Abstract
Amino acid Asp-351 in the ligand binding domain of estrogen receptor α (ERα) plays an important role in regulating the estrogen-like activity of selective estrogen receptor modulator-ERα complexes. 4-Hydroxyta-moxifen is a full agonist at a transforming growth factor a target gene in situ in MDA-MB-231 human breast cancer cells stably transfected with the wild-type ERα. In contrast, raloxifene (Ral), which is also a selective estrogen receptor modulator, is a complete antiestrogen in this system. Because D351G ERα allosterically silences activation function-1 activity in the 4-hydroxytamox-ifen-ERα complex with the complete loss of estrogenlike activity, we examined the converse interaction of amino acid 351 and the piperidine ring of the antiestrogen side chain of raloxifene to enhance estrogen-like action. MDA-MB-231 cells were either transiently or stably transfected with Asp-351 (the wild type), D351E, D351Y, or D351F ERα expression vectors. Profound differences in the agonist and antagonist actions of Ral-ERα complexes were noted only in stable transfec-tants. The agonist activity of the Ral-ERα complex was enhanced with D351E and D351Y ERα, but raloxifene lost its agonist activity with D351F ERα. The distance between the piperidine nitrogen of raloxifene and the negative charge of amino acid 351 was critical for estrogen-like actions. The role of the piperidine ring in neutralizing Asp-351 was addressed using compound R1h, a raloxifene derivative replacing the nitrogen on its piperidine ring with a carbon to form cyclohexane. The derivative was a potent agonist with wild type ERα. These results support the concept that the side chain of raloxifene shields and neutralizes the Asp-351 to produce an antiestrogenic ERα complex. Alteration of either the side chain or its relationship with the negative charge at amino acid 351 controls the estrogen-like action at activating function 2b of the selective estrogen receptor modulator ERα complex.
Raloxifene (Ral)1 (see Fig. 1) is a polyhydroxyphenyl benzo-thiophene antiestrogen that has low estrogen agonist activity in the rodent uterus (1). The compound originally referred to as Ly156758 or keoxifene was abandoned for development as a treatment for breast cancer (2), because its bioavailability was less than 2% administered dose (3). However, the recognition that raloxifene maintains bone density (4, 5) and inhibits mammary carcinogenesis in the rat (6, 7) illustrates the concept of selective estrogen receptor modulation. Raloxifene is used for the prevention of osteoporosis in postmenopausal women (8), and treatment is associated with a reduced incidence of breast cancer (9).
Fig. 1. Structures of raloxifene and the derivative R1h used in structure-function studies.
Compound R1h is a raloxifene derivative that has a cyclohexane ring instead of a piperidine ring.
Tamoxifen is the prototype selective estrogen receptor modulator (SERM) that is used clinically for the treatment and prevention of breast cancer (10, 11) with the ability to maintain bone density in postmenopausal women (12). However, tamoxifen therapy is also associated with estrogen-like effects in the uterus with an increased incidence of endometrial cancer (13). Raloxifene is currently being compared with tamoxifen for the prevention of breast cancer in a large clinical trial called the Study of Tamoxifen and Raloxifene. Because raloxifene has fewer estrogen-like effects than tamoxifen in laboratory tests, the purpose of the Study of Tamoxifen and Raloxifene trial is to determine whether the decreased estrogenic properties of the Ral-ERα complex compared with the tamoxifen-ERα complex translate to improved efficacy as a breast cancer preventive and result in a decrease in the estrogen-like side effects observed with tamoxifen in the uterus. Clearly, an understanding of the factors that govern the estrogenic and antiestrogenic properties of SERM-ERα complexes will provide additional opportunities to improve targeted therapeutic agents in the future.
We have devised a mechanism-based assay system to classify estrogens, tamoxifen-like compounds, raloxifene-like compounds, and pure antiestrogens (14, 15). The assay is based on the ability of compounds to initiate transforming growth factor α (TGFα) gene transcription in MDA-MB-231 ERα-negative breast cancer cells stably transfected with the cDNAs for the wild type or D351Y ERα. The D351Y ERα was selected because it was discovered as a natural ERα mutation in a tamoxifen-stimulated breast tumor (16) and was subsequently found to enhance the estrogen-like action of antiestrogens (17, 18). The critical role of amino acid 351 to regulate the estrogen-like actions of SERMs was subsequently confirmed with the crystallization of the ligand binding domain (LBD) of the ERα with either raloxifene (19) or 4-hydroxytamoxifen (4-OHT) (20), the active metabolite of tamoxifen (21). The antiestrogenic side chain of both 4-OHT and raloxifene interacts with the surface aspartate at 351, but raloxifene is located so that it appears to shield and neutralize the carboxylic acid. Because raloxifene is a complete antiestrogen at the TGFα gene target in situ (22) whereas 4-OHT is a complete estrogen (23), we hypothesized that the negative charge at surface amino acid 351 could act as part of a docking site for coactivators involved in TGFα gene transcription induced by SERMs. We took two strategic approaches to illustrate the critical role of Asp-351 on the estrogen-like action of the 4-OHT-ERα complex. We first showed that 4-OHT-D351G ERα complexes completely lost their estrogen-like properties to induce TGFα mRNA level, but the complexes maintained antiestrogenic properties (24). Additionally, changing the dimethylaminoethoxy side chain of 4-OHT for an allyl carboxylic acid (GW7604, Glaxo Wellcome, Durham, NJ) also resulted in a loss of estrogen-like properties for the SERM-ERα complex, but again there was retention of antiestrogenic properties (25). We proposed that the carboxylic acid of GW7604 repelled Asp-351, thereby disrupting the surface charge to prevent coactivator binding (25).
Based on the principles established for the 4-OHT-ERα complex (24, 25) and crystallographic structures of antiestrogen-bound LBD of ERα (19, 20), we address the structure-function relationships of the Ral-ERα complex in this study. We have tested the hypothesis that a surface-negative charge at amino acid 351 is necessary for the transcription of TGFα by replacing the aspartic acid with the longer glutamic acid in the Ral-ERα complex. Alternatively, we would predict that the change of the D351Y for a D351F would change the Ral-ERα complex from an estrogen-like to an exclusively antiestrogenic complex. The other component of the equation is the ligand. We have used a novel raloxifene derivative, compound R1h (Fig. 1), that has a cyclohexane ring in the side chain so that the surface-negative charge at Asp-351 cannot be neutralized but could be shielded instead. This would test whether shielding the negative charge at amino acid 351 or neutralization is the principal reason for the silencing of the Ral-ERα activation functions.
