Abstract
Background
Pneumococci could evade pneumococcal conjugate vaccines (PCV) by modifying, mutating, or deleting vaccine-serotype capsule genes or by downregulating capsule production. We sought to assess whether pneumococci that are nontypeable (NT) by the Quellung reaction truly lack capsule genes or are failing to produce capsule in vitro.
Methods
We applied multilocus sequence typing and a microarray for detection of pneumococcal polysaccharide capsule biosynthesis genes to NT carriage (children aged <5 years; years 1997–2000, 2006–2008) and NT invasive disease (IPD) (all ages; years 1994–2007) isolates from Native American communities.
Results
Twenty-seven of 28 (96.4%) NT IPD isolates had sequence types (STs) typically found among typeable IPD isolates and contained whole or fragments of capsule genes that matched known serotypes; 1 NT-IPD isolate had a profile resembling NT carriage isolates. Forty-nine of 76 (64.5%) NT carriage isolates had STs that typically lack capsule genes and were similar to NT carriage isolates found globally.
Conclusions
This is the first documentation of IPD from an NT strain confirmed to lack all known capsule genes. Most NT IPD isolates have or had the capacity to produce capsule, whereas a majority of NT carriage isolates lack this capacity. We found no evidence of pneumococcal adaptation to PCV7 via downregulation or deletion of vaccine-serotype capsule genes.
Streptococcus pneumoniae (pneumococcus) is the leading vaccine-preventable cause of morbidity and mortality among children aged <5 years [1]. Pneumococal disease prevention through pneumococcal conjugate vaccine (PCV) use among infants and children is a major area of public health focus globally. Pneumococci are categorized into serogroups and serotypes based on the immunological similarities of their polysaccharide capsule. The Quellung reaction, a standard method to serotype pneumococcal isolates, uses anti-sera against serotype-specific polysaccharide capsule [2]. However, there are pneumococcal isolates that cannot be typed by this method due to lack of capsule production; these are termed nontypeable (NT) pneumococci.
There are some ecological advantages for pneumococci to not produce capsule. A process called phase variation has been described in the pneumococcus, in which transparent variants show greater adherence to epithelial cell walls than opaque variants [3, 4]. Transparent variants decrease their expression of capsule and increase their expression of surface attachment proteins, assisting in attachment to epithelia and supporting colonization of the nasopharynx. In contrast, opaque variants are more associated with invasive disease and express higher levels of capsule, which conceal immunogenic surface proteins from the host. Lack of capsule production can occur by either downregulating gene expression of existing functional capsule polysaccharide biosynthesis (cps) genes or by the acquisition of genetic lesions, such as single nucleotide polymorphisms (SNPs), or deletion of cps genes, which renders capsule polysaccharide biosynthesis non-functional. Most genes required for capsule polysaccharide biosynthesis are located in a genomic locus flanked by dexB and aliA genes and include shared and serotype-specific gene sequences [5–7].
Most published research on NT pneumococci has been from conjunctivitis outbreaks from which NT strains are routinely isolated [8–13]. A small number of NT pneumococcal carriage studies have also been conducted [14, 15]. Two distinct groups of NT carriage isolates have been described using the multilocus sequence typing (MLST) method [8, 14–16]. One group contains a full complement of cps genes for a particular serotype, and the rest of the genome is closely related to known serotypeable lineages. The other group is not closely related to serotypeable pneumococci and contains no functional capsule genes. The NT pneumococci of the second group are common in conjunctivitis outbreaks and appear regularly among carriage isolates. It is generally believed that all pneumococci causing invasive pneumococcal disease (IPD) contain functional capsule genes and that IPD from strains that do not have the ability to produce capsule does not occur; however, there are extremely limited genetic data on NT IPD isolates. A small MLST analysis of 7 NT IPD and 20 NT carriage isolates identified 2 distinct groups, 1 related to “encapsulated” serotypeable strains, and the other related to NT strains typically found in conjunctivitis outbreaks [16].
