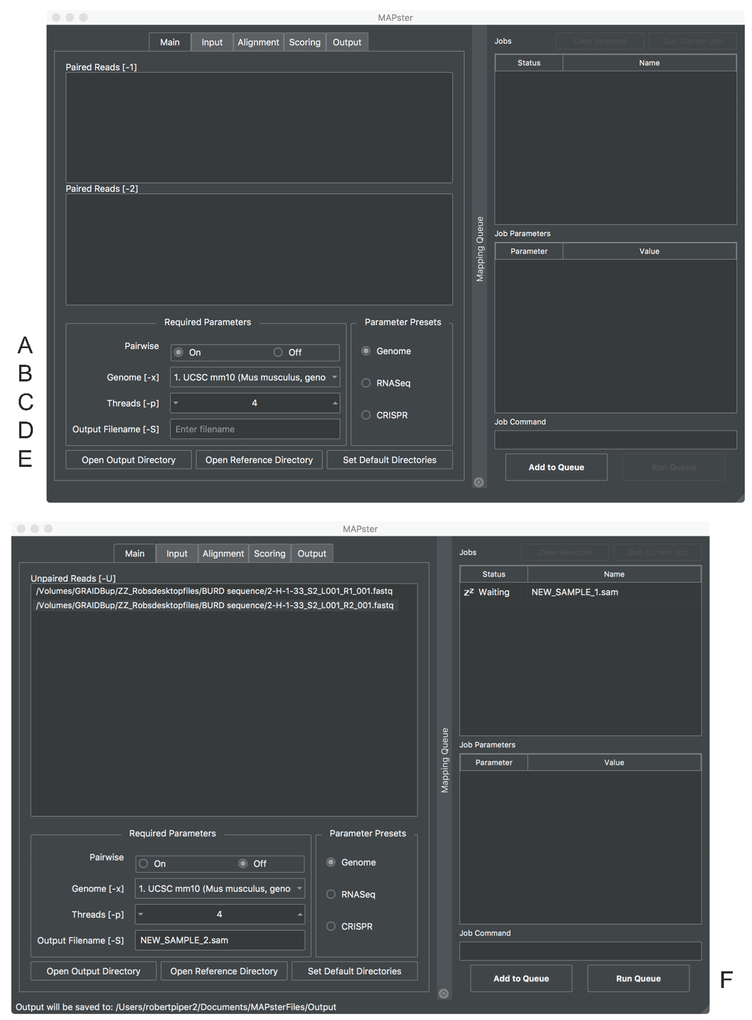
Figure 1: The MAPster interface.
Screen shot of the main window of MAPster. The boxes for entering required files and formats are shown. Turn “Pairwise’ (A) off to treat sequence files as single-end reads. The reference genome is selected with the ‘Genome’ menu bar (B). The number of processors used by HISAT2 is selected with the ‘Threads’ menu (C). The new sample name can be typed into ‘Output Filename’ text window. The directory for the output files can be designated in (E). Below is a window showing the queueing of single-end read files. After sample has been added to the queue, mapping can be initiated with the [Run Queue] button (F).