EXPERIMENTAL PROCEDURES
Cells and Biochemicals
MDA-MB-231 ERα-negative human breast cancer cells were obtained from American Type Culture Collection (Manassas, VA). Stable transfectants generated in MDA-MB-2321 cells were grown in phenol-red free minimal essential medium supplemented with 5% 3× dextran-coated charcoal-stripped calf serum, 2 mM gluta-mine, 6 ng/ml bovine insulin, 100 units/ml penicillin, 100 µg/ml streptomycin, and non-essential amino acids. 500 µg/ml G418 was added in the medium for stable transfectants. Cells were grown at 37 °C in a 5% CO2 incubator.
17β-Estradiol (E2) and 4-OHT were purchased from Sigma. Raloxifene and compound R1h were generous gifts from Eli Lilly and Company (Indianapolis, IN).
Plasmids
pSG5HEGO, a wild type ERα expression vector, was a generous gift from Professor Pierre Chambon (Institut de Genetique et de Biologie Moleculaire et Cellulaire, CNRS/INSERM, Universite Louis Pasteur, College de France, Strasbourg, France). pSG5HETO, a D351Y ERα expression vector, and pGEX-ASRC-1, an expression for GST-ΔSRC-1 fusion protein, were described previously (17, 24). Point mutations at Asp-351 were introduced using a QuikChange™ site-directed mutagenesis kit (Stratagene, La Jolla, CA) using pSG5HEGO as the template. The sequences of mutated ERα cDNAs were confirmed by sequencing analysis (ABI automated sequencer).
Transient Transfection
MDA-MB-231 cells were grown in phenol-red free minimal essential medium containing 5% 3× dextran-coated charcoal-stripped calf serum for 4 days and then transiently transfected with estrogen receptor expression vectors (1 µg/cuvette), vitellogenin A2ERE3-luciferase reporter (1 µg/cuvette), and pCMV-β-galactosidase (0.2 µg/cuvette) by electroporation (950 microfarads, 320 V) using a Bio-Rad Gene Pulser II (Hercules, CA). Luciferase and β-galactosidase activities were measured as described previously (17).
Stable Transfection
Asp-351, D351Y, and D351G stable transfectants were described previously (17, 24, 26). MDA-MB-231 cells were electroporated with 10 µg of ERα expression vectors and 0.5 µg of pBK-CMV (Stratagene, La Jolla, CA) to generate D351F and D351E stable transfectants. Neomycin resistant clones (10 clones/transfec-tants) were screened and characterized for ERα expression using Northern and Western blot analyses and hormone binding assays. Clones with comparable levels of ERα were chosen for the further study.
Western Blot Analysis
For ERα Western blot analysis, 25 µg of whole cell lysate were separated on a 7.5% SDS-PAGE. Anti-ERα polyclonal antibody G20 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The level of β-actin was determined as a loading control (β-actin antibody AC-15, Sigma). Anti-rabbit or anti-mouse IgG conjugated with horseradish peroxidase (Sigma) was used to visualize bands using an ECL kit (Amersham Biosciences, Inc.). ERα band densities relative to β-actin from at least three separate experiments were quantitated using ImageQuant (Molecular Dynamics, Sunnyvale, CA). For measuring TIF2 protein levels in the stable transfectants, fifty micrograms of nuclear extracts were loaded and separated on a 6% SDS-PAGE. Anti-GRIP1 polyclonal antibody (C-20) was purchased from Santa Cruz Biotechnology.
Ligand Binding Assay
Ligand binding assays were performed following a modified procedure (27). For saturation binding assays, the stable transfectants were incubated with increasing concentrations of 3H-labeled E2 (46 Ci/mmol, Amersham Biosciences, Inc.) for 2 h at room temperature to obtain total binding. To determine nonspecific binding, each concentration of 3H-labeled E2 was competed with 400-fold excess of radioactive inert diethylstilbestrol. The specific binding was obtained by subtracting the nonspecific binding from the total binding. For competition binding assays, the stable transfectants were incubated with 1 nM 3H-labeled E2 with increasing concentrations of raloxifene or compound R1h for 2 h at room temperature. Each binding assay was repeated at least three times, and the Kd values for E2 and IC50 for raloxifene or compound R1h were calculated using GraphPad Prism (GraphPad Prism Software, Inc., San Diego, CA).
Northern Blot Analysis
TGFα mRNA levels were assessed by Northern blot analyses as described previously (23). pS2 probe was generated by reverse transcription-PCR. The primers used are 5′-AT-GGCCACCATGGAGAACAAGGTG-3′ (sense) and/or 5′-CTAAAAT-TCACACTCCTCTTCTGG-3′ (antisense). β-actin mRNA levels were detected as the loading controls. The band densities were quantitated using ImageQuant. Reverse transcription-PCR for pS2 and β-actin were described previously (28). DNA assays were described previously (25).
GST Pull-down Assay
GST pull-down assays were performed as previously described (29, 30). 35S-Labeled wild type and mutated ERα were made using an in vitro transcription-coupled translation system (Promega, Madison, WI).
Statistics Analysis
The data from ligand binding assays, Northern blot, and Western blot analyses were analyzed by ANOVA followed by Dunnett’s multiple comparison test using GraphPad Prism.
RESULTS
Comparison of Transient and Stable Transfection in Assessment of Agonist Activities of SERMs
Transient transfection assays have been widely used to study transcriptional activity of ERα using a reporter gene under the control of estrogen responsive elements (EREs) in MDA-MB-231 ERα-negative human breast cancer cells (31–33). However, the agonist activity of 4-OHT is relatively low. We have developed an assay system using MDA-MB-231 cells stably transfected with ERα cDNA and an endogenous TGFα gene as the reporter in which SERMs show significantly higher agonist activities (18, 23). As shown in Fig. 2, 4-OHT or raloxifene had minimum agonist activity in MDA-MB-231 cells transiently transfected with the wild type (Fig. 2A) or D351Y ERα (Fig. 2B). In contrast, 4-OHT exhibited strong agonist activity with the wild type and D351Y ERα (Fig. 2, A and B), and raloxifene became an agonist with D351Y ERα (Fig. 2B) using stable transfectants with a TGFα gene target in situ. Based on these data, we decided to employ the stable transfection approach methodology to interrogate the ralox-ifene-ERα complexes.
Fig. 2. Comparison of agonist activities of 4-OHT and raloxifene in transient and stable transfection systems.
