Fig. 4.
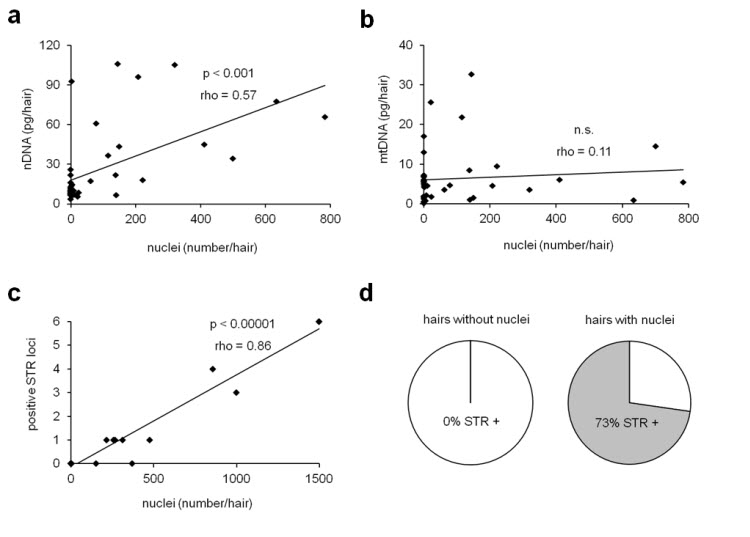
Analysis of single hairs by in situ labeling of DNA, DNA quantification and STR typing. Samples were prepared by cutting 2 cm from the distal end of single hairs of different individuals. The hairs were stained for DNA, and nuclei were counted under the microscope. Subsequently, the dye was removed by washing and the same hairs were then subjected to DNA extraction and quantification of nuclear and mitochondrial DNA by real-time PCR. Spearmańs rank correlation coefficient (rho) and the significance of correlation were determined for the frequency of DNA-positive nuclei and nuclear DNA (a) and the frequency of DNA-positive nuclei and mitochondrial DNA (b). In a separate experiment STR typing was performed on 9 hairs lacking nuclei and 11 hairs containing more than 8 nuclei per mm. The correlation between the total number of nuclei labeled in situ and the number of positive STR loci is shown in panel (c). All hairs that lacked Hoechst-positive nuclei did not yield positive STR results whereas 73% of hairs with Hoechst-positive nuclei yielded at least one positive STR result. The pie charts illustrate the portions of all hairs investigated (d). rho, Spearman’s rank correlation coefficient; n.s., not significant
