Abstract
Purpose
Metastatic castration-resistant prostate cancer (CRPC) is the lethal form of the disease. Many groups have performed mutational or immunohistochemistry (IHC) testing in metastatic CRPC to identify treatment targets. However, the frequency with which mutational or IHC data have an impact on clinical decision making and the outcomes of molecularly guided therapy in CRPC are largely unknown. We report our institution’s experience with mutational and IHC testing in patients with metastatic CRPC and its impact on clinical decision making and patient outcomes.
Methods
Between 2012 and 2015, 59 patients with CRPC underwent metastatic tissue biopsies and were genotyped with a 37–cancer gene panel in a Clinical Laboratory Improvement Amendments–certified laboratory. PTEN expression by IHC testing was also measured in 35 of these samples. A retrospective chart review was performed to determine whether the genomic information was acted upon and the outcome of patients whose treatment was guided by molecular testing.
Results
Forty-six of 59 patients with CRPC (78.0%) had biopsies with adequate tumor for mutational testing. Thirty-one of 46 subjects (67.4%) had mutations identified by sequencing. Of the 35 patients with CRPC whose biopsies were evaluated for PTEN expression by IHC testing, 13 had PTEN loss. Two patients had treatment on the basis of molecular testing, and one of these subjects had greater tumor control with molecularly guided therapy than his immediate prior therapy.
Conclusion
Targeted sequencing and IHC can identify clinically informative molecular abnormalities in CRPC. Despite this, a small minority of patients in our series underwent therapies guided by mutational or IHC testing. Actionability of abnormalities identified in metastatic CRPC may be improved with access to clinical trials, insurance approval for unapproved uses of existing anticancer drugs, and larger gene sequencing panels that include more frequently mutated genes.
INTRODUCTION
Prostate cancer is the second-leading cause of cancer-related death in men in the United States, and more than 26,000 men are predicted to die as a result of prostate cancer in 2017.1 Nearly all of these deaths occur as a result of metastatic tumors. The principal treatment of metastatic prostate cancer is androgen-deprivation therapy or other treatments that disrupt the function of the androgen receptor (AR) protein. Despite these treatments, progression to lethal castration-resistant prostate cancer (CRPC) is nearly universal. Once a tumor becomes resistant to AR-targeted therapy, few effective treatment options remain. This underscores the urgent need to identify important molecular targets that may guide not only drug development but also clinical decision making for patients.
Technological advances in sequencing have led to the identification of molecular abnormalities present in cancers. Moreover, tumor mutational testing for clinical decision making is standard for several cancer types, including melanoma and adenocarcinoma of the lung.2–4 Hospitals across the country are increasingly implementing tumor-sequencing platforms to help guide therapy for patients with advanced cancers.
Several studies have reported the frequency of abnormalities detected with mutational testing in advanced cancers.5 In CRPC, recent work demonstrates that these tumors share many aberrations in oncogenic pathways found in other solid tumors.6,7 Indeed, the majority of patients with CRPC have alterations in PTEN or phosphatidylinositol 3-kinase (PI3K).8,9 Importantly, both PI3K and Akt inhibitors are in clinical testing. Thus, there is a strong rationale to test for abnormalities in PTEN, PI3K, and other alterations that may be targeted with specific therapies. However, molecular testing is not routinely performed in patients with CRPC, and the utility of targeted molecular testing in this disease is unknown. Moreover, there is limited information on the frequency with which molecular testing guides therapy, as well as limited information on the outcomes of patients whose treatment was guided by molecular testing.
In this article, we describe our experience with mutational testing and PTEN immunohistochemistry (IHC) in a Clinical Laboratory Improvement Amendments (CLIA)–certified laboratory to guide therapy in men with metastatic CRPC. We highlight not only the frequency of abnormalities detected but also the frequency with which these abnormalities were acted upon. Finally, we present outcomes of the patients whose treatment was guided by molecular testing.
METHODS
Biopsies and subsequent mutational profiling and IHC testing were performed under an Oregon Health & Science University institutional review board (IRB)–approved metastatic tissue collection protocol. Inclusion criteria were men 18 years of age orolder with histologically confirmed prostate adenocarcinoma and radiographic evidence of metastatic disease amenable to image-guided biopsy. In addition, the patient’s disease had to be progressing in the setting of testosterone levels < 50 ng/dL within 28 days before biopsy. Progression was defined as soft tissue progression by Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, bone scan progression by the presence of at least two new lesions, symptomatic progression in an area of radiological-evident disease, or prostate-specific antigen progression defined as levels > 2 ng/mL that has risen on at least two measurements separated by at least 1 week. Those who received first-generation oral antiandrogens (flutamide, bicalutamide, nilutamide) as their most recent systemic therapy before biopsy had to have progressed after at least 4 weeks of antiandrogen discontinuation. Patients had to have an Eastern Cooperative Oncology Group performance status of 0 to 3, a platelet count ≥ 75,000/μL, prothrombin time or international normalized ratio and activated partial thromboplastin time < 1.5 times the upper limit of normal, and the ability to discontinue anticoagulation for at least 1 week before tumor biopsy. Patients without prior orchiectomy continued medical castration therapy while they were participating in the study. All patients signed an informed consent form before biopsy.
