Figure 6. Propagating variant effects on networks.
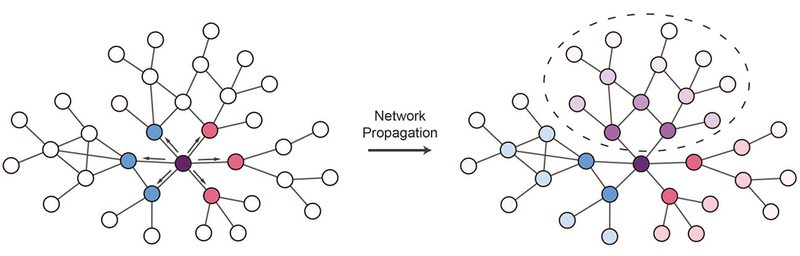
Variants can be used as signal sources for network propagation in order to identify network neighborhoods affected by variants. Edgetic effects can be used to constrain network propagation according to the effects of variants on specific protein interactions. On the left side of this schematic, two variants to the purple node affect interactions with different subsets of partners (indicated by blue and pink nodes respectively). Network propagation can be used to implicate network regions likely to be affected by each variant, and these can be contrasted to identify regions perturbed by both variants that could explain shared phenotypes (right network, circled purple shaded nodes), or regions affected specifically by each variant (right network, blue and pink shaded regions) which could help explain pleiotropic effects.