MDA-MB-231 cells were transiently or stably transfected with wild type (A) or D351Y ERα (B) expression vector as described under “Experimental Procedures.” Transient transfection results are shown in solid bars (left y axis). Stable transfection data are presented in open bars (right y axis). The cells were treated with ethanol vehicle (EtoH), 1 nm E2, 1 µm 4-OHT, or 1 µm Ral for 24 h.
Fig. 4. Determination of agonist or antagonist activities of raloxifene in stable transfectants by measuring induction of TGFα or pS2 mRNA.
A, cells expressing wild type (Asp-351) or D351E ERα were treated with ethanol vehicle (control) or increasing concentrations of Ral (0.01, 0.1,1, 10, 100, or 1000 nM) for 24 h. TGFα mRNAlevels were measured by Northern blot analysis. β-actin was used as RNA-loading control. Quantitative results (TGFα/β-actin) from three independent Northern blots were presented as the mean ±S.D. B, TGFα mRNA levels were measured and compared in cells expressing D351Y and D351F ERα cells by Northern blot analysis. C, the cells were treated with 10 nM E2 or in combination of increasing concentrations of Ral (0.01, 0.1, 1, 10, 100, or 1000 nM) for 24 h. TGFα mRNA levels were measured by Northern blot analysis. D, expression of pS2 and TGFα induced by E2 or Ral were compared. The concentration of E2 and Ral were 10 µM and 1 µm, respectively. Equal RNA loading for Northern blot (N.B) was ensured by measuring β-actin mRNA levels (data not shown). The data in C are D are a representative of three experiments. RT, reverse transcription.
Generating Stable Transfectants in MDA-MB-231 Cells
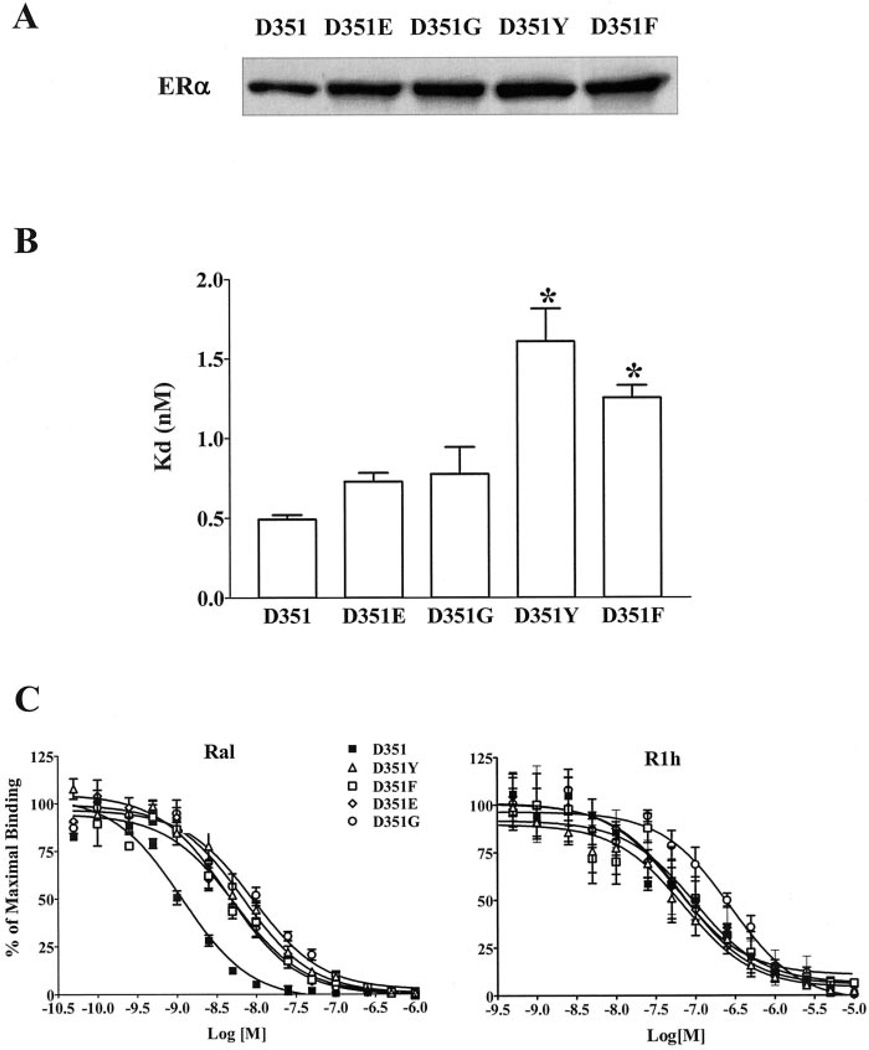
To study the role of amino acid 351 in the agonist activity of raloxifene, we constructed the wild type (26), D351Y (17), D351G (24), D351E, and D351F ERα expression vectors and generated stable transfectants in MDA-MB-231 cells (see “Experimental Procedures”). All of the stable transfectants expressed ERα at similar levels as determined by the Western blots (Fig. 3A). The binding affinities to E2 were similar in cells expressing the wild type, D351E, and D351G ERα. D351Y and D351F ERα had significantly lower binding affinities for E2 compared with the wild type ERα (p < 0.05) (Fig. 3B). The binding affinities to raloxifene and compound R1h were also assessed using competitive binding assays. The results are presented in Fig. 3C. Compared with the wild type ERα, D351G and D351Y had significantly higher IC50 values for raloxifene (p < 0.01). All of the wild type and mutant ERα have relatively lower binding affinities for R1h compared with those for raloxifene (Table I).
Fig. 3. Characterization of mutated ERα stably expressed in MDA-MB-231 cells.
A, the ERα expression levels in these stable transfectants were shown by Western blot analysis. B, binding affinities (Kd) for E2 were calculated from saturation binding assay data using Prism 3.0 and are presented as the mean ± S.E. The saturation binding assays for ERα stable transfectants were done at least four times. C, competition binding assay results for Ral and R1h are shown. Each experiment was repeated at least five times.
Table I.
IC50 values (nm) for raloxifene and compound R1h measured by competitive binding assays
| Asp-351 | D351E | D351G | D351Y | D351F | |
|---|---|---|---|---|---|
| Ral | 0.95 ± 0.15 | 4.35 ± 0.56 | 6.93 ± 1.19a | 7.70 ± 1.20a | 4.87 ± 1.09 |
| R1h | 69.47 ± 19.72 | 58.94 ± 7.74 | 272.3 ± 49.37a | 76.08 ± 15.16 | 109.1 ± 3.24 |
Compared to IC50 of wild type ERα (Asp-351), p < 0.01.