Metastatic tissue was collected by computed tomography or ultrasound-guided biopsies performed at Oregon Health & Science University in accordance with the standard operating procedure and institutional standards, with the goal of minimizing patient risk. Biopsies were fixed in 10% neutral-buffered formalin for 8 hours and then transferred to 70% ethanol, paraffin embedded, and subsequently stained with hematoxylin and eosin. All hematoxylin and eosin–stained slides were examined by a pathologist (G.V.T.), who reviewed tumor content and decided if the sample was adequate for molecular testing. All bone biopsies received surface decalcification before microtome sectioning. Briefly, the cut surface of the paraffin block was immersed in Overnight Bone Decalcifier solution for 5 minutes, rinsed in water, and then sectioned.
G.V.T. generated clinical reports, which were given to the treating clinician within 6 weeks. These reports included a discussion of the clinical significance and potential actionability of the findings on the basis of type of aberration, existing agents, or ongoing clinical trials. Actionable is defined on the basis of the 2017 joint Association for Molecular Pathology, ASCO, and College of American Pathologists guidelines.10 Mutations were grouped according to evidence-based variant categorization into three tiers, ie, Tier 1: Variants of Strong Clinical Significance; Tier II: Variants of Potential Clinical Significance; and Tier III: Variants of Unknown Clinical Significance. Most reported variants were categorized as Tier II and were considered actionable on the basis of there being one or more clinical trials that included the gene in question as a biomarker, or the existence of preclinical data indicating that the variant might be predictive of response to a potential therapy. Tier III variants were not regarded as actionable. Finally, IRB approval was obtained to review the charts of all of the patients in this series to determine whether the molecular findings were acted upon and the treatment outcomes of patients who received molecularly guided therapy.
The Data Supplement provides detailed descriptions of nucleic acid sequencing and PTEN IHC methods.
RESULTS
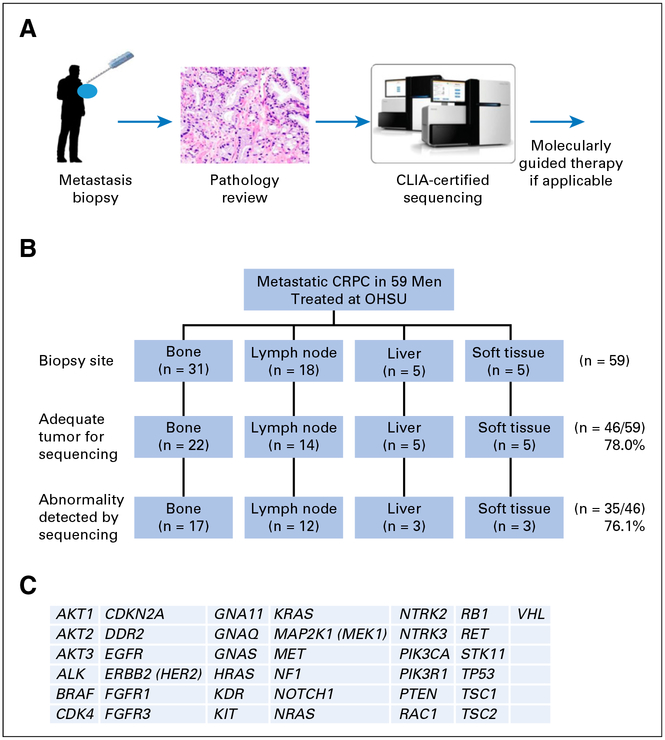
Between 2012 and 2015, 59 patients with CRPC were enrolled and biopsied through our IRB-approved metastatic tissue collection protocol described in Methods. Table 1 summarizes the baseline characteristics of the patients. A schematic for biopsy collection and tumor testing is shown in Fig 1A. A variety of tissue sites were biopsied, but bone and lymph node were the most common sites (Fig 1B). Obtaining sufficient tumor from metastatic bony lesions is challenging.11–13 However, we found that there was adequate tumor for sequencing in 71% of bone specimens. Seventy-eight percent of lymph node specimens had adequate tumor for sequencing, and 100% of liver and soft tissue specimens had adequate tumor for sequencing. In total, 46 of 59 patients with CRPC (78.0%) had biopsies with adequate tumor to submit for GeneTrails mutational testing (Fig 1C).