Pneumococcal conjugate vaccines, the current approach to disease prevention in infants and children, are capsule-based products. Establishing whether NT pneumococci, especially those found in IPD, lack capsule gene loci or are simply failing to produce capsule in vitro is essential. Four percent of IPD isolates obtained from infants across the United States were NT by Quellung in 2004 [17]. Downregulation of meningococcal serogroup C capsule production has been described in serogroup C meningococci isolated from populations who had received conjugate meningococcal serogroup C vaccine in the United Kingdom between 1999 and 2001 [18]. Capsule expression in other meningococcal serogroups remained constant during the time period. The adaptive abilities of the pneumococcus through recombination and horizontal gene transfer with other pneumococci or related species in the nasopharynx may give it the capacity to adapt to vaccine pressure by switching or deleting serotype-specific capsule gene loci.
Analysis of NT pneumococci is particularly important in Navajo and White Mountain Apache communities in which there are high rates of pneumococcal carriage and disease and high PCV7 coverage [19, 20]. Three percent (95% confidence interval [CI], 1.7%–4.1%) of IPD isolates obtained from Navajo children and adults between 2001 and 2006 [19] and 5% (95% CI, 4.2%–5.7%) of pneumococcal carriage isolates from Navajo and White Mountain Apache children collected between 2006 and 2008 were NT [20]. In contrast, only 1.88% (95% CI, .1%–.5%) of carriage isolates from children aged <7 years were NT in a study conducted in Massachusetts [21].
This study characterized pneumococci that were NT by the Quellung reaction from IPD and carriage samples collected from Navajo and White Mountain Apache communities. We aimed to describe their relatedness using MLST, to determine the presence of cps genes by microarray, and to assess any significant changes in these genes that may have occurred since introduction of PCV7. We expected the IPD isolates to contain genes for capsular biosynthesis and otherwise closely resemble serotypeable isolates by MLST. Conversely, we expected the carriage isolates to lack capsule genes and be comprised of sequence types that are typically NT.
METHODS
Pneumococcal carriage isolates were collected from Navajo and White Mountain Apache children aged <5 years who were enrolled in a PCV7 efficacy trial (1997–2000) [22] or enrolled in a study of nasopharyngeal (NP) colonization within families after routine use of PCV7 (2006–2008) [20]. The IPD isolates were obtained from population-based active surveillance of clinical microbiology laboratories serving these same communities [19]. Methods to obtain the NP and IPD specimens are described elsewhere [19, 23]. Morphology and alpha-hemolysis were the initial criteria used to select colonies from inoculated trypticase soy agar plates with 5% sheep blood agar and gentamicin (Becton Dickinson). Pneumococcus was confirmed by optochin sensitivity and bile solubility assays. If pneumococcus was isolated from the sample, serotype was assessed using the standard Quellung reaction.
We defined isolates as being NT if the isolate could not be typed by the Quellung reaction on 2 attempts for IPD isolates and a single attempt for NP isolates. From the latter longitudinal NP study, this analysis only included 1 NT carriage isolate per household to avoid over-representing NT strains circulating within households. All NT IPD isolates identified from 1994 through 2007 from Navajo and White Mountain Apaches of all ages were included in the analysis [19].
MLST
Pneumococci were prepared for MLST and sequence types determined by MLST using described methods [24, 25]. Polymerase chain reaction products were sequenced using a Prism 3730 × I Genetic Analyzer (Applied Biosystems). The raw sequences were analyzed using Molecular Evolutionary Genetics Analysis 4 software (http://www.megasoftware.net). Alleles and sequence types were assigned using the online MLST database (http://spneumoniae.mlst.net). The eBURST algorithm (version 3, http://eburst.mlst.net) was used to group sequence types into clonal complexes (CCs) composed of closely related sequence types [26]. To determine whether the sequence type was previously associated with a particular serotype(s), we searched the MLST database [27].
Molecular Serotyping
All IPD isolates and a random subset of carriage isolates were characterized by a microarray assay to determine the presence of known cps genes [28]. The microarray was designed by the Bacterial Microarray Group at St. George’s University of London (BμG@S; http://bugs.sgul.ac.uk/) and manufactured on the Agilent SurePrint 8 × 15 K platform (Agilent Technologies) [28]. The microarray included reporters to represent all cps genes involved in capsule polysaccharide biosynthesis of the 91 serotypes known to date [5, 29, 30]. The serotype was determined by the combination of cps genes found to be present in the isolate by microarray analysis. Pneumococcal samples for microarray were prepared using Qiagen DNeasy Mini Spin Column kits per manufacturer’s instructions.
