G26. These results confirm the progressive genomic divergence caused by drift and selection. This is also consistent with the proportion of total phenotypic variation explained by the additive effects of genes (i.e., narrow sense heritability: h2) to be 40% for each line as implemented in the mixed model analysis tools SOLAR and WOMBAT (Ren et al. 2013). The genetic parameters described here were derived under the guidance of Jun Li (University of Michigan).
Divergence of Disease Risks
The results of a diverse set of collaborative experiments performed starting with G10 animals are in accordance with the ETH. That is, the LCRs proved to be susceptible and the HCRs were found to be resistant to the development of numerous disease risks including diminished longevity. Segregation of 12 diverse health features with selection for low capacity for energy transfer is listed in Table 1. Several of these studies served as landmark events in the development of the models. Wisloff et al. (2005) were the first to show that the LCR rats score high on risk factors that constitute the metabolic syndrome; the smaller aerobic capacity was also associated with decreases in the amounts of transcription factors required for mitochondrial biogenesis and in the amounts of oxidative enzymes in skeletal muscle. Lessard et al. (2011) showed that exercise training reversed the impairments to glucose and lipid metabolism observed in the skeletal muscle of sedentary LCRs, whereas increasing the expression of β2-AR, Nur77, GLUT4, UCP3, and FAT/CD36. Koch et al. (2011) showed that HCRs live about 6 months longer than LCR rats when both are sedentary throughout their life span; this was the first biological showing that high aerobic capacity could associate with increased longevity in the sedentary condition. In 2015, Karvinen et al. (2015) came to the conclusion that, in both rat and man, lifelong exercise does not increase longevity. In agreement with Koch et al. (2011), they found HCRs live significantly longer than LCR rats for the sedentary condition. When exposed to the exercise of voluntarily running on wheels from 9 months of age until death, however, both strains lived 16% shorter lives relative to sedentary. For man, there was no difference in longevity for twins who were disparate for physical activity across life. In 2015, Overmyer et al. (2015) used metabolomic and proteomic profiling to show that HCRs efficiently oxidize fatty acids (FAs) and branched-chain amino acids (BCAAs), sparing glycogen and reducing accumulation of short- and medium-chain acylcarnitines. HCR mitochondria have reduced acetylation of mitochondrial proteins within oxidative pathways at rest, and there is rapid protein deacetylation with exercise, which is greater in HCRs than in LCRs. Fluxomic analysis of valine degradation with exercise shows a functional role of differential protein acetylation in HCRs and LCRs. Overmyer’s data suggest that efficient FA and BCAA usage contribute to high intrinsic exercise capacity and the health and longevity benefits associated with enhanced fitness.
Table 1.
Disease risks (LCRs relative to HCRs)
| Metabolic syndrome (Wisloff et al. 2005) | ↓ Hippocampal neurogenesis (Wikgren et al. 2012) | Exercise and longevity (Karvinen et al. 2015) |
| Low physical activity (Novak et al. 2010) | Fatty liver disease (Morris et al. 2014) | ↑ Susceptibility to infectivity (Feng et al. 2015) |
| Response to training (Lessard et al. 2011) | Alzheimer’s neurodegeneration (Choi et al. 2014) | ↓ Metabolic flexibility (Overmyer et al. 2015) |
| Premature aging (Koch et al. 2011) | ↑ Vulnerability to ventricular fibrillation (Hoydal et al. 2014) | Inducible cancer (H Thompson, unpubl.) |
As predicted by the “energy transfer hypothesis” (ETH), disease risks segregated with selection for low-capacity runners (LCRs) and resistance to disease segregated with selection for high-capacity runners (HCRs).
A THEORETICAL FOUNDATION
For completeness, we felt compelled to connect a fundamental set of ideas that were explanatory for operation of the ETH. We wanted a path that was objective as based on principles that underlie physics and chemistry. Our search for a central explanation of the ETH led us to align ideas borrowed from others, including (1) general results from nonequilibrium thermodynamics (Ilya Prigogine), and (2) specific targeted biological results (Hans Krebs and Peter Mitchell) to account for the ETH and the high complexity of life-forms.
(1) Arguments from nonequilibrium thermodynamics: It is generally accepted that life reflects extremely ordered, open, nonequilibrium, complex systems that have formed “spontaneously” over a course, termed evolution. In contrast with this biological state of order is the familiar idea that the evolution of a physico-chemical system leads to an equilibrium condition of maximal disorder. That is, the Second Law of Thermodynamics is the trend for concentrated energy to disperse with time as measured by entropy (S). S is generally related to the heat term (δQ) by
where dS is the change in entropy, δQ is the heat added to the system, and T is temperature. The most pertinent concept is that, for isolated (closed) systems, a process occurs only if it increases the total entropy of the system. The apparent contradictions between biological order and the laws of physics for closed systems cannot be resolved within the methods of equilibrium statistical mechanics.
Starting in the 1960s, Ilya Prigogine (1978) initiated explanation for the origin and maintenance of far-from-equilibrium systems. He developed a model that accounted for how (under specialized circumstances) order could arise from chaos (i.e., order from disorder). His model describes mathematically how open systems that are highly unstable, far from equilibrium, and fluctuating nonlinearly will spontaneously organize to a higher level of complexity and order. Prigogine named these systems “dissipative structures” because they persist in an open exchange of energy with the generally entropic universe to dissipate or work off the products of their instabilities.
Prigogine (1978) provided an extended (and simplified) version of the Second Law that applies to both isolated and open systems:
where deS is the flow of entropy caused by exchanges with the surroundings (environment) and diS is the entropy production as a result of processes inside the system, such as diffusion, chemical reactions, and heat conduction (internal metabolism). That is, living systems are dependent on outside energy fluxes (deS) to maintain their organization and dissipate energy gradients to carry out self-organizing processes. A possible overall change in entropy for the universe is depicted visually (Fig. 2) as a sigmoidal relationship across time from the big bang (lowest entropy) to thermodynamic equilibrium (highest entropy). Equilibrium (“heat death”) is a probable outcome in which the universe has no free energy remaining and by definition can no longer perform work.