Table 1.
Baseline Characteristics of 59 Patients With Metastatic Castration-Resistant Prostate Cancer
| Characteristic | Value |
|---|---|
| Age (years) | |
| Median | 70 |
| Range | 49–87 |
| Time from initial diagnosis of prostate adenocarcinoma to biopsy (years) | |
| Median | 5 |
| Interquartile range | 2–8.5 |
| Total Gleason score (%) | |
| 6 | 7.5 |
| 7 | 30.2 |
| 8 | 18.9 |
| 9 | 37.7 |
| 10 | 5.7 |
| ECOG performance status (%) | |
| 0 | 55.9 |
| 1 | 44.1 |
| > 2 | 0 |
| PSA at time of biopsy (ng/mL) | |
| Median | 25.75 |
| Interquartile range | 12.8–138.4 |
| No. of prior regimens for CRPC (%) | |
| 1 | 6.8 |
| 2 | 44.1 |
| 3 | 22.0 |
| ≥ 4 | 27.1 |
| Life-extending therapies for metastatic CRPC received before biopsy (%) | |
| Abiraterone acetate | 28.8 |
| Enzalutamide | 10.2 |
| Sipuleucel-T | 8.5 |
| Docetaxel | 6.8 |
Abbreviations: CRPC, metastatic castration-resistant prostate cancer; ECOG, Eastern Cooperative Oncology Group; PSA, prostate-specific antigen.
Fig 1.
Overview of the biopsy and clinical testing workflow. (A) Schema of biopsy and testing. (B) Biopsy sites of the samples used for Clinical Laboratory Improvement Amendments (CLIA)–approved sequencing and PTEN immunohistochemistry testing. (C) List of 37 genes tested for mutation in the CLIA-certified GeneTrails Solid Tumor Genotyping Panel. CRPC, castration-resistant prostate cancer; OHSU, Oregon Health & Science University.
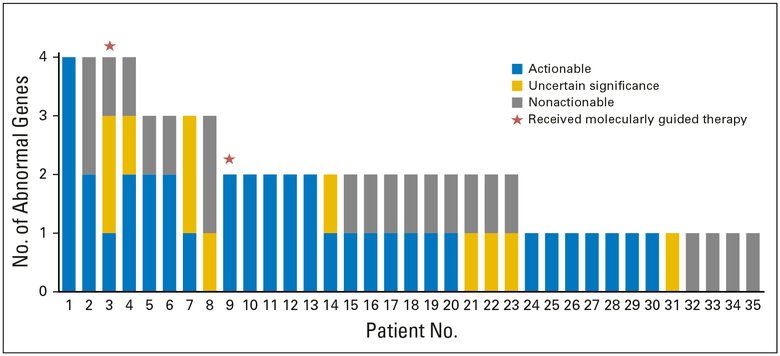
Of the 46 patients who had sufficient tumor for mutational testing, 31 (67.4%) had one or more mutational abnormalities identified by sequencing (Fig 2, Appendix Table A1). The median number of mutations found per patient was two (Fig 2). Among the 31 patients with at least one mutational abnormality, 60 mutations were identified by sequencing. Of 31 patients, 18 had at least one potentially actionable mutation, seven had at least one mutation of uncertain significance, and six had only nonactionable mutations.
Fig 2.
Mutational frequency among 35 patients with metastatic castration-resistant prostate cancer whose tumors underwent GeneTrails testing.
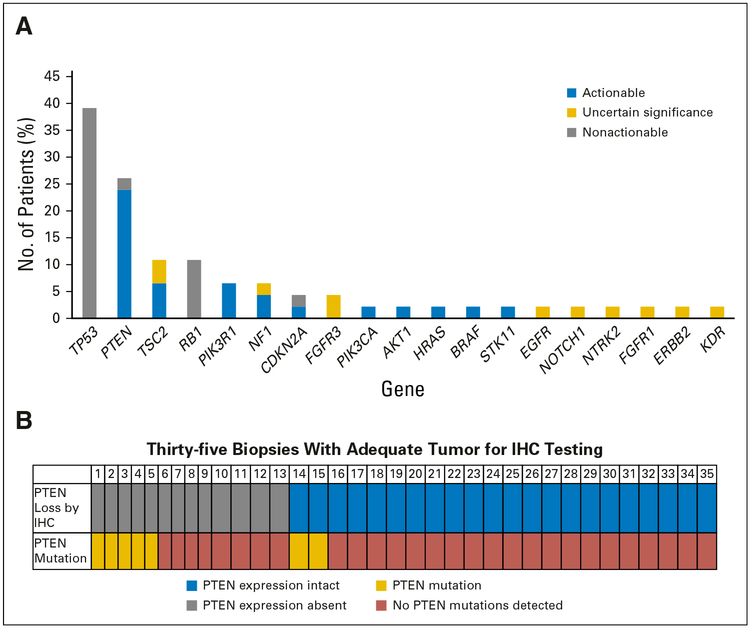
We also determined the frequency with which the mutational abnormalities were present in the patient cohort. The most frequently mutated genes were TP53 (18 of 46; 39.1%) and PTEN (12 of 46;26.1%; Fig 3A). We also examined PTEN expression in 35 metastatic biopsies from these subjects by IHC testing. PTEN loss was present in 13 of 35 patients (Fig 3B). Of note, PTEN mutations by sequencing were identified in seven of the 35 patients for whom PTEN IHC testing was performed. When considering mutational testing plus PTEN IHC testing in the 35 patients for whom both were performed, an abnormality in either mutational testing or PTEN IHC testing was present in 15 of 35 patients. There was a trend toward PTEN mutations and loss of PTEN protein expression by IHC testing (Fisher’s exact test two-tailed P value .075; Fig 3B).
