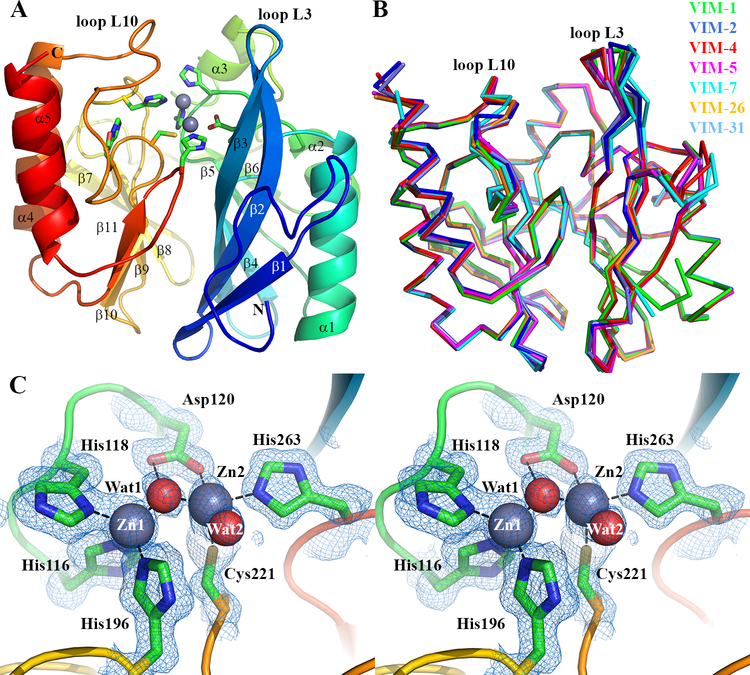
Figure 1. Crystal Structure of VIM-1.
A. View of the crystal structure of di-zinc VIM-1 showing the overall fold and location of active site. Protein main chain is colour-ramped from blue (N-) to red (C-terminus). Active site residues are rendered as sticks; zinc ions as grey spheres. Secondary structure elements are labeled. B. Superposition of di-zinc VIM-1 with structures of other VIM enzymes (VIM-2, pdb 4BZ3; VIM-4, pdb 2WHG; VIM-5, pdb 5A87; VIM-7, pdb 4D1T; VIM-26, pdb 4UWO; VIM-31, pdb 4FR7; coloured as shown). C. Stereoview of di-zinc VIM-1 active site. Electron density shown is 2|Fo| - |Fc| Φcalc contoured at 1.5 sigma. This Figure was created using PyMol (www.pymol.org).