Figure 2.
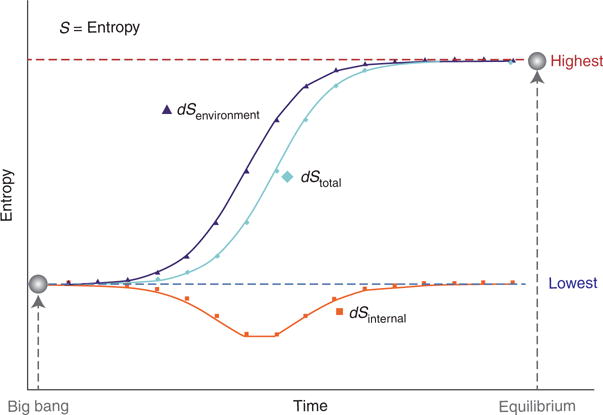
Visualization of possible changes in entropy (dS) across time from the big bang (lowest entropy) to equilibrium (highest entropy). dS = deS = diS, where deS is the flow of entropy caused by exchanges with the surroundings (environment), and diS is the entropy production as a result of processes inside the system, such as diffusion, chemical reactions, and heat conduction (internal metabolism) (Prigogine 1978).
Energy dissipation leading to organization seems to be the fundamental property of matter in response to an external energy transfer as exemplified by cyclogenesis, nucleosynthesis, and heliotropism. A cyclone is a meteorological system driven by a temperature contrast between the hot tropical sea surface and the cold top of the tropopause in which ordered convection cells form that mediate more efficient dissipation of energy (Fig. 3A) (Ozawa and Shimokawa 2015). Stellar nucleosynthesis is the process that forms new atomic nuclei from the accretive fusion of preexisting protons and neutrons. Each step toward a heavier element requires a higher energy transfer such that nucleosynthesis can be considered an evolutionary process of making order from disorder (Fig. 3B) (Langanke and Wiescher 2001). Heliotropism, also known as solar tracking, is the daily or seasonal motion of plant parts in response to the direction of the sun. Tracking the sun maximizes the amount of direct solar radiation received that can enhance reproductive success by increasing pollination, fertilization success, and seed development leading to enhanced energy dissipation (Atamian et al. 2016). This drive toward organization for energy dissipation is at the heart of evolution and accounts for the “catastrophe of complexity” (Bortz 2015). That is, at every moment in time atoms and molecules are organizing for avenues for greater energy transfer (Prigogine 1978). But this organization is navigating in a constantly changing environment such that “strategies” are constantly in flux and do not leave a simple trail.
Figure 3.
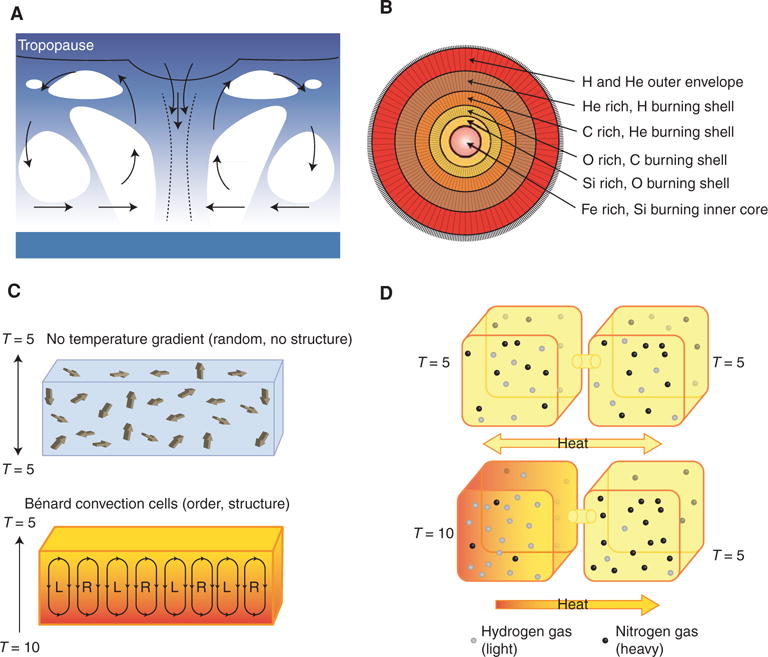
Energy dissipation leading to organization that can dispel energy even more effectively seems to be the fundamental property of matter in response to an external energy transfer. Four examples are easy to envision. (A) A schematic cross section of a tropical cyclone. A cyclone is a meteorological system driven by a temperature contrast between the hot tropical sea surface and the cold top of the tropopause in which ordered convection cells form that mediate more efficient dissipation of energy. (B) Stellar nucleosynthesis is the process within stars that forms new atomic nuclei from the accretive fusion of preexisting protons and neutrons. Depicted is the layering of element formation. As temperature increases toward the center of a star, heavier elements are formed. Iron (Fe) is formed at the center of stars and elements heavier than Fe require the higher energy of supernovae for formation. (C) Bénard cell formation is a type of motion that occurs in a thin liquid layer between two horizontal plates when a heat flow is imposed from below (T2). When the heat flux reaches a critical value (T1 < T2), the fluid becomes coherent and convective. Structured (ordered) coherent hexagonal-shaped cells emerge that increase the rate of heat transfer and gradient destruction in the system (disorder). (D) Thermo-diffusion. Consider a closed system containing hydrogen (H) and nitrogen (N). At uniform temperature (T5 = T5), there will be a random distribution of these two gases. If a temperature gradient is imposed (T10 > T5), hydrogen will move to the hotter side and nitrogen to the cooler. Here, entropy production is associated with a heat flow–producing disorderand it is also simultaneouslyassociated with a more ordered condition of the gases.