Fig 3.
(A) Most frequent abnormal findings by gene mutation. (B) Frequency of PTEN loss by immunohistochemistry (IHC) and mutational testing.
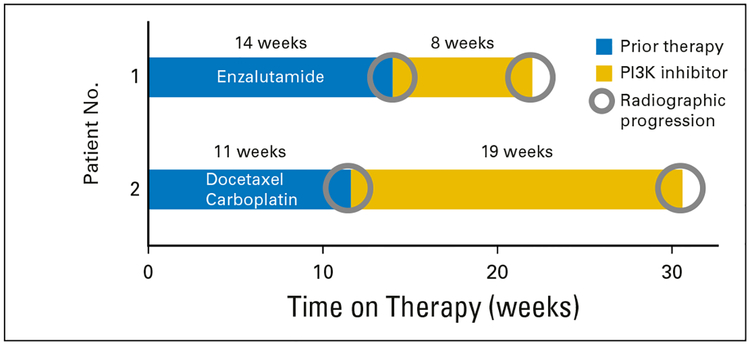
We also examined the frequency with which potentially actionable alterations were actually acted upon. Two of the 18 patients with a potentially actionable abnormality had treatment guided by mutational and IHC testing. In both cases, these patients had mutational abnormalities in the PTEN/PI3K pathway.
Patient 1 was 77 years of age, had metastatic CRPC, and experienced clinical and disease progression after 14 weeks of enzalutamide therapy. A biopsy of his iliac bone was performed, and testing revealed a PTEN p.I33fs*11 somatic mutation and loss of PTEN protein expression by IHC. On the basis of these findings, he was enrolled in a PI3K inhibitor clinical trial. However, he developed clinical and disease progression 8 weeks later (Fig 4).
Fig 4.
Swimmer plots illustrating patients’ time on treatment of the therapy given immediately before molecular testing and time on treatment with a phosphatidylinositol 3-kinase (PI3K) inhibitor that was guided by molecular testing. Radiographic progression was the reason for treatment discontinuation in all cases.
Patient 2 was 67 years of age and had metastatic CRPC; his prostate-specific antigen level was decreasing in the setting of disease progression. Biopsy of a pelvic mass revealed somatic mutations of FGFR3, NOTCH1, and RB1; PTEN loss was found by IHC testing. He was also found to have neuroendocrine carcinoma on histologic analysis. Subsequently, he was treated with docetaxel and carboplatin for four cycles but developed clinical and disease progression by 11 weeks. On the basis of PTEN loss, as shown by IHC testing of his biopsy specimen, he was enrolled in a clinical trial of a PI3K inhibitor. His symptoms of pain improved, and his scans showed stable disease for 19 weeks before clinical and radiographic progression was eventually determined (Fig 4).
DISCUSSION
Advances in next-generation sequencing have improved our ability to identify tumor genomic alterations. Increasingly, this information is reaching clinicians who are faced with making treatment decisions for patients. Metastatic CRPC is no exception, but there are few reports on CLIA-certified mutational testing in CRPC. Furthermore, there is little information on the frequency with which mutations are acted upon and the outcomes of those interventions. In this article, we describe our 3-year experience with mutational and IHC testing and the outcomes of molecularly guided therapy in patients whose tumors underwent this testing.
