Figure 3.
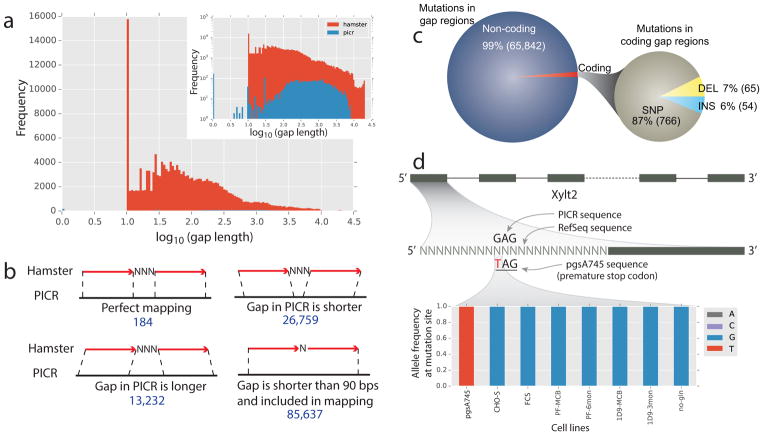
Important variants are located in sequence gaps in previous assemblies. (a) >95% of sequence gaps were filled in the PICR metassembly (inset shows the log frequency of gaps to highlight the low frequency of PICR gaps not visible in the normal histogram). (b) The missing sequence in gaps in the RefSeq assembly was identified by aligning RefSeq sequence flanking the gaps to the PICR sequence.
(c) Across 13 cell lines, we found 65,842 SNP and indel mutations in the RefSeq gap regions, and 1.3% of these were found in coding regions. (d) A legacy CHO cell line, pgsA745, identified Xylt2 as the glycosyltransferase responsible for the first step in glycosaminoglycan biosynthesis, as this cell line is deficient in glycosaminoglycan biosynthesis. Because of a gap in the RefSeq assembly, only in the new PICR metassembly can the causal variant be identified. A G->T mutation introduces an early stop codon in exon 1, resulting in a loss in Xylt2 activity. The genotype is shown for a variety of CHO cell lines [Lewis et al., 2013], [Feichtinger et al., 2016], [van Wijk et al., 2017], with only pgsA745 showing the early stop codon.