Analysis of PCV7 Adaptation
To assess whether downregulation of PCV7 serotype capsule genes has occurred more frequently since introduction of PCV7, we compared the proportion of PCV7 serotypes identified by microarray among the NT pneumococci before and after PCV7 introduction using a Fisher’s exact test. If this mode of adaptation to PCV7 was occurring, we would expect to see an increased proportion of PCV7 serotypes detected by microarray among the NT pneumococci since introduction of the vaccine. We also compared the proportion of sequence types associated with PCV7 serotypes [27] before and after PCV7 introduction, again using a Fisher’s exact test. An increase in sequence types associated with PCV7 serotypes among the NT strains after PCV7 introduction would suggest adaptation to vaccine pressure via reduction in capsule production. All statistical analyses were performed in STATA 9.1.
RESULTS
Sequence Type Analysis and Clustering Unique to Carriage Isolates
A total of 104 NT pneumococci were analyzed by MLST, 28 from IPD episodes and 76 from NP carriage events. The sequence types (STs) of the isolates, year and source of isolation, and serotypes commonly associated with that particular ST according to the MLST online database are found in the Appendix. Among the 76 carriage isolates, 25 STs were identified. Forty-nine of the 76 (64.5%) NT carriage isolates were sequence types ST344 (n = 17), ST448 (n = 13), ST1054 (n = 9), ST1186 (n = 2), and ST2011 (n = 8), which are almost always identified as NT strains [26]. Thirteen novel STs were identified among the carriage isolates. Within these novel STs, strains 345, 417, 1418–08, and 2141–08 also each had 1 novel allele sequence that had not been previously identified in the MLST database.
Among the 28 NT IPD isolates, 16 STs were identified. Sequence types 191 (n = 4) and 227 (n = 5) were the 2 most common STs found among these NT IPD isolates, and they were not found among the NT carriage isolates. Sequence types 191 and 227 are commonly found among typeable IPD isolates from this population and others [27, 31]. Only 1 IPD strain (strain 1618) was identified as ST1054, which is an ST typically associated with NT carriage strains [15]. A sequence type for IPD strain 1617 could not be determined. The primers failed to work for the aroE and spi alleles after multiple attempts, and the other alleles were novel, suggesting this isolate was actually not a pneumococcus, but a closely related species.
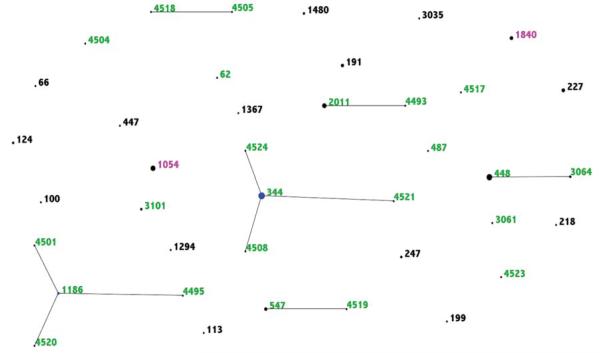
The eBURST diagram in Figure 1 illustrates the relationship and differences between STs found in IPD and carriage. Only ST1840 and ST1054 were found in both IPD and carriage. There were 2 ST1840 IPD isolates and 3 ST1840 carriage isolates. This ST is generally associated with serotype 16F and NT strains. Sequence type 1054 is generally associated with NT carriage isolates, and 9 of 10 of our ST1054 isolates were indeed from carriage. However, 1 ST1054 isolate was found in IPD, and the microarray analysis revealed this isolate did not contain a cps locus, as typical to ST1054 NT carriage isolates. Sequence types found among IPD isolates did not form any CCs with other STs found in this NT dataset, whereas there were 6 CCs identified among our carriage isolates. Of the 6 founders of these CCs, ST344, ST448, ST1186, and ST2011 are types that are generally only associated with NT strains and not found among strains with capsule [14, 15].
Figure 1.
eBURST comparison of nontypeable pneumococcal isolates showing the relationship and differences between sequence types (STs) found in invasive pneumococcal disease (IPD) and carriage. Sequence types in black were only found in IPD; STs in green were only found in carriage; and STs in pink were found in both IPD and carriage.