Obtaining adequate tumor from patients with metastatic CRPC, particularly those with bone metastases, has been notoriously difficult.11–13 However, nearly 80.0% of our biopsy specimens had sufficient DNA for tumor sequencing, including 71.0% of bone biopsy specimens. Factors that contribute to successful biopsies have been reviewed elsewhere, but include experience of the radiologists performing these procedures, biopsy site selection, and judicious decalcification process and use of amplicon-based next-generation sequencing panels.11–13
When we instituted our tissue collection protocol, little was known about the mutational landscape of metastatic CRPC. Therefore, we used a panel performed in a CLIA-certified laboratory that included 37 genes frequently mutated in solid tumors. Importantly, results of mutational testing were available within 6 weeks (meantime, 14 days), and all results were shared with the patients and treating providers.
Sixty-seven percent (31 of 46) of patients were found to have mutations. TP53 was the most commonly mutated gene, followed by PTEN, TSC2, and RB1. TP53 mutations of all classes were reported as nonactionable. PTEN was inactivated by loss of expression or mutation in 19 of 46(41.3%) patient biopsy specimens, which is consistent with other reports.8,14 Preclinical studies suggest that RB1-mutant tumors do not respond to CDK4/6 inhibitors, and all of the RB1 aberrations we found were predicted to be loss of function. However, these RB1 aberrations were reported as nonactionable because there are currently no effective strategies to target tumors with mutant retinoblastoma protein. TSC2 mutations were regarded as potentially actionable because of the predicted activation of mammalian target of rapamycin signaling and possible increased sensitivity to mammalian target of rapamycin inhibitors such as everolimus. Forty-eight percent (22 of 46) of tested cases had mutations involving the PI3K pathway, 4.3% (two of 46) involving RAF kinases, and 4.3% (two of 46) involving CDK inhibitors, which is similar to previous studies.7
Other potentially actionable mutations identified included targets of existing anticancer therapies such as EGFR, ERBB2, and KIT. RAS/RAF pathway abnormalities and CDK mutations were also seen in a subset of patients; there were agents targeting these pathways in clinical testing or approved for melanoma treatment at the time patients were enrolled in our biopsy protocol. Despite 31 of 46 patients (58%) having a theoretically actionable mutation, true actionability was limited because only two of 46 of our patients (4%) were prescribed agents that could block the identified gene alteration. Possible reasons include the lack of information on the importance of these mutations in CRPC and the lack of insurance coverage to prescribe these medicines for off-label uses in patients with CRPC. This lack of insurance coverage is probably because of the absence of positive basket trials showing a benefit of molecularly guided therapy versus standard therapies.15 Moreover, lack of nearby or onsite clinical trials with these agents may have also had an impact on the ability of patients to receive these drugs. Finally, many of the patients in our study had not exhausted all of the approved therapies for metastatic CRPC. This was also probably a contributing factor to why so few patients with theoretically actionable molecular alterations received molecularly guided therapy.
PI3K/AKT/PTEN pathway abnormalities were quite common in our cohort, and both subjects who received molecularly guided therapy received a PI3K inhibitor. The first patient progressed immediately (ie, as of the first assessment), and his progression-free survival with molecularly guided therapy was nearly identical to that with his immediate prior therapy. The other patient who received a PI3K inhibitor had stable disease for 19 weeks, which exceeded the progression-free survival of his immediate prior therapy. This second individual also had symptomatic improvement, which he did not experience with his immediate prior therapy. There are several potential reasons for the differences in response. First, although each patient had an abnormality in the same pathway, they received different PI3K inhibitors. In addition, it was unclear whether the PTEN/PI3K abnormalities in these tumors led to activation of this pathway in both tumors. Moreover, we did not have on-treatment biopsies or progression biopsies during PI3K inhibitor treatment, so we are unable to determine whether the drugs adequately hit the target in each subject. Finally, prior work has shown that PI3K inhibition leads to reciprocal feedback and activation of the AR pathway.16 Indeed, the subject who did not respond to PI3K inhibition had an adenocarcinoma, whereas the patient who seemed to derive greater benefit from PI3K inhibitor treatment had an AR-negative neuroendocrine tumor. Thus, it is possible that the latter patient did not have adaptive AR activation in response to PI3K inhibition. Finally, because our targeted panel only examined 37 genes for mutations, we cannot rule out the possibility that the first, nonresponding patient’s tumor had additional genomic abnormalities that were more consequential than his PIK3CA mutation and PTEN loss. Functional testing in tumor avatars from metastatic biopsies may help clarify the importance of molecular alterations found in tumor biopsy specimens, and we are currently developing these cultures from metastatic biopsy samples. Finally, it is unclear if intrapatient tumor heterogeneity with clones that did not harbor a PIK3CA mutation or PTEN loss also contributed to disease progression in the first subject.