Agonist Activity of Raloxifene Is Influenced by Amino Acid 351
Our previous study (24) has shown that a loss of the negative charge at amino acid 351 abolishes the agonist activity of 4-OHT. In the current study, we explored further the role of the negative charge at amino acid 351 in the agonist activity of raloxifene. In Fig. 4A, we compared the agonist activity of raloxifene in cells expressing the wild type versus D351E ERα. In the wild type ERα (19), a strong hydrogen bond (H-bond) (2.7 Å, 180°) is formed between Asp-351 and the piperidine nitrogen. We hypothesized that the H-bond between Asp-351 and the nitrogen on piperidine ring of raloxifene holds the side chain of raloxifene at a position that shields and neutralizes the negative charge on Asp-351. This may explain why raloxifene does not exhibit any agonist activity with wild type ERα. However, when Asp-351 was replaced by Glu-351, which has a longer negatively charged side chain, raloxifene became an agonist and induced the TGFα mRNA level in a concentration-dependent manner. The partial agonist activity of raloxifene with D3531Y ERα (22) is shown in Fig. 4B. Interestingly, Phe-351, which has a phenyl ring instead of a phenolic ring on its side chain, completely abolished the agonist activity of raloxifene. These results indicate that an exposed phenolic hydroxyl at amino acid 351 enhances the agonist activity of raloxifene.
Although raloxifene had no agonist activity in cells expressing D351G and D351F ERα (Fig. 4, A and B), antiestrogenic activity was retained. We treated the cells expressing wild type, D351F, or D351G ERα with increasing concentrations of raloxifene in combination with 10 nM E2. Raloxifene inhibited E2-induced TGFα mRNA levels in concentration-dependent manners in the wild type and D351F–expressing cells (Fig. 4C). Because of the decreased binding affinity of D351G ERα to raloxifene (Fig. 3C), raloxifene only inhibited the effects of 10 nM E2 at 1 µm. Similar results were obtained when 1 nM E2 was used (data not shown). Therefore, a specific amino acid at 351 is not required for the antagonist activity of raloxifene, which is consistent with what has been previously reported in a transient transfection system (33).
We also determined the expression of pS2 gene (34) mediated by wild type or mutated ERα in the absence or presence of 10 nM E2 or 1 µm raloxifene. To our surprise, pS2 gene expression was only induced significantly by E2 in wild type and D351Y ERα-containing cells determined by Northern blot analysis and reverse transcription-PCR (Fig. 4D).
The Piperidine Ring of Raloxifene Plays an Important Role in Its Antagonist Activity
As we mentioned earlier, the strong H-bond between Asp-351 and the piperidine nitrogen of raloxifene is responsible for holding the side chain of raloxifene in such a disfavored conformation that shields and neutralizes the negative charge on Asp-351. Therefore, raloxifene behaves as an antagonist with wild type ERα in our system. To explore the possibility that only steric hindrance is responsible for antiestrogenic action, we used the compound R1h from Eli Lilly and Company. R1h has a cyclohexane ring on its side chain (Fig. 1) and has been shown to have minimal antagonist activity in vivo (35). Unlike raloxifene (Fig. 4A), R1h produced a concentration-related increase in TGFα mRNA (Fig. 5A). R1h also induced TGFα mRNA levels in D351E–expressing cells (Fig. 5B). However, compound R1h did not increase TGFα mRNA levels in D351G, D351F, or D351Y ERα stable transfectants. These results indicated that the piperidine ring is required for the antagonist activity of raloxifene.
Fig. 5. Agonist activity of compound R1h.
A, the cells expressing the wild type ERα were treated with ethanol vehicle (Control) or increasing concentrations of R1h for 24 h. Top panel is a representative Northern blot. Bottom panel shows the quantitative results (mean ±S.E.) from three independent Northern blots. B, the cells expressing the wild type, D351E, D351Y, D351F, or D351G ERα were treated with either Control or 1 µm R1h for 24 h. The equal loading of total RNA was ensured by β-actin level. This experiment was repeated three times. C, the levels of pS2 mRNA were measured by Northern blot analysis in wild type ERα-expressing cells treated with control, 10 nM E2, 1 µm Ral, or 1 µm R1h for 24 h. D, stimulatory effect of R1h on MCF-7 cell growth was measured by DNA assay. The cells were treated for 5 days with vehicle control, 1 µM E2, 1 µm Ral, 1 µm R1h, 1 µm ICI 182,780, or a combination of E2 and Ral, R1h and ICI 182,780, E2 and ICI 182,780.
To determine whether compound R1h also activates other ERα-regulated genes, we measured pS2 mRNA levels in the wild type ERα containing stable transfectants treated with vehicle control, 1 nM E2, 1 µm raloxifene, or 1µm R1h for 24 h. As shown in Fig. 5C, R1h induced pS2 mRNA level significantly (compared with the control, p < 0.01), which was confirmed by reverse transcription-PCR (data not shown).
To further characterize the agonist activity of R1h, we also conducted growth assays to determine the stimulatory effect of R1h on the growth of ERα-positive human breast cancer cells. As shown in Fig. 5D, R1h stimulated the growth of MCF-7 cells (compared with the control, p < 0.05), which was blocked by 1 iutM ICI 182,780. Similar results were obtained in T47D cells (data not shown).
Raloxifene and Compound R1h Did Not Induce AF-2 Coac-tivator-ERα Interaction
To investigate whether the agonist activities of raloxifene and compound R1h are because of an increase in AF-2 function, we performed GST-ASRC-1 pulldown assays using 35S-labeled ERα. As shown in Fig. 6A, E2 induced the wild type, D351E, and D351Y ERα binding to SRC-1, which is consistent with previous reports (33, 36). However, the mutant ERα-raloxifene complexes did not induce SRC-1 binding despite estrogen-like action. Actually, Ral had an inhibitory effect on the basal ERa-SRC-1 interaction. Thus, R1h (Fig. 6B) did not adopt a conformation that induced a significant interaction of the wild type ERα and SRC-1.
Fig. 6. Hormone-dependent interaction of ERα with SRC-1 in vitro.