Presence of cps Genes in NT IPD Isolates
The microarray analysis of the 28 NT IPD isolates revealed that 20 isolates had cps gene content fully matching the capsule gene loci of known serotypes, whereas 8 had capsule gene content that only partially matched capsule gene loci of known serotypes (Table 1). The serotypes predicted by microarray corresponded to the serotypes associated with the MLST analysis in all but 1 case (strain 1634). This strain was ST218, which is generally associated with serotype 12F; however, serotype 7F capsule genes were found on the microarray. Strain 1626 had an ST199, typically associated with serotype 19A; however, the microarray analysis identified capsule genes for both serotype 19A and serotype 37. The intensity measured for serotype 19A was much stronger, suggesting a serotype 19A sample was mixed or contaminated with serotype 37 during the isolation or culturing process. We failed to determine the sequence type for strain 1617, as previously noted; however, the microarray identified the presence of capsule genes typically associated with serotype 16A. ArrayCGH analysis of the S. pneumoniae genome backbone also present on the array indicated that strain 1617 might be a closely related Streptococcus species, such as Streptococcus mitis, and not actually an NT pneumococcus (data not shown). The isolate lacked all known shared pneumococcal capsule genes for initiation and modulation, so it likely did not produce any capsule, or it may have had divergent homologues with equivalent function present that were not detected by the current microarray version.
Table 1.
Array Serotype, Source, Year of Isolation, Sequence Type (ST), and Associated Serotypes of Nontypeable (NT) Pneumococcal Isolatesa
| Strain ID | Source | Year | Array Serotype | ST | Associated Serotypesb |
|---|---|---|---|---|---|
| 1620 | IPD | 1990 | 9N | 66 | 9N, 19F, 14 |
| 1614 | IPD | 1994 | 18C | 113 | 18C |
| 1613 | IPD | 1994 | 7F | 191 | 7F |
| 1610 | IPD | 1994 | 4 | 247 | 4 |
| 1611 | IPD | 1994 | 4 | 247 | 4 |
| 1616 | IPD | 1995 | 14 | 124 | 14 |
| 1615 | IPD | 1995 | 7F | 191 | 7F |
| 1612 | IPD | 1995 | 5 | 3035 | 5 |
| 1617 | IPD | 1995 | NT (non-pneumoniae): 16A-like but wzg, wzh, wzd, wze, wchA, rmlD absent or divergent |
Failed | NA |
| 1618 | IPD | 1998 | NT: only glf present | 1054 | NT |
| 1619 | IPD | 1999 | 14 | 124 | 14 |
| 1626 | IPD | 2002 | 19A (92%) + 37 (8%) | 199 | 19A and others |
| 1636 | IPD | 2002 | NT: only rmlB, rmlD, rmlA, rmlC present | 227 | 1 |
| 1635 | IPD | 2003 | 7F | 191 | 7F |
| 1625 | IPD | 2003 | 37 | 447 | 37, 11 |
| 1622 | IPD | 2003 | NT: 22A-like but wcwC and wcwA absent or divergent | 1294 | 22F |
| 1627 | IPD | 2003 | 33A/F | 1367 | 33F |
| 1623 | IPD | 2003 | 16F | 1840 | 16F |
| 1628 | IPD | 2004 | 1 | 227 | 1 |
| 1621 | IPD | 2004 | NT: only rmlB, rmlD, rmlA, rmlC present | 227 | 1 |
| 1629 | IPD | 2004 | NT: only rmlB, rmlD, rmlA, rmlC present | 227 | 1 |
| 1633 | IPD | 2004 | 8 | 1480 | 8 |
| 1631 | IPD | 2005 | 8 | 1480 | 8 |
| 1632 | IPD | 2005 | NT: 16F-like but wzg gene absent or divergent | 1840 | 16F |
| 1624 | IPD | 2006 | 7F | 191 | 7F |
| 1634 | IPD | 2006 | 7F | 218 | 12F |
| 1630 | IPD | 2006 | NT: only rmlB, rmlD, rmlA, rmlC present | 227 | 1 |
| 1637 | IPD | 2007 | 33A/F | 100 | 33F |
| 2566-06 | NP | 2006 | NT: only glf present | 448 | NT |
| 2537-06 | NP | 2006 | 35F | 487 | 35F |
| 2489-06 | NP | 2006 | NT: no cps genes detected | 2011 | NT |
| 2563-06 | NP | 2006 | 35A | 4505 | None |
| 0900-07 | NP | 2007 | NT: only glf present | 344 | NT |
| 2009-07 | NP | 2007 | 34 | 547 | 34 |
| 1944-07 | NP | 2007 | 6C | 3101 | 6A |
| 1878-08 | NP | 2008 | NT: only glf present | 1054 | NT |
| 1927-08 | NP | 2008 | 35A | 3026 | 35A |
Abbreviations: IPD, invasive pneumococcal disease; NA, not available; NP, nasopharyngeal.