Our panel included only 37 genes, which was another limitation of our study. This panel was not specific for CRPC, and many of the mutational abnormalities tested were not actionable. It is probable that the limited number of actionable genes in this panel also led to a low rate of molecularly guided therapy. Since adoption of the panel used in this study, Robinson et al7 have performed exome sequencing of metastatic CRPC tumors and identified additional genes frequently altered in metastatic CRPC. In that report, Robinson et al7 reported that 89% of patients with CRPC harbored a clinically actionable mutation. However, upon examination of the mutations that were identified, it is uncertain what proportion of these identified alterations could truly be used to guide therapy. Indeed, 63% of patients had genetic alterations of the AR pathway; however, it is unclear which therapies should be used in men who harbor these genetic alterations of the AR pathway. Moreover, several of the other genetic alterations described in that report are not readily targetable (ie, WNT pathway, cell cycle pathway, p53 alterations). Similarly, we report finding a mutation in 67% (31 of 46) of our patients, with 58% (18 of 31) harboring at least one potentially actionable mutation (ie, a drug was either approved or in clinical testing to block that genetic alteration). Importantly, our study did not investigate alterations in AR because this gene was not part of our Solid Tumor GeneTrails panel. Subsequent work has demonstrated a low frequency of AR mutations, including activating AR F877L mutations in this patient population.17 Moreover, Robinson et al reported that a high proportion of tumors among patients with CRPC (19.3%) harbored aberrations in DNA repair pathway genes such as BRCA2, BRCA1, or ATM. Subsequently, Mateo et al18 published the results of a phase II study demonstrating the preliminary antitumor activity of olaparib for patients with metastatic prostate cancer. Those patients whose tumors harbored mutations or copy number loss of these and other related DNA repair genes seemed to have the greatest benefit.18 On the basis of the frequency with which these abnormalities are found in CRPC, we anticipate that we would have identified 10 additional subjects with an actionable alteration if our panel had included mutational and copy number testing for genes in the DNA repair pathway. Recently, we expanded our mutational panel to include DNA repair genes with the hope that we identify patients who may benefit from poly (ADP-ribose) polymerase inhibitor or platinum-based therapy.
Importantly, even with more comprehensive sequencing panels, many hurdles related to access to therapy must be overcome to make molecular testing truly actionable. Both of the patients in our cohort who had therapy guided by molecular testing results enrolled in clinical trials with investigational agents, and one patient traveled out of state several hundred miles to receive his therapy. For many patients, travel outside the immediate area is not an option, and access to investigational agents is therefore problematic if a trial is not open locally. One solution to this is single-patient investigational new drug applications for compassionate use, although that process is time consuming and requires dedicated staff to assist with these requests. Tumors from other patients had molecular abnormalities that could have been targeted with an existing approved agent. However, none received these therapies. Insurance company disapproval was one reason; thus a critical aspect of making molecular abnormalities actionable is insurance approval for unapproved uses of existing anticancer drugs.
We are in a new era of cancer treatment, wherein molecular testing of tumors is commonplace. Our study provides important information on the need to not only have comprehensive molecular panels but also the complexities of implementing molecularly guided therapy. We will not truly be able to implement precision cancer care for patients through molecular testing until issues of access to drugs that target the identified abnormalities can be resolved.
Acknowledgments
Support
Supported by a Stand Up To Cancer–Prostate Cancer Foundation Prostate Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0409). This research grant is made possible by the generous support of the Movember Foundation. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research (7465sc). Other support includes Department of Defense Synergistic Idea Award (W81XWH-13–1–0420), National Institutes of Health/National Cancer Institute R01 awards (R01 CA178610, R01 CA169172), and Cancer Center Support Grant (P30 CA069533).
APPENDIX
Table A1.