Order from disorder can also be observed from two small-scale laboratory studies: (1) Bénard cell formation is a type of motion that occurs in a thin liquid layer between two horizontal plates when a heat flow is imposed from below (Fig. 3C). The initial heat flow through the system is transmitted through collisions between neighboring molecules (conductance). When the heat flux reaches a critical value the system fluid becomes coherent and convective. Structured (ordered) coherent hexagonal-shaped cells emerge that increase the rate of heat transfer and gradient destruction in the system (disorder). (2) Thermodiffusion, also known as thermophoresis or the Ludwig–Soret effect, was first described by Carl Ludwig in 1856. Consider a closed system containing hydrogen and nitrogen (Fig. 3D). At uniform temperature, there will be a random distribution of these two gases. If a temperature gradient is imposed, hydrogen will move to the hotter side and nitrogen to the cooler. Here, entropy production is associated with a heat flow–producing disorder and it is also simultaneously associated with a more ordered condition of the gases.
These considerations lead to the view that both the origin and evolution of biological systems are paths for energy transfer via dissipative structures that lead to the development of ordered systems that will dispel energy even more effectively. Per unit mass, living things, such as a grass lawn, most certainly produce more entropy relative to inanimate objects such as rocks (Whitfield 2007). Critically, Dewar et al. (2006) have shown that features of the biomolecular motor ATP synthase have evolved in conformity with the statistical selection principles of maximum entropy production (MEP). ATP synthase displays four properties predicted by these statistical selection principles of MEP and we rephrase from Dewar: (1) an optimal angular position for the ATP-binding transition close to experimentally derived values, (2) an inverse association between the optimal gearing ratio and the proton motive force, (3) optimal operation at an inflection point in the curve of ATP synthesis rate versus protomotive force that would enable rapid metabolic control, and (4) a high optimal conversion efficiency of free energy. Dewar et al. (2006) declare, “our results suggest a statistical interpretation for the evolutionary optimization of ATP synthase function.”
(2) For specificity at the integrative level of biological energy transfer, we borrowed and used targeted information from two broad-scale contributions that helped to shape the initial platform for our ETH. First, Hans Krebs discovered that the function of the citric acid cycle (TCA) is to convert the energy released during the combustion of acetate into the energy transferred to the pyrophosphate bonds of ATP. Importantly, he also defined the concept that metabolic cycles evolved for greater efficiency of energy transfer. The 1981 paper from Baldwin and Krebs entitled “The Evolution of Metabolic Cycles” focused our attention toward joining ideas from evolution and thermodynamics; this was pivotal for our progress. Second, Peter Mitchell (Mitchell 1961) discovered the chemiosmotic mechanism of ATP formation that occurs during cellular respiration in mitochondria. This mechanism is largely outlined in his 1961 paper entitled “Coupling of Phosphorylation to Electron and Hydrogen Transfer by a Chemi-Osmotic Type of Mechanism.” This paper ends with the assertion, “the underlying thesis of the hypothesis put forward here is that if the processes that we call metabolism and transport represent events in a sequence, not only can metabolism be the cause of transport, but also transport can be the cause of metabolism.” Mitchell’s equating of transport and metabolism led us to theorize that a transport step (diffusion of hydrogen ion) coupled to a transformation step (phosphorylation of ADP) might represent an evolutionary feature connecting the inanimate (diffusion) with the animate (metabolism) (Fig. 4).
Figure 4.
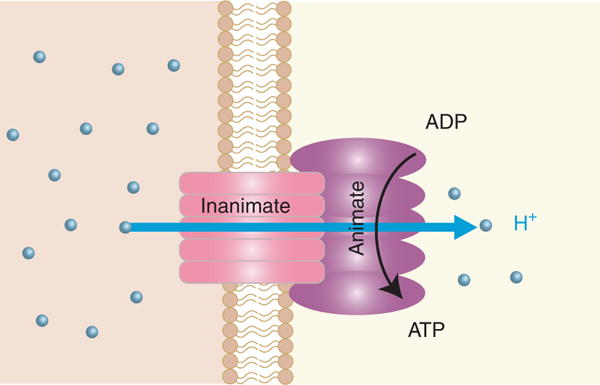
In 1961, Peter Mitchell published his landmark manuscript entitled “Coupling of Phosphorylation to Electron and Hydrogen Transfer by a Chemi-Osmotic Type of Mechanism.” In the last paragraph of this paper, he declares that transport and metabolism may be conceived of as the “same process.” We extended this idea to speculate that the H+ diffusion can be considered an “inanimate” step within evolution that couples to the “animate” metabolic formation of ATP from ADP.
The contributions of Krebs and Mitchell define critical features for the molecular specification of energy transfer for motion of mass at the atomic, molecular, cellular, tissue, and organ levels of organization. Implicit is the argument that all biological function derives from the property of motion (Lipowsky and Klumpp 2005) as mediated via the coordinated operation of molecular motors. We synthesized the ideas of Prigogine, Krebs, and Mitchell to formulate two statements that constitute a theoretical base for the ETH:
Evolution was underwritten by obligatory energy-dissipating mechanisms (entropy) and,
Emergence of complexity was coupled to the high energetic nature afforded by atmospheric oxygen.
Consistent with these statements, it is known that reduction of oxygen provides for close to the largest possible energy transfer per electron exchange. Recall that, among the 85 stable elements of the universe only nine are more electronegative on the Pauling scale than carbon and thus able to serve as an acceptor of electrons from carbon-based fuel substrates (Catling et al. 2005). Fluorine ranks 1 and oxygen 2 in electronegativity. In addition, new evidence suggests that differences in atmospheric oxygen are explanatory for the large-scale biological events of insect gigantism, polar gigantism, and Romer’s gap in vertebrate and arthropod terrestrialization as reviewed by Koch and Britton (2008).
UNSUPERVISED EXPLORATION FOR DISEASE
The wide signal difference for exercise capacity between LCRs and HCRs (Fig. 1A) afforded us a means for another test of the ETH. The second test was to determine whether disease features segregated with molecular pathways are known to mediate capacity for energy transfer. This was accomplished by application of unsupervised (and thus presumably unbiased) “omic” methodologies.