A, 35S-labeled wild type or mutated ERα-translated in vitro was incubated with GST-ΔSRC-1 immobilized on glutathione-Sepharose in the presence of ethanol vehicle (Control), 1 nm E2, or 1 µm Ral at 4 °C for 2 h. After extensive washing, the bound ER·SRC-1 complexes were eluted by glutathione and separated on a 7.5% SDS-PAGE. B, the pull-down assay was done with the wild type ERα and GST-ΔSRC-1 in the presence of Control, 1 nm E2, or 1 µm R1h.
An increased concentration of TIF2 can apparently boost the agonist activity of 4-OHT (33). Therefore, it would be reasonable to hypothesize that the MDA-MB-231 cells expressing D351E and D351Y ERα might by chance express higher levels of TIF2. We measured the TIF2 protein levels in our stable transfectants using Western blot analysis. However, all transfectants had similar levels of TIF2 protein (data not shown).
Agonist or Antagonist Activity of Raloxifene Was Not Attributed to Increased or Decreased ERα Levels
Increased levels of ERα could contribute to agonist activities of raloxifene or R1h in D351Y and D351E or the wild type ERα expressing cells, respectively. By contrast, premature destruction of ERα could enhance antiestrogen action. To exclude these possibilities, we determined the ERα levels in transfectants treated with differ-ent ligands for 24 h. Compared with vehicle control, 1 nM E2 down-regulated ERα levels as we would expect, however, neither raloxifene nor compound R1h dramatically affected ERα levels in transfectants (Fig. 7).
Fig. 7. Western blot analysis of the effects of different ligands on the wild type or mutated ERα protein levels.
The cells were treated with ethanol vehicle (Control), 1 nm E2, 1 µm Ral, or 1 µm R1h for 24 h. 25 µg of whole cell lysates were loaded and separated on a 7.5% SDS-PAGE. The levels of ERα were determined by Western blot analysis using anti-ERα antibody G20. Even loading was ensured by β-actin levels measured using anti-β-actin antibody. Each Western blot was repeated at least three times.
DISCUSSION
We have interrogated the structure-function relationships of the Ral-ERα complex that regulates TGFα gene expression in MDA-MB-231 breast cancer cells stably transfected with mutant ERα cDNAs. We demonstrate that the relationship between the amine of the piperidine ring of raloxifene and the surface Asp-351 is critical to program the estrogenic/antiestrogenic properties of the Ral-ERα complex. These data support and advance the body of recent information that describes the structure-function relationships of SERM-ERα complexes (24, 25, 33, 37).
In earlier studies, we chose to focus our initial research strategy only on breast cancer cells that are ERα-negative but stably transfected with cDNAs for the wild type and mutant receptors (17, 26). We employed an endogenous TGFα gene as a complex target to monitor the estrogenic and antiestrogenic action of SERMs and pure antiestrogen ICI 182,780. Although the methodology is time-consuming and rather labor-intensive, there are two observations that have been critically important to validate the use of the stable transfection model for structure relationship studies of ERα ligand interactions. Firstly, it is clear from transient transfection studies (17, 33, 37) (Fig. 2) that the discovery of the natural mutant D351Y ERα in a tamoxifen-stimulated breast tumor (16) would have been impossible, because there is no powerful induction of an artificial reporter gene activity with a simple promoter system by D351Y ERα when E2 and 4-OHT are compared. Secondly, E2 and tamoxifen are equally effective in promoting the growth of newly resistant tamoxifen-stimulated tumors in athymic mice (38, 39), however, 4-OHT does not induce ERE-luciferase activity through the wild type ERα to the same extent as E2 in transient transfection systems (33, 37) (Fig. 2). The development of TGFα as a gene target in situ has resulted in the development of an assay system that can classify estrogens, SERMs, and pure antiestrogens based on structure-function relationships and specific mutations at amino acid 351 (22, 40, 41).
The D351Y ERα changes raloxifene from an antagonist to a partial agonist in the stable transfection system (17, 18). These biological data illustrated the importance of amino acid 351 to program the actions of SERMs. However, the critical role of Asp-351 in the molecular mechanism of action of raloxifene was not clear until the x-ray crystallographic structure of Ral-bound LBD showed that Asp-351 forms a H-bond with the piperidine ring of the antiestrogenic side chain of raloxifene (19). Indeed, this area of interaction between the side chain of antiestrogens and the ER had previously been defined as the “antiestrogenic region” (42, 43).
The H-bond has a stabilizing effect on raloxifene binding, and this is illustrated in our competitive binding studies. When Asp-351 is mutated to Glu-351 and Tyr-351, the distance between amino acid 351 and the nitrogen of piperidine is unlikely to favor the formation of H-bond. Gly-351 and Phe-351 cannot form a H-bond with the side chain of raloxifene. As shown in Fig. 3B, all four ERα with mutations at amino acid 351 had relatively higher IC50 values for raloxifene, although only binding affinities of D351G and D351Y ERα for raloxifene were significantly lower (p < 0.05). Compound R1h has a cyclohexane ring (Fig. 1) that cannot form a H-bond with Asp-351, and consequently the IC50 for R1h was significantly higher than that for raloxifene with the wild type receptor. Thus, the H-bond between amino acid 351 and the side chain of raloxifene is important for the higher binding affinity of raloxifene.
An examination of the surface structure of Ral-ERα (19) shows that a strong H-bond (2.7 Å, 180°) formed between Asp-351 and the piperidine nitrogen of raloxifene forces the piperidine ring into an awkward high energy gauche position so that the bulky side chain of raloxifene can shield and neutralize the Asp-351 on the receptor surface (Fig. 8). In this form, raloxifene has no agonist activity (Fig. 4A) and behaves as a complete antagonist with the wild type ERα (Fig. 4C). This finding correlates with the crystal structure result that shows helix 12 is repositioned and sits in the hydrophobic groove to block AF-2 coactivator binding (19). However, when a glutamic acid with a longer negative side chain was introduced into amino acid 351, the distance between Glu-351 and the nitrogen of the piperidine ring is longer and produces only a weak H-bond (3.5–5.0 Å, 130° calculated using Insight II) (Fig. 9A). In this model, the negative charge of Glu-351 would be exposed on the receptor surface and available to bind an as yet unidentified coactivator at the AF2b site (Fig. 8) (36). We have hypothesized that the longer distance between the nitrogen of the diethylaminoethoxy side chain of 4-OHT and Asp-351 results in the residual charge being exploited by coactivator binding in the AF2b site to enhance estrogen-like properties (24). Therefore, it was not surprising that raloxifene became an agonist with the D351E ERα and induced TGFα mRNA level in a concentration-dependent manner (Fig. 4A). Raloxifene exhibits partial agonist activity with D351Y ERα that has a negative oxygen on the phenolic group. Interestingly, raloxifene lost its agonist activity with D351F ERα (Fig. 4B) and had no agonist activity with D351G ERα (24) (Fig. 8). These data further support our hypothesis that an exposed negative charge at amino acid 351 is required for the agonist activities of SERMs. However, the negative charge is not necessary for the antagonist property of raloxifene (Fig. 4C) (24), which is consistent with the study with 4-OHT by Anghel et al. (33).
