Figure 1.
Proteomic data acquisition, manipulation and verification.
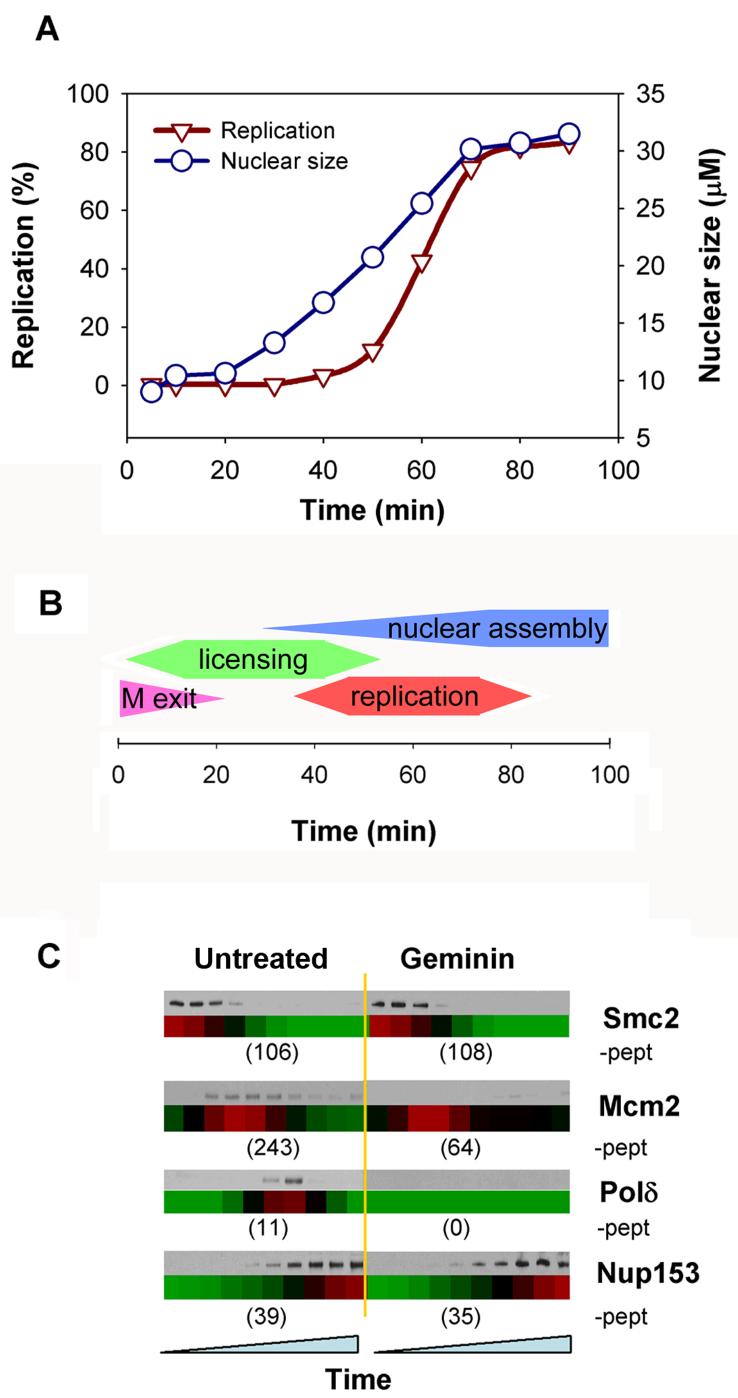
(A) Sperm chromatin was incubated in egg extract and the efficiency of DNA replication was monitored by [α32P] dATP incorporation. The mean size of ∼20 randomly selected nuclei at each timepoint is also shown. (B) Cartoon of cell cycle events occurring during incubation of sperm nuclei in Xenopus egg extract, showing the approximate timing of mitotic exit, origin licensing (Mcm2-7 loading), nuclear assembly and DNA replication. (C) Chromatin was isolated in parallel to the mass spectrometry samples and immunoblotted for the indicated proteins. Below are corresponding heat maps derived from mass spectrometry (red highest, green lowest, black intermediate). The total number of peptides identified for each protein across the entire timecourse is also shown.