The laboratory of Heikki Kainulainen (University of Jyväskylä) investigated skeletal muscle gene expression–phenotype relationships to connect aerobic endurance capacity with metabolic disease risk factors. The study compared 12 HCRs and 12 LCRs from generation 18 of the selection that differed by 615% for maximal treadmill endurance running capacity (Kivela et al. 2010). On average, LCRs were heavier and had increased blood glucose, insulin, and triglycerides compared with HCRs. HCRs were higher for resting metabolic rate, voluntary wheel running activity, serum high-density lipoproteins, muscle capillarity, and mitochondrial area.
RNA samples from gastrocnemius muscle of all 24 rats were evaluated with genome-wide Illumina Sentrix RatRef-12 BeadChip (BD-27-303) that contained 22,523 probes. Data were analyzed with R software and normalized with quantiles normalization with a bioconductor. Statistically significant differences for genes between HCR and LCR rats were tested using t-statistics and a Benjamini–Hochberg algorithm controlling false discovery rate (FDR). Adjusted P values <0.05 were taken as statistically significant. As discussed in Kivela et al. (2010),
Functional enrichment testing was performed using 4 different methods. First, the clustering of differentially expressed genes into functional groups and their significance of overrepresentation among the groups was estimated with the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/home.jsp) (Dennis et al. 2003; Huang et al. 2009). Genes were clustered according to enrichment of Gene Ontology (GO) terms within differentially expressed genes (P <0.05). The statistical significance of clusters was estimated by modified Fisher exact P value. As an alternative to DAVID, we used LRpath (Sartor et al. 2009) to identify functional groups with significantly higher levels of differential expression [http://lrpath.ncibi.org/]. As opposed to DAVID, LRpath allows the data to remain on a continuous scale and, in doing so, allows identification of both functional groups that contain many genes with low differential expression and functional groups that contain few genes with very high differential expression. LRpath uses logistic regression to calculate P values for GO terms and KEGG pathways, and Benjamini–Hochberg algorithm is used to control the FDR. In addition, Reactome Skypainter (http://www.reactome.org/) was used to determine which biological events are statistically overrepresented in our differentially expressed gene set. For a given list of genes, Skypainter can identify common events (functions and pathways) for these genes. Further, GSEA (www.broad.mit.edu/gsea/; Subramanian et al. 2005) was applied for functional clustering of all genes without a priori filtering of the expression data. In the analysis, predetermined curated and motif gene sets of MSigDB database (www.broad.mit.edu/gsea/msigdb/) were utilized to compare HCR and LCR phenotypes.
The genome-wide microarray analysis revealed 239 known or predicted genes that were differently expressed between HCR and LCR lines (n = 12 HCR + 12 LCR, FDR <0.05). One hundred twenty-six of the genes were up-regulated in HCR lines versus LCR lines, and 113 genes were down-regulated. The expression data analyzed with the four different clustering methods (DAVID, Reactome Skypainter, LRPath, and GSEA) all produced similar results. The most enriched gene clusters that came up in all analyses were related to mitochondria and lipid metabolism. Functional clustering of the differentially expressed genes according to their GO annotations with DAVID revealed that many of the genes were related to mitochondria, carboxyl acid metabolism, lipid metabolism, oxidoreductase activity, immune response, and protein metabolism (Table 2) (rephrased from Kivela et al. 2010).
Table 2.
GO categories that were overrepresented among the significantly differentially expressed genes between HCRs and LCRs by functional clustering with DAVID
| Category | ↑ | ↓ |
|---|---|---|
| Mitochondriona | 7 | |
| Carboxyl acid metabolisma | 9 | |
| Lipid and lipoprotein metabolisma | 7 | |
| Lipid catabolisma | 3 | |
| Oxidoreductase activitya | 7 | |
| Immune responsea | 8 | |
| Protein metabolismb | 16 | |
| Attachment of cytoskeleton proteinsa | 6 |
Data from Kivela et al. 2010, with permission, from the Federation of American Societies for Experimental Biology © 2010.
Values indicate number of genes.
GO, Gene Ontology; HCRs, high-capacity runners; LCRs, low-capacity runners; ↑, gene expression higher in HCRs than in LCRs; ↓, gene expression lower in HCRs than in LCRs.
False discovery rate (FDR) P < 0.01.
FDR P < 0.05.
To relate genotypes and phenotypes, the most significant gene clusters obtained from the GSEA analysis were further correlated to physiological and biochemical parameters. GSEA, a statistical technique that relies on the principle that genes act as groups in a coordinated manner and not in isolation, was used to identify the subsets of genes that are congruently regulated within a given gene set. Centroids (centroid is the mean expression of the coregulated genes within a subset) were found to correlate with numerous systemic metabolic and physiological parameters. That is, the expression of genes in OXPHOS, TCA cycle, PPAR signaling, and STAT3 pathways were significantly correlated with voluntary wheel running, insulin sensitivity, blood triglycerides, and fatty acids. Overall, these mRNA expression signatures that link aerobic endurance capacity to metabolic disease risk factors are fully consistent with the ETH.
SELECTION FOR RESPONSE TO TRAINING (LRT AND HRT LINES)
Operationally, one’s current exercise capacity can be considered as the sum of (1) an intrinsic component that operates in the sedentary (non-trained state), and (2) an extrinsic component that follows as an adaptive response that accrues from all activity above the sedentary state (Bouchard et al. 2000). Evolution of adaptational capacity for enhanced energy transfer is clearly consistent with the selection principles of “maximum entropy production” (Dewar et al. 2006) and the ETH. Of recent interest are reports showing that there is wide variation for response to exercise training whereby some individuals experience no improvement (or even declines), whereas others show large gains (Vollaard et al. 2009; Timmons et al. 2010; Keller et al. 2011; Timmons 2011; Bouchard et al. 2012). To complement the LCR/HCR intrinsic rat model, we initiated (in 2001) large-scale bidirectional selection to develop lines of low-response trainers (LRTs) and high-response trainers (HRTs), once again, using the N:NIH genetically heterogeneous stock of rats as the founder population (Fig. 5A).
Figure 5.