Fig. 8. Surface structures around amino acid 351 of Ral-bound LBDs of ERα.
A structural model of dimeric human ERα bound to raloxifene was derived from the Protein Data Bank (code 1ERR) (19) by removing all water molecules with the exception of the ordered water-forming H-bond with the O3 of raloxifene, adding hydrogens and minimizing in the consistent valance force field (CVFF) using Discover (Accelrys, San Diego, CA). Mutant receptors were constructed using Biopolymer (Accelrys) to replace Asp-351 with Gly, Glu, Phe, or Tyr and to obtain a minimum energy rotomer for the mutant side chain. The results were visualized using Insight II (Accelrys).
Fig. 9. Computer modeling of the LBDs of ERα occupied by Ral or R1h.
A, relationships between the side chain of Asp-351 or Glu-351 with the piperidine of raloxifene. The nitrogen in the piperidine of raloxifene forms a H-bond with Asp-351 (left panel). When Asp-351 is replaced by a glutamic acid, the positions of both the piperidine and the side chain of Glu-351 changes (middle panel). As a result, the distance between the nitrogen in the piperidine of raloxifene and the side chain of Glu-351 is longer (3.5–5.0 Å) (right panel). Thus, the negative charge of Glu-351 is exposed on the surface. B, a comparison of Ral-Asp-351 versus R1h–Asp-351 (left panel), Ral-Tyr-351 versus R1h–Tyr-351 (right panel). Because the cyclohexane of R1h cannot form a hydrogen bond with Asp-351, the side chain of R1h assumes a more extended conformation. As a result, the negative charge of Asp-351 is exposed, and R1h–ERα is an agonist. When Asp-351 is replaced by a tyrosine, Tyr-351 partially occupies the space filled by the piperidine of raloxifene in wild type ERα. Thus, the piperidine of raloxifene is displaced and the side chain of raloxifene is allowed to assume an extended conformation. Ral-D351Y ERα is a partial agonist. However, the cyclohexane interacts with Tyr-351, and R1h–D351Y ERα does not display any agonist activity.
The notion that an exposed negative charge at amino acid 351 is necessary for the agonist activity of raloxifene was further supported by using compound R1h. R1h is a raloxifene derivative (Fig. 1) that cannot form a H-bond with Asp-351 to neutralize its negative charge, because the piperidine ring is replaced by cyclohexane. Without the H-bond the side chain of R1h would not be held in the same position to shield Asp-351 as raloxifene. In fact, the side chain of R1h is in a more extended conformation (Fig. 9B) so that the negative charge of Asp-351 will be exposed on the surface. In fact, R1h was an agonist at the TGFα gene using wild type ERα (Fig. 5A), indicating that the cyclohexane is unable to interfere with coactivator binding at AF2b. Interestingly, R1h exhibited no agonist activities with either D351G or D351F ERα (Fig. 5B), which suggests the requirement for an appropriate charge at 351.
These data support the view that R1h would be a Type II estrogen. Type II estrogen receptors require the collaboration of AF2b and AF-1 to form an estrogen-like transcription unit. This finding contrasts with the Type I estrogens that use AF-2 and AF-1 synergistically (41). However, an unexpected result was obtained with R1h and the mutant D351Y ERα. As anticipated, R1h was not an estrogenic complex with D351Y ERα. Computer modeling (Fig. 9B) helps to interpret our finding. When R1h binds in the mutant ERα, the cyclohexane ring on the side chain of R1h has a hydrophobic interaction with Tyr-351. As a result, Tyr-351 is not available to form the AF2b site for coactivators to produce an agonist response at the TGFα gene expression (Fig. 5B). Taken together, we believe that the exposed negative charge at amino acid 351 is important to form AF2b (24, 36) (Fig. 8), and shielding and neutralizing the negative charge is critical for antagonist activity of raloxifene.
It was reported that TIF2 overexpression amplifies 4-OHT-mediated transcriptional activity of D351Y ERα (33), and GRIP1 has a weak interaction with D351Y ERα in the presence of 4-OHT and enhances the agonist activities of 4-OHT and raloxifene with the D351Y ERα (37). All stable transfectants expressed similar levels of TIF2 protein (data not shown), thus changes in this coactivator levels are not likely to be the reason for the different agonist activity of Ral-ERα complexes. To further investigate the possible role of p160 coactivators in the agonist activity of the Ral-ERα complex, we performed pulldown assays with GST-ASRC-1 to determine ERa-SRC-1 interaction. E2 induced ERα, associating with SRC-1 as reported previously (24, 36, 44–46). Raloxifene did not induced SRC-1 binding to D351E and D351Y ERα (Fig. 6A), despite the fact that raloxifene was either an agonist or a partial agonist with the mutant receptors (Fig. 4, A and B). Additionally, R1h did not induce an ERα-SRC-1 interaction (Fig. 6B), despite the fact that R1h is a strong agonist with the wild type ERα (Fig. 5A). Conversely, we determined whether the level of ERα was affected by raloxifene or R1h. Our hypothesis was that antagonist actions might be correlated with a decreased ERα level or a loss of ERα, but agonist actions might be correlated with an increase in ERα level. However, this was not the case (Fig. 7).
Our goal was to focus on the modulation of the TGFα gene through an analysis of the structure-function relationships of both the ligand and ERα in the Ral-ERα complex. However, the recent report that estradiol induced pS2 gene expression in MDA-MB-231 cells ectopically expressing wild type ERα (47) raised the possibility of expanding our targeted studies. We confirm that the levels of both TGFα and pS2 mRNA can be increased with E2 in MDA-MB-231 cells stably transfected with the wild type ERα (Figs. 4D and 5C). Additionally, the raloxifene derivative R1h has estrogenic activity at both the TGFα and pS2 genes and can increase cell proliferation in ERα-positive human breast cancer cell lines MCF-7 (Fig. 5D) and T47D (data not shown). R1h also exhibited a strong estrogenic activity in T47D:C4:2 ERα-negative cells (48) cotransfected with type ERα expression vector and vitellogenin A2-ERE3-luciferase reporter plasmid (data not shown). These data illustrated that the critical role of the side chain of raloxifene to block estrogen-like effects in the SERM-ERα complex and reinforce previous conclusions about the importance of the appropriate side chain for antiestrogen actions (49, 50).
