Figure 1.
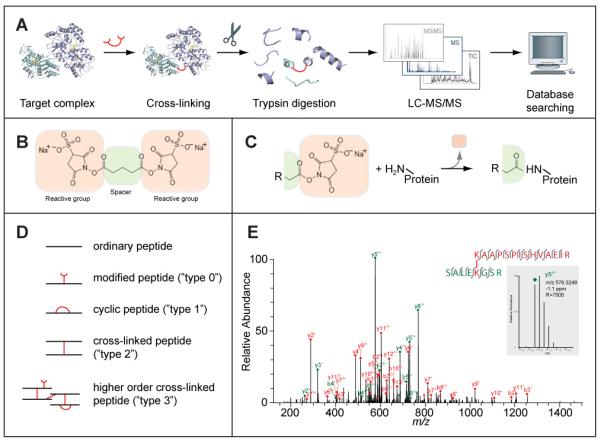
(A) Outline of the cross-linking/mass spectrometry process. A target complex is cross-linked in solution and digested with trypsin into peptides. The peptides are analysed by liquid chromatography coupled high-resolution mass spectrometry (LC-MS/MS) to obtain high-resolution masses and fragment masses (high/high) for cross-linked peptides. The fragmentation spectra of all peptides are subjected to database searching to identify cross-linked peptides. As an optional step, cross-linked peptides can be enriched before their LC-MS analysis. (B) A typical cross-linker, here Bis[Sulfosuccinimidyl] glutarate (BS2G), is composed of two reactive groups on either end separated by a spacer. This cross-linker reacts with primary amines (lysine side chain, protein N-terminus). Others target thiols (cysteine side chain) or activate carboxylic acids (aspartate, glutamate, protein C-terminus) for reaction with primary amines. (C) Reaction of a cross-linker with a primary amine. Part of the cross-linker, the leaving group, is replaced by the primary amine to form a covalent bond between the spacer and the amine. In this case, a peptide bond is formed. R can stand for either the rest of the cross-linker or may contain another protein, if the cross-linker had already reacted on its other end. (D) Peptides types that can be observed after cross-linking and trypsin digestion. (E) High resolution fragmentation spectrum of a cross-linked peptide obtained on an LTQ-Orbitrap mass spectrometer (adapted from (Chen et al., 2010)). Fragment peaks are annotated in red or green, depending on the peptide that fragmented and following the naming convention for peptides (y: C-terminal fragment, b: N-terminal fragment, both as a result of dissociating the peptide bond in the peptide back bone, followed by the number of amino acids included in the fragment and the charge of the fragment). All observed fragments are also indicated as bond cleavages between amino acids in the two cross-linked peptides. In this case, virtually all possible fragments of the peptide pair have been matched and virtually all peaks have been annotated resulting in a high-confidence identification of this cross-link. The inset shows a zoom onto one fragment peak (m/z 576, 3248) which matched with -1.1 ppm to the proposed peptide sequence. The high resolution of the spectrum (R 7505 for this peak) allows clear separation of the isotope peaks and consequently assignment of the fragment's charge state.