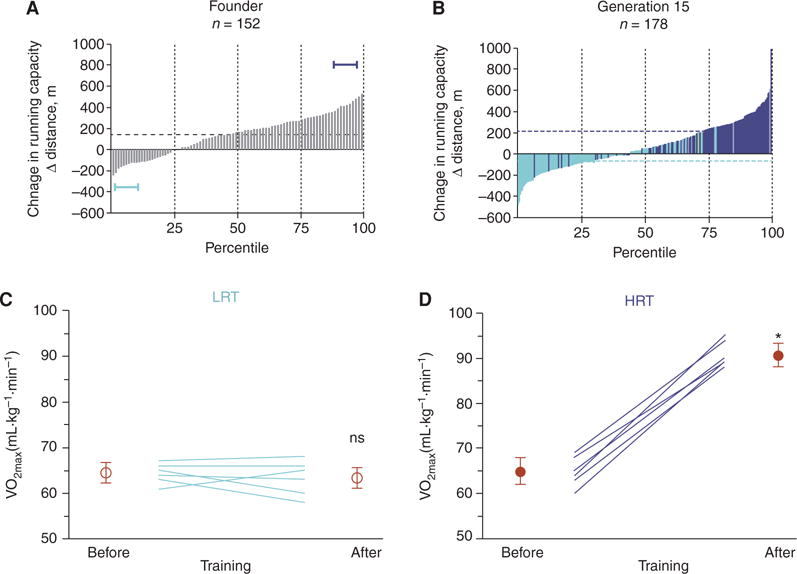
Selective breeding for low and high training response to treadmill running. (A) Rats were artificially selected for the change in running capacity in response to 8 weeks of training. Depicted is the training response for each of the 152 founder population rats. The horizontal brackets indicate the lowest and highest 10th percentile animals that were used as founders to start the low-response trainer (LRT) and high-response trainer (HRT) selected lines. (From Koch et al. 2013; reprinted, with permission, from the authors.) (B) The changes in running capacity with training for LRTrats (light colored bars) and HRTrats (dark bars) after 15 generations of training. Selection had caused marked divergence in training response between the lines. (C) Maximal oxygen consumption (VO2max) measured before and after 8 weeks of high-intensity aerobic interval training in LRTrats. Training failed to produce VO2max changes in LRTs. (D) High-intensity training produced on average a 40% increase in VO2max in HRTs. (From Wisloff et al. 2015; reprinted, with permission; from Elsevier © 2015.)
Development of the Trainer Models
Maximal treadmill running distance was tested before and after standardized aerobic treadmill training over an 8-week period (three exercise sessions per week). Response to training was calculated as the change in maximal treadmill running capacity with training. After 15 generations of selection (n = 3114 rats), HRT rats improved on average 223 m as a result of exercise training, whereas exercise capacity declined 65 m in LRTrats given the same absolute training environment (Koch et al. 2013).
Figure 5B shows the distribution of training-induced changes in running capacity for the founder population and after 15 generations of selection. The narrow-sense heritability (h2) for the change in distance run was 0.10 + 0.02 (Hegmann and Possidente 1981). We predicted that h2 would be smaller for the trainer models relative to the intrinsic, not necessarily because of less additive genetic variance, but because of a larger influence of environmental variance. That is, intrinsic capacity is estimated from three acute runs over 5 days, whereas training capacity is estimated over 70 days. This represents a 14-fold difference in time as an environmental factor, and h2 is inversely related to environmental variance. Recall that h2 = VA/VG + VE), where VA additive genetic variance, VG genetic variance, and VE = environmental variance.
LRT rats display diminished cardiac and metabolic responses to training. Wisloff et al. (2015) evaluated cardiac function in LRT and HRT rats before and after 8 weeks of high-intensity interval training on a treadmill. In response to training, the LRTs showed no improvement in VO2max (Fig. 5C), whereas the HCRs increased VO2max by 40% (Fig. 5D). At the cardiomyocyte level, HRT rats displayed positive remodeling for numerous measures of morphometrics, contractile dynamics, and calcium cycling, whereas LRT cardiomyocytes presented with either no remodeling or actual negative remodeling for these measures of function. From microarray data (left ventricular free wall), we also performed a high-throughput functional annotation analysis (using the DAVID database) to identify enriched biological themes among the differentially expressed genes (DEGs) (Wisloff et al. 2015). In the sedentary condition, a gene set was mapped to a 3 serine-related activity term that was enriched fivefold in HRTs relative to LRTs. Among these, the gene with the greatest differential (threefold higher in HRTs relative to LRTs) was kallikrein related peptidase 12, a serine protease predicted to be a strong effector of cell growth and response. In the trained condition, DEGs formed gene sets that were identified with terms pertaining to cell adhesion. Genes up-regulated in HRTs included the cadherin-associated protein catenin and members of the integrin and metalloproteinase-disintegrin families; all are critically important in regulating angiogenesis, neurogenesis, and tissue development.
Lessard et al. (2013) discovered that, in response to training, LRT rats showed pronounced metabolic dysfunction characterized by insulin resistance and increased adiposity, showing that the “exercise-resistant” phenotype segregates with disease risk. Low responders also had impaired exercise-induced angiogenesis in skeletal muscle, increased stress/inflammatory signaling, and altered transforming growth factor b signaling that was characterized by hyper-phosphorylation of a novel exercise-regulated phosphorylation site on SMAD2. These experiments discovered key transducers for low exercise training response that may represent novel targets for the treatment of metabolic disease.
TRANSLATIONAL VALUE OF THE MODELS
Major effort has gone into defining disease risks that are much stronger in the LCR relative to the HCR rats (Table 1). Translationally, this provides substrate to test for efficacy of reversal of the disease processes in the LCRs in response to altered behaviors and drugs. For example, the complexity of Alzheimer’s dementia (AD) has been particularly challenging for finding a drug treatment: The LCR-HCR models offer promise for two reasons. First, in humans, those that are active as elders had 60% lower odds of developing AD compared with those in a sedentary group (Rovio et al. 2005). Second, a recent study provides evidence of a neurodegenerative process in the hippocampus of aged LCR rats, consistent with those seen in age-related dementing illnesses such as AD (Choi et al. 2014). Thus, the LCR rats represent promising substrate for testing the efficacy of antidementia drugs within the context of complex causation. This same scenario could be applied to numerous other contrasting conditions contained in the LCR-HCR rat models including metabolic syndrome, fatty liver disease, obesity, aging, and longevity.