To provide evidence that the difference in agonist activities of Ral-ERα complexes in this study is not unique to TGFα gene, we also measured the effects of Ral and R1h on pS2 gene expression in our stable transfectants and on vitellogenin A2-ERE3-luciferase activity in transient transfection assays in T47D:C4:2 cells. The results from transient transfection assays (data not shown) are consistent with our results in this study, suggesting that agonist activities of Ral·D351E, Ral·D351Y, R1h·Asp-351, and R1h·D351E ERα complexes are not only limited to regulating TGFα gene. However, we were surprised to find that, despite the fact that the structure-function relationships of SERMs were parallel at TGFα and pS2 target genes with wild type ERα, the activation of pS2 gene was not induced by E2 with ERα mutants. Recent studies (51–53) illustrate that the DNA sequence can influence the folding and potential of ER for transcriptional activation. A future examination of the promoters for multiple estrogen-related genes with a spectrum of mutant ERs might identify the relationship between EREs, ligands, and complex conformation to decipher both estrogen specificity and SERM action.
In summary, we have studied the interaction of amino acid 351 and the antiestrogenic side chain of raloxifene (Table II). Estrogen-like actions are enhanced by extending the distance between the nitrogen of the piperidine ring and the negative charge on amino acid 351. A change of D351E, which increases the interactive distance from 2.7 Å in Ral·Asp-351 (19) to 3.5–5.0 Å in Ral·D351E, increases the estrogen-like action of raloxifene. This result is comparable to moving the side chain of 4-OHT 1 Å further away from Asp-351, which enhances also estrogen-like actions (24). Conversely, removing the neutralizing charge of the piperidine by substituting a cyclohexane again results in increased estrogen-like actions. Clearly, subtle changes in the relationship of the antiestrogenic side chain and the AF2b site have profound effects on the estrogen-like action of the SERM-ERα complexes.
Table II.
Estrogenic activities of raloxifene and compound R1h in the stable transfectants
| Asp-351 | D351E | D351G | D351Y | D351F | |
|---|---|---|---|---|---|
| Ral | − | + | − | + | − |
| R1h | + | + | − | − | − |
Footnotes
This work was supported by in part by the Specialized Program of Research Excellence Grant CA89018-01, the Avon Products Foundation, and the Lynn Sage Breast Cancer Research Foundation.
The abbreviations used are: Ral, raloxifene; SERMS, selective estrogen receptor modulators; ERα, estrogen receptor α; TGFα, transforming growth factor α; 4-OHT, 4-hydroxytamoxifen; ERE, estrogen response element; AF, activation function; AF2b, activation function 2b; LBD, ligand binding domain; GST, glutathione S-transferase; ANOVA, analysis of variance; mRNA, messenger RNA.
REFERENCES
- 1.Black LJ, Jones CD, Falcone JF. Life Sci. 1983;32:1031–1036. doi: 10.1016/0024-3205(83)90935-9. [DOI] [PubMed] [Google Scholar]
- 2.Buzdar AU, Marcus C, Holmes F, Hug V, Hortobagyi G. Oncology (Basel) 1988;45:344–345. doi: 10.1159/000226637. [DOI] [PubMed] [Google Scholar]
- 3.Snyder KR, Sparano N, Malinowski JM. Am. J. Health Syst. Pharmacol. 2000;57:1669–1678. [PubMed] [Google Scholar]
- 4.Jordan VC, Phelps E, Lindgren JU. Breast Cancer Res. Treat. 1987;10:31–35. doi: 10.1007/BF01806132. [DOI] [PubMed] [Google Scholar]
- 5.Black LJ, Sato M, Rowley ER, Magee DE, Bekele A, Williams DC, Cullinan GJ, Bendele R, Kauffman RF, Bensch WR, Frolik CA, Termine JD, Bryant HU. J. Clin. Invest. 1994;93:63–69. doi: 10.1172/JCI116985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottardis MM, Jordan VC. Cancer Res. 1987;47:4020–4024. [PubMed] [Google Scholar]
- 7.Anzano MA, Peer CW, Smith JM, Mullen LT, Shrader MW, Logsdon DL, Driver CL, Brown CC, Roberts AB, Sporn MB. J. Natl. Cancer Inst. 1996;88:123–125. doi: 10.1093/jnci/88.2.123. [DOI] [PubMed] [Google Scholar]
- 8.Ettinger B, Black DM, Mitlak BH, Knickerbocker RK, Nickelsen T, Genant HK, Christiansen C, Delmas PD, Zanchetta JR, Stakkestad J, Gluer CC, Krueger K, Cohen FJ, Eckert S, Ensrud KE, Avioli LV, Lips P, Cummings SR. J. Am. Med. Assoc. 1999;282:637–645. doi: 10.1001/jama.282.7.637. [DOI] [PubMed] [Google Scholar]
- 9.Cummings SR, Eckert S, Krueger KA, Grady D, Powles TJ, Cauley JA, Norton L, Nickelsen T, Bjarnason NH, Morrow M, Lippman ME, Black D, Glusman JE, Costa A, Jordan VC. J. Am. Med. Assoc. 1999;281:2189–2197. doi: 10.1001/jama.281.23.2189. [DOI] [PubMed] [Google Scholar]
- 10.Early Breast Cancer Trialists’ Collaborative Group. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 11.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. J. Natl. Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 12.Love RR, Mazess RB, Barden HS, Epstein S, Newcomb PA, Jordan VC, Carbone PP, DeMets DL. N. Engl. J. Med. 1992;326:852–856. doi: 10.1056/NEJM199203263261302. [DOI] [PubMed] [Google Scholar]
- 13.Assikis VJ, Neven P, Jordan VC, Vergote I. Eur. J. Cancer. 1996;32:1464–1476. doi: 10.1016/0959-8049(96)00184-0. [DOI] [PubMed] [Google Scholar]
- 14.Levenson AS, Jordan VC. Eur. J. Cancer. 1999;35:1628–1639. doi: 10.1016/s0959-8049(99)00183-5. [DOI] [PubMed] [Google Scholar]