Sorted by source, year of isolation, and ST.
Per Multilocus Sequence Typing online database.
Strain 1622 (ST1294) and strain 1632 (ST1840) had partial capsule genes of the serotype predicted by MLST; however, there were also absent or divergent genes rendering these strains different from the cps gene content of known serotypes. Specifically, strain 1622’s array serotype was 22A-like but was missing or had divergent HG48 wcwC and HG63 wcwA genes, which code for transferases [7]. Strain 1632’s array serotype was 16F and only differed at or was missing the HG0 wzg gene. The wzg gene is an initiation housekeeping gene that is shared by all serotypes [5]. Lack of the wzg gene is associated with lack of capsule synthesis [15].
Due to incomplete cps gene loci, an array serotype was indeterminable for 5 isolates. One of these, NT IPD strain 1618 (ST1054), was indeterminable because there was only 1 known capsule gene detected, glf, which is a result typical of NT pneumococci from carriage [16]. We believe this strain would be unable to produce capsule and is therefore a true NT IPD isolate. This isolate was collected from a blood culture of an infant aged 7 months with a diagnosis of bacteremia without focus. Strains 1621, 1629, 1630, and 1636 were ST227, which is associated with serotype 1. However, these 4 strains only had the rhamnose biosynthesis genes rmlB, rmlD, rmlA, and rmlC present, a region present in serotype 1 but also in a number of other serotypes. These isolates lacked capsule genes required for initiation, synthesis, and transport, and therefore they likely could not produce capsule in their current state.
Absence of cps Genes in a Majority of NT Carriage Isolates
We analyzed a sample of our NT carriage isolates by microarray to confirm the lack of known cps genes in STs that are typically NT and to determine if STs associated with specific serotypes had any known capsule genes present. A sample of NT carriage isolates was selected for microarray analysis based on our MLST results (Appendix). We randomly chose 1 ST344, ST448, ST1054, and ST2011 because these types have been consistently identified as NT strains in this population [30] and in other studies [14, 15]. We also chose 4 NT carriage isolates with STs that are generally associated with a particular serotype and 1 isolate with a novel ST. The analysis of the carriage isolates revealed that the typically NT pneumococci (ie, ST344, ST448, ST1054, and ST2011) either had only glf present or no known capsule genes (Table 1). Therefore none would be able to produce capsule. The STs that were associated with specific serotypes had complete cps genes present for the expected serotype, suggesting that they were either not expressed in vitro or were not detected by Quellung.
No Evidence of Pneumococcal Adaptation to PCV7 by Downregulation or Deletion of Capsule Genes
The MLST and microarray analysis of our NT isolates do not show evidence that pneumococci in this population are adapting to PCV7 pressure by capsule loci deletion or downregulation, although the analysis was limited in power. Among NT IPD isolates from prior to routine PCV7 introduction in 2000, 45% had PCV7 serotype capsule genes by microarray and 55% had PCV7-associated STs, whereas none of the NT IPD isolates after PCV7 introduction (2001–2007) had PCV7 serotype capsule genes or STs. Furthermore, no NT carriage isolate had a PCV7 ST or microarray serotype regardless of the isolation date. This analysis provides additional evidence that the vast majority of NT IPD isolates resemble serotypeable IPD isolates in the makeup of their genetic material. We infer they were likely expressing capsule in vivo and were therefore eliminated from invasive disease and carriage after PCV introduction.
DISCUSSION
This study has described 4 groups of NT isolates in a defined population: (1) isolates that lack known cps gene sequences (or only had the glf gene present); (2) isolates that have partial cps loci; (3) isolates with complete cps loci when compared with known serotypes; and (4) isolates that are not likely to be pneumococci (strain 1617). We consider the first 2 types to be truly NT pneumococci because they are deficient in the ability to produce capsule, whereas those of the third type likely retain the capability of capsule production but are not expressing capsule in vitro. Identifying non-pneumococci via microarray is not uncommon in pneumococcal carriage studies ( J. Hinds, personal communication) and is important because these related species may also contribute cps or antimicrobial resistance genes to the gene pool available for genetic exchange in the nasopharynx. However, isolate purification or contamination must be ruled out before exploring the possibility that this non-pneumococcal isolate had the potential to cause invasive disease.
