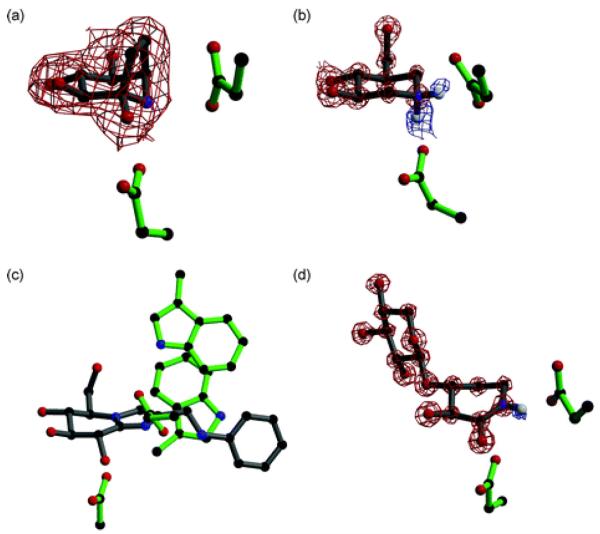
Fig. 4.
(a) Calystegine (8) in complex with a family 1 β-glucosidase (PDB code 2CBV94); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. Observed electron density (for the maximum likelihood weighted 2Fobs−Fcalc map, contoured at 1σ) is shown for calystegine, showing it binds in a similar orientation to isofagomine. (b) Cellobio-derived form of isofagomine (9) in complex with a family 5 endoglucanase (PDB code 1OCQ101); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. Observed electron density for the maximum likelihood weighted 2Fobs−Fcalc map, contoured at 2.5σ, is shown in red and for the Fobs−Fcalc map, contoured at 2.1σ, is shown in blue. The ‘difference’ density shows the presence of two hydrogen atoms on the nitrogen atom of isofagomine. (c) Phenylaminomethyl-substituted glucoimidazole in complex with a family 3 β-d-glucan glucohydrolase (PDB code 1X39133); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. The two tryptophan residues in the active site are proposed to make hydrophobic interactions with the phenyl ring of the inhibitor, but this interaction has not been observed in all enzyme complexes with substituted imidazole inhibitors. (d) Xylobio-derived isofagomine lactam in complex with a family 10 xylanase (PDB code 1OD8137); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. Observed electron density for the maximum likelihood weighted 2Fobs−Fcalc map, contoured at 4σ, is shown in red and for the Fobs−Fcalc map, contoured at 1.8σ, is shown in blue. The difference density shows the presence of a hydrogen atom on the nitrogen atom of the isofagomine lactam, indicating it exists as the amide tautomer and not the iminol as originally proposed.