Specific Molecular Abnormalities Identified and PTEN Expression Status
| Patient No. | Biopsy Site | Sample Identifier | Loss of PTEN Expression by IHC Testing (indicated by X) | Abnormality |
|---|---|---|---|---|
| 1 | Lymph node | DTB-OH-Ol 1 | TP53 S241T (T3). EGFR 5′ splice variant (T3). | |
| 2 | Bone | DTB-OH-O14 | X | |
| 3 | Bone | DTB-OH-022 | BRAF p.G469A (T2). | |
| 4 | Bone | DTB-OH-024 | X | PTEN p.I33fs*ll (T2). |
| 5 | Lymph node | DTB-OH-039 | TSC2 E1366A (T2). | |
| 6 | Liver | DTB-OH-040 | X | PTEN DEL L318fs*3 (T2). TP53 P190L (T3). RBI copy number loss (T3). |
| 7 | Lymph node | DTB-OH-043 | X | RBI R787Stop (T3). |
| 8 | Bone | DTB-OH-064 | TSC2 p.L448V (T2). | |
| 9 | Bone | DTB-OH-073 | TP53 p.Y205C and p.R273fs*73 (T3). | |
| 10 | Bone | DTB-OH-077 | TSC2 pE1005Q and p.R1056T (T2). | |
| 11 | Lymph node | DTB-OH-O8O | PTEN p.R130Q (T2). | |
| 12 | Bone | DTB-OH-O88 | TP53 p.R282fs*24 (T3). | |
| 13 | Bone | DTB-OH-089 | X | |
| 14 | Bone | DTB-OH-O91 | TP53 pE270V(T3). HRAS p.Q61K(T2). CDKN2A copy number loss (T3). | |
| 15 | Bone | DTB-OH-092 | X | TP53 p.G245S (T3). |
| 16 | Liver | DTB-OH-097 | X | NF1 p.D372A(T3). |
| 17 | Bone | DTB-OH-099 | X | |
| 18 | Lymph node | DTB-OH-102 | TP53 p.D48fs*75 (T3). NE1 p.H1163fs*31 (T3). | |
| 19 | Bladder | DTB-OH-103 | X | FGFR3 p.V3 83M (T3). RB1 p.Y424X (T3). NOTCH1 p.N2040S (T3). |
| 20 | Lymph node | DTB-OH-111 | TP53 p.V143M (T3). CDKN2A copy number loss (T3). KDR p.N793S (T3). | |
| 21 | Bone | DTB-OH-115 | TP53 copy number loss (T3). PIK3CA amplification (T2). RBI copy number loss (T3). TSC2 p.ClOOOY (T2). | |
| 22 | Lymph node | DTB-OH-129 | X | PTEN copy number loss (T2). |
| 23 | Lymph node | DTB-OH-132 | X | |
| 24 | Lymph node | DTB-OH-135 | X | PTEN copy number loss (T2). TP53 copy number loss (T3). |
| 25 | Bone | DTB-OH-141 | PTEN copy number loss (T2). TP53 copy number loss (T3). NF1 p.D283Y (T3). | |
| 26 | Bone | DTB-OH-149 | X | PTEN p.R233* (T2). TP53 copy number loss (T3). PIKR1 p.S229L (T2). STK11 p.S404F (T3). |
| 27 | Lymph node | DTB-OH-151 | TP53 p.R273H and copy number loss (T3). NTRK2 p.R808* (T3). | |
| 28 | Bone | DTB-OH-159 | AKT1 pE17K(T2). | |
| 29 | Lymph node | DTB-OH-167 | PTEN pE98R (T2). EGF’R copy number loss (T3). ERBB2 p.VI1811 (T3). | |
| 30 | Perianal | DTB-OH-176 | PTEN pE157* and copy number loss (T2). TP53 copy number loss (T3). | |
| 31 | Bone | DTB-OH-181 | RBI copy number loss (T3). | |
| 32 | Bone | DTB-OH-187 | PTEN copy number loss (T2). TP53 copy number loss (T3). | |
| 33 | Lymph node | DTB-OH-190 | TP53 copy number loss (T3). FGF’Rl p.S140L (T3). | |
| 34 | Bladder | DTB-OH-200 | PTEN copy number loss (T2). TP53 p.R337L (T3). PIK3R1 splice site (T3). TSC2 p.R208Q (T2). |
Abbreviation: IHC, immunohistochemistry.
Footnotes
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Molecular Testing in Patients With Castration-Resistant Prostate Cancer and Its Impact on Clinical Decision Making
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Derrick L. Tao
No relationship to disclose
Shawna Bailey
Research Funding: Janssen Research & Development (Inst), Medivation (Inst), Millennium (Inst), Aragon Pharmaceuticals (Inst), Bristol-Myers Squibb (Inst), OncoGenex (Inst), Astellas Scientific & Medical Affairs (Inst), Bayer (Inst), Myriad Genetics (Inst), SOTIO (Inst), Sanofi (Inst), Zenith Epigenetics (Inst), Boehringer Ingelheim Pharmaceuticals (Inst), Merck (Inst)
Tomasz M. Beer
Stock and Other Ownership Interests: Salarius Pharmaceuticals
Consulting or Advisory Role: AstraZeneca, Churchill Pharmaceuticals, Dendreon, Janssen Biotech, Janssen Oncology, Janssen Research & Development, Johnson & Johnson, Janssen Japan, Roche
Research Funding: Astellas Pharma (Inst), Bristol-Myers Squibb (Inst), Dendreon (Inst), Janssen Research & Development (Inst), Medivation (Inst), OncoGenex (Inst), SOTIO (Inst), SOTIO, Theraclone Sciences (Inst), Boehringer Ingelheim (Inst)
Erik Foss
Research Funding: Coinvestigator for a research grant from Moximed for magnetic resonance imaging of potential and postoperative patients undergoing KineSpring knee implant surgery (an orthopedic implant designed to alleviate degenerative joint pain).