We conclude that this collection of NT IPD isolates fundamentally differs from the majority of NT carriage pneumococcal isolates in this population and others. Only 1 NT IPD isolate, strain 1618 (ST1054), had an MLST and array profile that suggests true NT IPD and resembles NT carriage isolates. All other STs identified among the NT IPD isolates have routinely been found among serotypeable IPD isolates in this population [31]. Most of the NT IPD isolates studied also have complete cps genes present for known serotypes, suggesting that capsule production was simply downregulated in vitro, thereby resulting in a negative Quellung reaction. A few NT IPD isolates were either missing or had divergent cps genes as identified by the microarray and appear to be remnants of known serotypes. No pneumococci isolated after routine PCV7 introduction had PCV7 serotype genes by microarray, mirroring the reduction observed in PCV7 serotype IPD rates by Quellung [19].
In contrast, the NT carriage strains we tested were comprised of either isolates with complete cps gene loci, which were likely downregulated in vitro, or isolates with either no known capsule genes present or only glf, both typical of truly NT pneumococci isolates. Both carriage groups have STs typically found among the other serotypeable carriage isolates from this population [31] and include STs that have been described in other NT carriage studies [14–16]. We did not find any NT carriage isolate with only a partial cps gene sequence; however, the small number of carriage isolates we analyzed by microarray limits this information.
The few isolates with partial known cps loci were unique to the IPD isolates and likely lacked specific genes required to produce serotype-specific antigen targeted in the Quellung reaction. The missing serotype 1 genes in strains 1621, 1629, 1630, and 1636 (all ST227) are usually located in a contiguous region in serotype 1 that is flanked by 2 insertion sequence elements (IS1167), suggesting that the deletion seen in all 4 strains may have been transposase or recombination mediated. The isolates were collected between 2002 and 2006 from 3 adults and 1 child living in 3 geographically distinct areas. The presence of insertion sequence elements and their diverse geographies and time periods suggest the isolates may have emerged independently. Further exploration would be necessary to determine whether these deletions may have occurred in vivo or in vitro.
Some NT isolates may simply be identified as such because of imperfect pneumococcal typing methods. Clarifying the likelihood that isolates are truly NT will improve the quality of pneumococcal typing efforts that are critical to pneumococcal disease surveillance and vaccine development. There may also be explanations for why isolates with complete cps loci were NT, technical errors aside. Single nucleotide polymorphisms in the gene sequence have been shown to truncate cps gene products, which in turn prevent capsule production [32]. These isolates are NT by the Quellung reaction, but the gene sequences are likely similar enough to be detected on the microarray. Small deletion(s) or SNPs could be identified by sequencing and compared with known complete cps gene sequences for a specific serotype; their effect on cps gene expression could be assessed by transcriptional analysis by microarray; and their effect on cps production could be measured by an enzyme-linked immunosorbent assay [4].
Impact and Future of NT Pneumococci
This study affirms the widespread dissemination of NT pneumococcal STs that lack capsule gene loci in carriage and the near absence of these particular STs in IPD. It does appear that true NT IPD can occur, albeit very rarely. Capsule is clearly not required for survival in and transmission between the nasopharynx and other mucosal membranes, such as the conjunctiva. Our study found no evidence of pneumococcal adaptation to PCV7 by downregulation or deletion of PCV7 serotype capsule genes after 8 years of routine vaccine use, although our analysis was limited in power. We confirmed that there were no STs or array serotypes associated with PCV7 serotypes in the vaccine era among the NT IPD or carriage isolates, mirroring what has been observed among the isolates identified as vaccine serotype by conventional means [20, 31].
Nontypeable IPD isolates identified in future studies should be further analyzed beyond serotyping to determine what capsule genes are present and which appear necessary for invasive disease. Although rare in this study, NT IPD isolates with incomplete capsule gene loci would also require characterization of capsule products and assessment of whether deletions occurred in vivo or in vitro postisolation. Distinguishing the role of specific capsule genes could help direct additional disease prevention efforts and identify adaptive mechanisms to current ones.
