Fig 3.
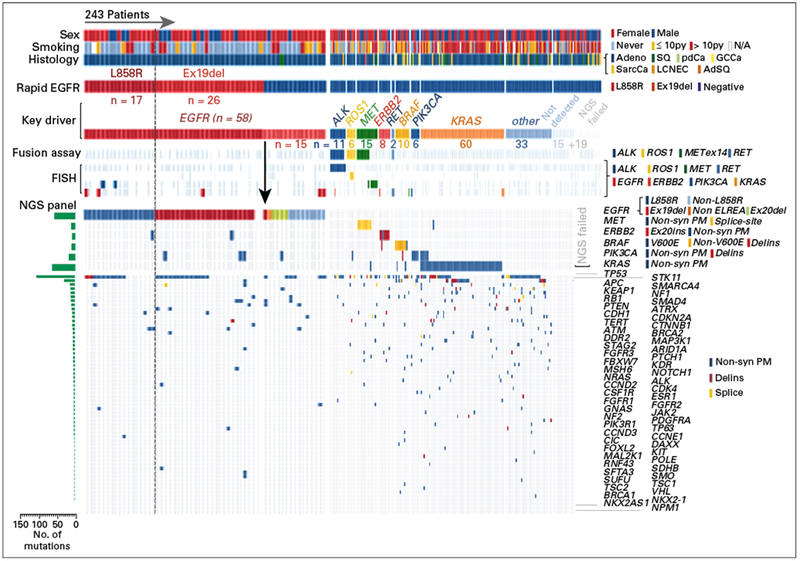
Integration of molecular-genetic testing in 243 patients with non–small-cell lung cancer who underwent rapid EGFR genotyping. The heatmap portrays clinicopathologic features (top three rows), rapid EGFR results, and key molecular drivers along with the results of next-generation sequencing (NGS)–based fusion detection, fluorescence in-situ hybridization (FISH), and NGS panel results. Key findings include (1) an isolated false-negative rapid EGFR result (arrow), (2) the inability of the rapid EGFR test to detect EGFR mutations at other residues, (3) identification of at least one underlying driver mutation in more than 50% of all tested cases by using the integrated molecular diagnostic approach, and (4) the association between clinicopathologic features and certain key drivers (eg, never-smoking women with adenocarcinoma and EGFR v > 10 pack-year smoking history and KRAS). Adeno, adenocarcinoma; AdSQ, adenosquamous; Delins, insertion/deletion; Exl9del, exon 19 deletion; Ex20ins, exon 20 insertion; GCCa, giant cell carcinoma; LCNEC, large-cell neuroendocrine carcinoma; N/A, not applicable; Non-syn, nonsynonymous; PM, point mutation; py, pack year; SarcCa, sarcomatoid carcinoma; SQ, squamous.