Arguments point to the use of reductionist paths as a major problem for research and development (R&D) of drugs for treatment of complex diseases. “Eroom’s law” (Moore’s spelled backward; Scannell et al. 2012) is the observation that drug discovery is becoming slower and more expensive over time, a trend first observed in the 1980s (Scannell et al. 2012). Eroom’s law is a paradox because improvements in technology such as high-throughput screening, biotechnology, combinatorial chemistry, and computational drug design should have advanced drug discovery.
Jack Scannell and Jim Bosley have attempted to explain Eroom’s law (Scannell and Bosley 2016). Their main objectives were to (1) provide quantitatively and historically plausible explanations, and (2) identify factors to which R&D efficiency is sensitive. To achieve this, they developed a quantitative decision–theoretical model of the R&D process and applied the rules to assess the probability of correct decisions. The model represents therapeutic candidates (e.g., putative drug targets, molecules in a screening library) within a “measurement space,” with candidates’ positions determined by their performance on a variety of assays (e.g., binding affinity, toxicity, in vivo efficacy) whose results correlate to a greater or lesser degree. They applied decision rules to segment the space and assess the probability of correct R&D decisions. Their central conclusion is that the limited creation and use of valid animal disease models may be the major constraint on R&D productivity. That is, too much reliance on reductionist molecular models that are devoid of the very complexity that embodies disease conditions is a fundamental flaw.
CONCLUDING REMARKS
We have developed a systematic approach to understand the extremes of both health and exercise capacity that is prospective, hypothesis-based, and underwritten by fundamental ideas from evolution and nonequilibrium thermodynamics. Our path originated in the 1980s from literature showing the strong statistical linkage between low exercise capacity and all-cause morbidity and mortality that is statistically robust yet mechanistically unresolved (Myers et al. 2002). This clinical association led us to formulate the ETH.
We used maximal treadmill running performance as a surrogate for maximal energy transfer. As a test of the energy transfer hypothesis, we found that two-way artificial selective breeding of rats for low and high innate endurance running capacity (Koch and Britton 2001) also yields rats that differ for numerous disease risks, including the metabolic syndrome, cardiovascular complications, neurodegeneration, cognitive decline, premature aging, and reduced longevity (Wisloff et al. 2005; Koch et al. 2012). Segregation of disease risks with selection for low exercise capacity represents an unbiased test of the ETH.
We formulated a fundamental explanation for the energy transfer hypothesis that addresses complexity and serves as a driver for new interpretation and hypothesis building (Koch and Britton 2008). We use three tenets: (1) the big bang was time zero for biological evolution, (2) all function derives from the property of motion (Lipowsky and Klumpp 2005) as mediated via the coordinated operation molecular motors, and (3) energy dissipation is the fundamental property of matter and that organization (order from disorder) occurs as embodied within the nonequilibrium thermodynamic ideas of Ilya Prigogine (1978). The accretive fusion of neutrons and protons (nucleosynthesis) to form nuclei of increasing atomic number accounts for the higher abundance of lighter elements in the universe and provided the substrate for biological evolution as a path for increased energy transfer. These ideas promote the attainment of enhanced energy transfer as the driving force underlying biological evolution. That is, evolution travels the path of capacity for energy transfer and more complexity equates with more energy transfer. We envision that the “chemiosmotic” diffusional movement of hydrogen ions across a membrane (à la Peter Mitchell [1961]) is the inanimate event that couples to phosphorylation of ADP to form ATP as the transitional animate event. As an extension, we propose that increased capacity for energy transfer segregates with health as a function of the fundamental pattern driving evolution.
A complex metabolism developed that contains variation for energy transfer as a result of natural selection operating in an open varying environment. Here, we took some of that resultant variation represented in the genetically heterogeneous stock of N:NIH rats and applied artificial selection for the extremely directed environments of low and high running capacity. Thus, we have separated genotypes for greater and lesser capacity for energy transfer. This also produced differences for immediacy of ATP availability and thus for contrasts in repair and function.
SPECULATION
One of the editorial reviewers asked us to “[p]redict what humans may be like physiologically in 50 years if we continue down the road of low fitness and high fatness.” We predict a continuance of the escalating prevalence of metabolic disease. The strong assortative mating association for body mass that gets transmitted to offspring will be hard to reverse at the epidemiological level (Jacobson et al. 2007). Such a parental driving force will be amplified via genetic, epigenetic, and shared environmental features. Our selected lines are driven by these same forces, only the mate selection was not “voluntary.”
Acknowledgments
The LCR-HCR rat model system was funded by the Office of Research Infrastructure Programs (ORIP) Grant P40OD021331 (to L.G.K. and S.L.B.) from the National Institutes of Health (NIH). We acknowledge the expert care of the rat colony provided by Lori Heckenkamp and Shelby Raupp. These rat models are maintained as an international resource with support from the Department of Anesthesiology at the University of Michigan, Ann Arbor, Michigan. Contact lgkoch@med.umich.edu or brittons @umich.edu if you have an interest in studying the rat models. We are grateful to Peter Mitchell, Bruce Walsh, Jonathon Flint, Richard Mott, and James Watson for illuminating insights on the development of ideas in this review.