Supplementary Material
Acknowledgments
The authors express their sincere appreciation to the children and adults from the Navajo and White Mountain Apache communities who participated in the studies. We gratefully acknowledge the tireless efforts of the Center for American Indian Health field staff who collected these data over many years and the dedicated support provided by Delois Jackson, Bob Gertz, and Dr Bernard Beall at the Centers for Disease Control and Prevention (CDC) for NP sample storage and serotyping. We also gratefully acknowledge the serotyping of the invasive strains by Karen Rudolph and Marcella Harker-Jones at the CDC Arctic Investigations Program, Anchorage, Alaska. We appreciate the MLST support provided by Cynthia Bishop and Dan Godoy at Imperial College and acknowledge the Wellcome Trust for funding the Bacterial Microarray Group at St. George’s, University of London. We also thank Marc Lipsitch at the Harvard School of Public Health and Lawrence Moulton at the Johns Hopkins School of Public Health for their helpful comments on this work.
Financial support. This work is part of the research of the PneumoCarr Consortium funded by the Grand Challenges in Global Health initiative, which is supported by the Bill & Melinda Gates Foundation, the Foundation for the National Institutes of Health, the Wellcome Trust, and the Canadian Institutes of Health Research. This study was supported by the Native American Research Centers for Health (U26IHS300013/03), a joint initiative between the National Institutes of Health and Indian Health Service to reduce native health disparities and build tribal autonomy in conducting health research. This study was also funded by the Centers for Disease Control and Prevention National Vaccine Program Office and the Thrasher Research Fund. W. P. H.’s contribution to the project was funded by the Royal Society (University Research Fellowship) and the National Institute of General Medical Sciences (award number U54GM088558).
Footnotes
Potential conflicts of interest. K. L. O. and M. S. have received grant support and/or honoraria from Wyeth Vaccines (now Pfizer), Sanofi-Pasteur, and Merck. W. P. H. has acted as an advisor to GlaxoSmithKline.
All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.O’Brien KL, Wolfson LJ, Watt JP, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Harvey RA, Champe PC, Fisher BD, Strohl WA. Microbiology. 2nd ed Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 3.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. Phase variation in pneumococcal opacity: Relationship between colonial morphology and nasopharyngeal colonization. Infect Immun. 1994;62:2582–9. doi: 10.1128/iai.62.6.2582-2589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JO, Weiser JN. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis. 1998;177:368–77. doi: 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- 5.Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol. 2007;189:7856–76. doi: 10.1128/JB.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavroidi A, Aanensen DM, Godoy D, et al. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol. 2007;189:7841–55. doi: 10.1128/JB.00836-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berron S, Fenoll A, Ortega M, Arellano N, Casal J. Analysis of the genetic structure of nontypeable pneumococcal strains isolated from conjunctiva. J Clin Microbiol. 2005;43:1694–8. doi: 10.1128/JCM.43.4.1694-1698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M, Turco JH, Zegans ME, et al. An outbreak of conjunctivitis due to atypical Streptococcus pneumoniae. N Engl J Med. 2003;348:1112–21. doi: 10.1056/NEJMoa022521. [DOI] [PubMed] [Google Scholar]
- 10.Buck JM, Lexau C, Shapiro M, et al. A community outbreak of conjunctivitis caused by nontypeable Streptococcus pneumoniae in Minnesota. Pediatr Infect Dis J. 2006;25:906–11. doi: 10.1097/01.inf.0000238143.96607.ec. [DOI] [PubMed] [Google Scholar]
- 11.Shayegani M, Parsons LM, Gibbons WE, Jr, Campbell D. Characterization of nontypable Streptococcus pneumoniae–like organisms isolated from outbreaks of conjunctivitis. J Clin Microbiol. 1982;16:8–14. doi: 10.1128/jcm.16.1.8-14.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho MG, Steigerwalt AG, Thompson T, Jackson D, Facklam RR. Confirmation of nontypeable Streptococcus pneumoniae–like organisms isolated from outbreaks of epidemic conjunctivitis as Streptococcus pneumoniae. J Clin Microbiol. 2003;41:4415–7. doi: 10.1128/JCM.41.9.4415-4417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker JH, Musher DM, Silberman R, Phan HM, Watson DA. Genetic relatedness among nontypeable pneumococci implicated in sporadic cases of conjunctivitis. J Clin Microbiol. 1999;37:4039–41. doi: 10.1128/jcm.37.12.4039-4041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sa-Leao R, Simoes AS, Nunes S, Sousa NG, Frazao N, de Lencastre H. Identification, prevalence and population structure of non-typeable Streptococcus pneumoniae in carriage samples isolated from preschoolers attending day-care centres. Microbiology. 2006;152(Pt 2):367–76. doi: 10.1099/mic.0.28596-0. [DOI] [PubMed] [Google Scholar]