- 15.MacGregor JI, Jordan VC. Pharmacol. Rev. 1998;50:151–196. [PubMed] [Google Scholar]
- 16.Wolf DM, Jordan VC. Breast Cancer Res. Treat. 1994;31:129–138. doi: 10.1007/BF00689683. [DOI] [PubMed] [Google Scholar]
- 17.Catherino WH, Wolf DM, Jordan VC. Mol. Endocrinol. 1995;9:1053–1063. doi: 10.1210/mend.9.8.7476979. [DOI] [PubMed] [Google Scholar]
- 18.Levenson AS, Catherino WM, Jordan VC. J. Steroid Bio-chem. Mol. Biol. 1997;60:261–268. doi: 10.1016/s0960-0760(96)00184-7. [DOI] [PubMed] [Google Scholar]
- 19.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JÅ, Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 20.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 21.Jordan VC, Collins MM, Rowsby L, Prestwich G. J. Endo-crinol. 1977;75:305–316. doi: 10.1677/joe.0.0750305. [DOI] [PubMed] [Google Scholar]
- 22.Levenson AS, Jordan VC. Cancer Res. 1998;58:1872–1875. [PubMed] [Google Scholar]
- 23.Levenson AS, Tonetti DA, Jordan VC. Br. J. Cancer. 1998;77:1812–1819. doi: 10.1038/bjc.1998.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacGregor Schafer J, Liu H, Bentrem DJ, Zapf JW, Jordan VC. Cancer Res. 2000;60:5097–5105. [PubMed] [Google Scholar]
- 25.Bentrem DJ, Dardes RC, Liu H, MacGregor-Schafer J, Zapf JW, Jordan VC. Endocrinology. 2001;142:838–846. doi: 10.1210/endo.142.2.7932. [DOI] [PubMed] [Google Scholar]
- 26.Jiang SY, Jordan VC. J. Natl. Cancer Inst. 1992;84:580–591. doi: 10.1093/jnci/84.8.580. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Wormke M, Safe SH, Bjeldanes LF. J. Natl. Cancer Inst. 1994;86:1758–1765. doi: 10.1093/jnci/86.23.1758. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Burdette JE, Xu H, Gu C, van Breemen RB, Bhat KP, Booth N, Constantinou AI, Pezzuto JM, Fong HH, Farnsworth NR, Bolton JL. J. Agric. Food Chem. 2001;49:2472–2479. doi: 10.1021/jf0014157. [DOI] [PubMed] [Google Scholar]
- 29.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 30.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraus WL, McInerney EM, Katzenellenbogen BS. Proc. Natl. Acad. Sci. U. S. A. 1995;92:12314–12318. doi: 10.1073/pnas.92.26.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekena K, Weis KE, Katzenellenbogen JA, Katzenellenbogen BS. J. Biol. Chem. 1997;272:5069–5075. doi: 10.1074/jbc.272.8.5069. [DOI] [PubMed] [Google Scholar]
- 33.Anghel SI, Perly V, Melancon G, Barsalou A, Chagnon S, Rosenauer A, Miller WH, Jr, Mader S. J. Biol. Chem. 2000;275:20867–20872. doi: 10.1074/jbc.M002098200. [DOI] [PubMed] [Google Scholar]
- 34.Jakowlew SB, Breathnach R, Jeltsch JM, Masiakowski P, Chambon P. Nucleic Acids Res. 1984;12:2861–2878. doi: 10.1093/nar/12.6.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grese TA, Sluka JP, Bryant HU, Cullinan GJ, Glasebrook AL, Jones CD, Matsumoto K, Palkowitz AD, Sato M, Termine JD, Winter MA, Yang NN, Dodge JA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14105–14110. doi: 10.1073/pnas.94.25.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Lee E-S, Reyes ADL, Zapf JW, Jordan VC. Cancer Res. 2001;61:3632–3639. [PubMed] [Google Scholar]
- 37.Webb P, Nguyen P, Valentine C, Weatherman RV, Scanlan TS, Kushner PJ. J. Biol. Chem. 2000;275:37552–37558. doi: 10.1074/jbc.M007435200. [DOI] [PubMed] [Google Scholar]
- 38.Gottardis MM, Jordan VC. Cancer Res. 1988;48:5183–5187. [PubMed] [Google Scholar]
- 39.Wolf DM, Jordan VC. Breast Cancer Res. Treat. 1994;31:117–127. doi: 10.1007/BF00689682. [DOI] [PubMed] [Google Scholar]
- 40.Schafer JI, Liu H, Tonetti DA, Jordan VC. Cancer Res. 1999;59:4308–4313. [PubMed] [Google Scholar]
- 41.Jordan VC, Schafer JM, Levenson AS, Liu H, Pease KM, Simons LA, Zapf JW. Cancer Res. 2001;61:6619–6623. [PubMed] [Google Scholar]
- 42.Lieberman ME, Gorski J, Jordan VC. J. Biol. Chem. 1983;258:4741–4745. [PubMed] [Google Scholar]
- 43.Tate AC, Greene GL, DeSombre ER, Jensen EV, Jordan VC. Cancer Res. 1984;44:1012–1018. [PubMed] [Google Scholar]
- 44.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Mol. Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 45.Kalkhoven E, Valentine JE, Heery DM, Parker MG. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valentine JE, Kalkhoven E, White R, Hoare S, Parker MG. J. Biol. Chem. 2000;275:25322–25329. doi: 10.1074/jbc.M002497200. [DOI] [PubMed] [Google Scholar]
- 47.Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. Endocrinology. 2001;142:4120–4130. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pink JJ, Bilimoria MM, Assikis J, Jordan VC. Br. J. Cancer. 1996;74:1227–1236. doi: 10.1038/bjc.1996.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordan VC, Gosden B. Mol. Cell. Endocrinol. 1982;27:291–306. doi: 10.1016/0303-7207(82)90095-8. [DOI] [PubMed] [Google Scholar]
- 50.Robertson DW, Katzenellenbogen JA, Hayes JR, Katzenellenbogen BS. J. Med. Chem. 1982;25:167–171. doi: 10.1021/jm00344a015. [DOI] [PubMed] [Google Scholar]
- 51.Klinge CM, Jernigan SC, Smith SL, Tyulmenkov VV, Kulakosky PC. Mol. Cell. Endocrinol. 2001;174:151–166. doi: 10.1016/s0303-7207(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 52.Klinge CM. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramsey TL, Klinge CM. J. Mol. Endocrinol. 2001;27:275–292. doi: 10.1677/jme.0.0270275. [DOI] [PubMed] [Google Scholar]