Footnotes
Editors: Juleen R. Zierath, Michael J. Joyner, and John A. Hawley
Additional Perspectives on The Biology of Exercise available at www.perspectivesinmedicine.org
References
- Atamian HS, Creux NM, Brown EA, Garner AG, Blackman BK, Harmer SL. Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science. 2016;353:587–590. doi: 10.1126/science.aaf9793. [DOI] [PubMed] [Google Scholar]
- Baldwin JE, Krebs H. The evolution of metabolic cycles. Nature. 1981;291:381–382. doi: 10.1038/291381a0. [DOI] [PubMed] [Google Scholar]
- Bonow RO, Borer JS, Rosing DR, Henry WL, Pearlman AS, McIntosh CL, Morrow AG, Epstein SE. Preoperative exercise capacity in symptomatic patients with aortic regurgitation as a predictor of postoperative left ventricular function and long-term prognosis. Circulation. 1980;62:1280–1290. doi: 10.1161/01.cir.62.6.1280. [DOI] [PubMed] [Google Scholar]
- Bortz WMII. Metabolic field (Schrodinger); an explanatory platform for biology: Based on lecture at Trinity College, Dublin, Ireland, July 18, 2012. Med Hypotheses. 2015;85:894–897. doi: 10.1016/j.mehy.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Rankinen T, Chagnon YC, Rice T, Perusse L, Gagnon J, Borecki I, An P, Leon AS, Skinner JS, et al. Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE family study. J Appl Physiol. 2000;88:551–559. doi: 10.1152/jappl.2000.88.2.551. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Blair SN, Church TS, Earnest CP, Hagberg JM, Häkkinen K, Jenkins NT, Karavirta L, Kraus WE, Leon AS, et al. Adverse metabolic response to regular exercise: Is it a rare or common occurrence? PLoS ONE. 2012;7:e37887. doi: 10.1371/journal.pone.0037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce RA, Blackmon JR, Jones JW, Strait G. Exercising testing in adult normal subjects and cardiac patients. Pediatrics. 1963;32:742–756. [PubMed] [Google Scholar]
- Catling DC, Glein CR, Zahnle KJ, McKay CP. Why O2 is required by complex life on habitable planets and the concept of planetary “oxygenation time”. Astrobiology. 2005;5:415–438. doi: 10.1089/ast.2005.5.415. [DOI] [PubMed] [Google Scholar]
- Choi J, Chandrasekaran K, Demarest TG, Kristian T, Xu S, Vijaykumar K, Dsouza KG, Qi NR, Yarowsky PJ, Gallipoli R, et al. Brain diabetic neurodegeneration segregates with low intrinsic aerobic capacity. Ann Clin Transl Neurol. 2014;1:589–604. doi: 10.1002/acn3.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey JD, Reilly P, Brown S, Bomze H, Cryer PE. Exercise training and plasma catecholamines in patients with ischemic heart disease. Am J Cardiol. 1978;42:372–376. doi: 10.1016/0002-9149(78)90930-x. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dewar RC, Juretic D, Zupanovic P. The functional design of the rotary enzyme ATP synthase is consistent with maximum entropy production. Chem Phys Lett. 2006;430:177–182. [Google Scholar]
- Dudley JW, Lambert RJ. 100 generations of selection for oil and protein in corn. In: Janick J, editor. Plant breeding reviews. Vol. 24. Wiley; Hoboken, NJ: 2010. pp. 79–110. [Google Scholar]
- Dworkin I, Palsson A, Birdsall K, Gibson G. Evidence that Egfr contributes to cryptic genetic variation for photoreceptor determination in natural populations of Drosophila melanogaster. Curr Biol. 2003;13:1888–1893. doi: 10.1016/j.cub.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Feng X, Maze M, Koch LG, Britton SL, Hellman J. Exaggerated acute lung injury and impaired antibacterial defenses during Staphylococcus aureus infection in rats with the metabolic syndrome. PLoS ONE. 2015;10:e0126906. doi: 10.1371/journal.pone.0126906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez NC, Kirkton SD, Howlett RA, Britton SL, Koch LG, Wagner HE, Wagner PD. Continued divergence in VO2max of rats artificially selected for running endurance is mediated by greater convective blood O2 delivery. J Appl Physiol. 2006;101:1288–1296. doi: 10.1152/japplphysiol.01527.2005. [DOI] [PubMed] [Google Scholar]
- Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res. 1984;8:477–479. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- Hoydal MA, Stolen TO, Johnsen AB, Alvez M, Catalucci D, Condorelli G, Koch LG, Britton SL, Smith GL, Wisloff U. Reduced aerobic capacity causes leaky ryanodine receptors that trigger arrhythmia in a rat strain artificially selected and bred for low aerobic running capacity. Acta Physiol (Oxf) 2014;210:854–864. doi: 10.1111/apha.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jacobson P, Torgerson JS, Sjostrom L, Bouchard C. Spouse resemblance in body mass index: Effects on adult obesity prevalence in the offspring generation. Am J Epidemiol. 2007;165:101–108. doi: 10.1093/aje/kwj342. [DOI] [PubMed] [Google Scholar]
- Karvinen S, Waller K, Silvennoinen M, Koch LG, Britton SL, Kaprio J, Kainulainen H, Kujala UM. Physical activity in adulthood: Genes and mortality. Sci Rep. 2015;5:18259. doi: 10.1038/srep18259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Vollaard NB, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, Britton SL, Bouchard C, Koch LG, Timmons JA. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol. 2011;110:46–59. doi: 10.1152/japplphysiol.00634.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Crow JF. On the maximum avoidance of inbreeding. Genet Res. 1963;4:399–415. [Google Scholar]
- Kivela R, Silvennoinen M, Lehti M, Rinnankoski-Tuikka R, Purhonen T, Ketola T, Pullinen K, Vuento M, Mutanen N, Sartor MA, et al. Gene expression centroids that link with low intrinsic aerobic exercise capacity and complex disease risk. FASEB J. 2010;24:4565–4574. doi: 10.1096/fj.10-157313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol Genomics. 2001;5:45–52. doi: 10.1152/physiolgenomics.2001.5.1.45. [DOI] [PubMed] [Google Scholar]