Brooke Beckett
No relationship to disclose
Alice Fung
Speakers’ Bureau: Guerbet
Bryan R. Foster
Honoraria: CIVCO Medical Solutions
Alexander Guimaraes
Honoraria: Philips Healthcare 2015 ($1,500); Siemens Medical Solutions 2014 ($2,000)
Speakers’ Bureau: Siemens Medical Solutions 2014
Travel, Accommodations, Expenses: Siemens Medical Solutions 2014, Philips Healthcare 2015
Jeremy P. Cetnar
No relationship to disclose
Julie N. Graff
Honoraria: Bayer, Astellas Medivation, Janssen Oncology
Consulting or Advisory Role: Exelixis
Research Funding: Sanofi (Inst), Medivation (Inst), Bristol-Myers Squibb (Inst), Merck Sharp & Dohme (Inst), Janssen Oncology (Inst)
Patents, Royalties, Other Intellectual Property: OncoResponse: Exceptional responders
Travel, Accommodations, Expenses: Bayer, Merck Sharp & Dohme, Clovis Oncology
Kristine M. Eilers
No relationship to disclose
Eric J. Small
Stock and Other Ownership Interests: Fortis Therapeutics, Harpoon Therapeutics
Honoraria: Janssen-Cilag
Consulting or Advisory Role: Fortis Therapeutics, Gilead Sciences
Research Funding: Janssen (Inst)
Christopher L. Corless
Honoraria: Roche
Consulting or Advisory Role: Roche, Thermo Fisher Scientific Biomarkers
Travel, Accommodations, Expenses: Roche, Thermo Fisher Scientific
George V. Thomas
No relationship to disclose
Joshi J. Alumkal
Consulting or Advisory Role: Astellas Pharma, Bayer
Research Funding: Aragon Pharmaceuticals (Inst), Astellas Pharma (Inst), Millennium (Inst), Novartis (Inst), Zenith Epigenetics (Inst)
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2016. CA Cancer J Clin 66:7–30, 2016 [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, Puzanov I, Kim KB, et al. : Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363:809–819, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, et al. : EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304:1497–1500, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, et al. : Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448:561–566, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Hainsworth JD, Meric-Bernstam F, Swanton C, et al. : Targeted therapy for advanced solid tumors based on molecular profiles: Early results from MyPathway, an open-label, phase IIa umbrella basket study. J Clin Oncol 34, 2016 (suppl; abstr LBA11511) [DOI] [PubMed] [Google Scholar]
- 6.Berger MF, Lawrence MS, Demichelis F, et al. : The genomic complexity of primary human prostate cancer. Nature 470:214–220, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson D, Van Allen EM, Wu YM, et al. : Integrative clinical genomics of advanced prostate cancer. Cell 161: 1215–1228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BS, Schultz N, Hieronymus H, et al. : Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng HH, Klemfuss N, Montgomery B, et al. : A pilot study of clinical targeted next generation sequencing for prostate cancer: Consequences for treatment and genetic counseling. Prostate 76:1303–1311, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MM, Datto M, Duncavage EJ, et al. : Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19:4–23, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKay RR, Zukotynski KA, Werner L, et al. : Imaging, procedural and clinical variables associated with tumor yield on bone biopsy in metastatic castration-resistant prostate cancer. Prostate Cancer Prostatic Dis 17:325–331, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spritzer CE, Afonso PD, Vinson EN, et al. : Bone marrow biopsy: RNA isolation with expression profiling in men with metastatic castration-resistant prostate cancer—Factors affecting diagnostic success. Radiology 269:816–823, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross RW, Halabi S, Ou SS, et al. : Predictors of prostate cancer tissue acquisition by an undirected core bone marrow biopsy in metastatic castration-resistant prostate cancer—A Cancer and Leukemia Group B study. Clin Cancer Res 11: 8109–8113, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Pourmand G, Ziaee AA, Abedi AR, et al. : Role of PTEN gene in progression of prostate cancer. Urol J 4:95–100, 2007 [PubMed] [Google Scholar]
- 15.Le Tourneau C, Delord JP, Gonçalves A, et al. : Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16:1324–1334, 2015 [DOI] [PubMed] [Google Scholar]
- 16.Carver BS, Chapinski C, Wongvipat J, et al. : Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19:575–586, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azad AA, Volik SV, Wyatt AW, et al. : Androgen receptor gene aberrations in circulating cell-free DNA: Biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res 21:2315–2324, 2015 [DOI] [PubMed] [Google Scholar]
- 18.Mateo J, Carreira S, Sandhu S, et al. : DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373:1697–1708, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]