- 15.Hanage WP, Kaijalainen T, Saukkoriipi A, Rickcord JL, Spratt BG. A successful, diverse disease-associated lineage of nontypeable pneumococci that has lost the capsular biosynthesis locus. J Clin Microbiol. 2006;44:743–9. doi: 10.1128/JCM.44.3.743-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hathaway LJ, Meier P Stutzmann, Battig P, Aebi S, Muhlemann K. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J Bacteriol. 2004;186:3721–9. doi: 10.1128/JB.186.12.3721-3729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poehling KA, Talbot TR, Griffin MR, et al. Invasive pneumococcal disease among infants before and after introduction of pneumococcal conjugate vaccine. JAMA. 2006;295:1668–74. doi: 10.1001/jama.295.14.1668. [DOI] [PubMed] [Google Scholar]
- 18.Maiden MC, Ibarz-Pavon AB, Urwin R, et al. Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J Infect Dis. 2008;197:737–43. doi: 10.1086/527401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weatherholtz R, Millar EV, Moulton LH, et al. Invasive pneumococcal disease a decade after pneumococcal conjugate vaccine use in an American Indian population at high risk for disease. Clin Infect Dis. 2010;50:1238–46. doi: 10.1086/651680. [DOI] [PubMed] [Google Scholar]
- 20.Scott JR, Millar EV, Lipsitch M, et al. Impact of long-term routine pneumococcal conjugate vaccine use on carriage and invasive disease potential in Native American communities. J Infect Dis. 2011;205:280–8. doi: 10.1093/infdis/jir730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanage WP, Huang SS, Lipsitch M, et al. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J Infect Dis. 2007;195:347–52. doi: 10.1086/510249. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien KL, Moulton LH, Reid R, et al. Efficacy and safety of seven-valent conjugate pneumococcal vaccine in American Indian children: group randomised trial. Lancet. 2003;362:355–61. doi: 10.1016/S0140-6736(03)14022-6. [DOI] [PubMed] [Google Scholar]
- 23.Millar EV, O’Brien KL, Zell ER, Bronsdon MA, Reid R, Santosham M. Nasopharyngeal carriage of Streptococcus pneumoniae in Navajo and White Mountain Apache children before the introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2009;28:711–16. doi: 10.1097/INF.0b013e3181a06303. [DOI] [PubMed] [Google Scholar]
- 24.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144(Pt 11):3049–60. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention [Accessed December 2008];Streptococcus laboratory protocol: lysate preparation. 2008 http://www.cdc.gov/ncidod/biotech/strep/protocol_emm-type.htm.
- 26.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. [Accessed March 2009];Streptococcus pneumoniae multi-locus sequence typing database. 2009 MLST.net. http://spneumoniae.mlst.net/
- 28.Hinds J, Gould K, Witney A, et al. Molecular serotyping of Streptococcus pneumoniae: A microarray-based genomic tool for isolate typing, detection of multiple carriage and surveillance of serotype replacement in vaccine trials. Program and abstracts of the 6th International Symposium on Pneumococci & Pneumococcal Diseases; Reykjavik, Iceland. Geneva, Switzerland: Kenes International; 2008. [Google Scholar]
- 29.Park IH, Pritchard DG, Cartee R, Brandao A, Brandileone MC, Nahm MH. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol. 2007;45:1225–33. doi: 10.1128/JCM.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park IH, Park S, Hollingshead SK, Nahm MH. Genetic basis for the new pneumococcal serotype, 6C. Infect Immun. 2007;75:4482–9. doi: 10.1128/IAI.00510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott JR, Hanage WP, Lipsitch M, et al. Pneumococcal sequence type replacement among American Indian children: A comparison of pre- and routine-PCV7 eras. Vaccine. 2011;30:2376–81. doi: 10.1016/j.vaccine.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Melchiorre S, Camilli R, Pietrantoni A, et al. Point mutations in wchA are responsible for non-typeability of two invasive Streptococcus pneumoniae isolates. Microbiology. 2012;158:338–44. doi: 10.1099/mic.0.054270-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.