- Koch LG, Britton SL. Aerobic metabolism underlies complexity and capacity. J Physiol. 2008;586:83–95. doi: 10.1113/jphysiol.2007.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Kemi OJ, Qi N, Leng SX, Bijma P, Gilligan LJ, Wilkinson JE, Wisloff H, Hoydal MA, Rolim N, et al. Intrinsic aerobic capacity sets a divide for aging and longevity. Circ Res. 2011;109:1162–1172. doi: 10.1161/CIRCRESAHA.111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Britton SL, Wisloff U. A rat model system to study complex disease risks, fitness, aging, and longevity. Trends Cardiovasc Med. 2012;22:29–34. doi: 10.1016/j.tcm.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Pollott GE, Britton SL. Selectively bred rat model system for low and high response to exercise training. Physiol Genomics. 2013;45:606–614. doi: 10.1152/physiolgenomics.00021.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinos P, Myers J, Kokkinos JP, Pittaras A, Narayan P, Manolis A, Karasik P, Greenberg M, Papademetriou V, Singh S. Exercise capacity and mortality in black and white men. Circulation. 2008;117:614–622. doi: 10.1161/CIRCULATIONAHA.107.734764. [DOI] [PubMed] [Google Scholar]
- Langanke K, Wiescher M. Nuclear reactions and stellar processes. Rep Prog Phys. 2001;64:1657–1701. [Google Scholar]
- Lessard SJ, Rivas DA, Stephenson EJ, Yaspelkis BB, III, Koch LG, Britton SL, Hawley JA. Exercise training reverses impaired skeletal muscle metabolism induced by artificial selection for low aerobic capacity. Am J Physiol Regul Integr Comp Physiol. 2011;300:R175–R182. doi: 10.1152/ajpregu.00338.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard SJ, Rivas DA, Alves-Wagner AB, Hirshman MF, Gallagher IJ, Constantin-Teodosiu D, Atkins R, Greenhaff P, Qi NR, Gustafsson T, et al. Resistance to aerobic exercise training causes metabolic dysfunction and reveals novel exercise-regulated signaling networks. Diabetes. 2013;62:2717–2727. doi: 10.2337/db13-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky R, Klumpp S. “Life is motion”: Multiscale motility of molecular motors. Physica A. 2005;352:53–112. [Google Scholar]
- Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- Morris EM, Jackman MR, Johnson GC, Liu TW, Lopez JL, Kearney ML, Fletcher JA, Meers GM, Koch LG, Britton SL, et al. Intrinsic aerobic capacity impacts susceptibility to acute high-fat diet-induced hepatic steatosis. Am J Physiol Endocrinol Metab. 2014;307:E355–E364. doi: 10.1152/ajpendo.00093.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J, Prakash M, Froelicher V, Do D, Partington S, Atwood JE. Exercise capacity and mortality among men referred for exercise testing. N Engl J Med. 2002;346:793–801. doi: 10.1056/NEJMoa011858. [DOI] [PubMed] [Google Scholar]
- Novak CM, Escande C, Burghardt PR, Zhang M, Barbosa MT, Chini EN, Britton SL, Koch LG, Akil H, Levine JA. Spontaneous activity, economy of activity, and resistance to diet-induced obesity in rats bred for high intrinsic aerobic capacity. Horm Behav. 2010;58:355–367. doi: 10.1016/j.yhbeh.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer KA, Evans CR, Qi NR, Minogue CE, Carson JJ, Chermside-Scabbo CJ, Koch LG, Britton SL, Pagliarini DJ, Coon JJ, et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 2015;21:468–478. doi: 10.1016/j.cmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa H, Shimokawa S. Thermodynamics of a tropical cyclone: Generation and dissipation of mechanical energy in a self-driven convection system. Tellus A. 2015;67:24216. [Google Scholar]
- Prigogine I. Time, structure, and fluctuations. Science. 1978;201:777–785. doi: 10.1126/science.201.4358.777. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev. 2000;80:135–172. doi: 10.1152/physrev.2000.80.1.135. [DOI] [PubMed] [Google Scholar]
- Ren YY, Overmyer KA, Qi NR, Treutelaar MK, Heckenkamp L, Kalahar M, Koch LG, Britton SL, Burant CF, Li JZ. Genetic analysis of a rat model of aerobic capacity and metabolic fitness. PLoS ONE. 2013;8:e77588. doi: 10.1371/journal.pone.0077588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovio S, Kareholt I, Helkala EL, Viitanen M, Winblad B, Tuomilehto J, Soininen H, Nissinen A, Kivipelto M. Leisure-time physical activity at midlife and the risk of dementia and Alzheimer’s disease. Lancet Neurol. 2005;4:705–711. doi: 10.1016/S1474-4422(05)70198-8. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Leikauf GD, Medvedovic M. LRpath: A logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Bosley J. When quality beats quantity: Decision theory, drug discovery, and the reproducibility crisis. PLoS ONE. 2016;11:e0147215. doi: 10.1371/journal.pone.0147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA. Variability in training-induced skeletal muscle adaptation. J Appl Physiol. 2011;110:846–853. doi: 10.1152/japplphysiol.00934.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NB, Nielsen S, et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–1496. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollaard NB, Constantin-Teodosiu D, Fredriksson K, Rooyackers O, Jansson E, Greenhaff PL, Timmons JA, Sundberg CJ. Systematic analysis of adaptations in aerobic capacity and submaximal energy metabolism provides a unique insight into determinants of human aerobic performance. J Appl Physiol. 2009;106:1479–1486. doi: 10.1152/japplphysiol.91453.2008. [DOI] [PubMed] [Google Scholar]
- Whitfield J. Survival of the likeliest? PLoS Biol. 2007;5:e142. doi: 10.1371/journal.pbio.0050142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikgren J, Mertikas GG, Raussi P, Tirkkonen R, Ayravainen L, Pelto-Huikko M, Koch LG, Britton SL, Kainulainen H. Selective breeding for endurance running capacity affects cognitive but not motor learning in rats. Physiol Behav. 2012;106:95–100. doi: 10.1016/j.physbeh.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- Wisloff U, Bye A, Stolen T, Kemi OJ, Pollott GE, Pande M, McEachin RC, Britton SL, Koch LG. Blunted cardiomyocyte remodeling response in exercise-resistant rats. J Am Coll Cardiol. 2015;65:1378–1380. doi: 10.1016/j.jacc.2015.01.041. [DOI] [PubMed] [Google Scholar]


