Abstract
The expression of spore-specific marker transcripts at different stages of the asexual life cycle of Saprolegnia parasitica was analyzed. One of the markers, designated puf1, was found to be expressed transiently upon each of several cycles of zoospore encystment and reemergence. The transcript is induced immediately upon zoospore encystment and is rapidly lost when a cyst is triggered to germinate. In nongerminating cysts, puf1 is maintained until a time point when the cysts can no longer be triggered to germinate and thus have become determined for zoospore reemergence. The results show that the cyst stage has two phases, of about equal duration, which are physiologically and transcriptionally distinct and that the transcriptional machinery of oomycetes is also active in nongerminating spores. puf1 encodes a putative mRNA binding protein belonging to a conserved class of proteins including the Drosophila melanogaster Pumilio protein, Caenorhabditis elegans FBF, and Saccharomyces cerevisiae Puf5, all of which are involved in regulation of gene expression by posttranscriptional mechanisms.
The order Saprolegniales includes many severe pests of plants and aquatic animals (5). Like other oomycetes, they have been studied little at the molecular level, and since they are not closely related to fungi (11), findings from fungi may not apply to the oomycetes (13). Another consequence of their evolutionary distance from higher fungi is that most fungicides developed against the latter are not active against oomycetes. The species in this study, Saprolegnia parasitica, is known mostly as a severe fish pathogen but has also been reported to infect crustaceans (7). Like most oomycetes it reproduces asexually as biflagellate swimming zoospores, which are also the main infective units in the parasitic species. Like in many other parasitic water molds, no sexual stage has been observed, suggesting that reproduction is exclusively asexual.
When the zoospore reaches a potential host, it may encyst and germinate. In many parasitic oomycetes, spores which have encysted at a substrate not promoting germination have the ability to undergo repeated zoospore emergence, meaning that a new zoospore is released from the cyst and continues searching for a potential host (3, 4). In contrast to fairly good knowledge at the physiological level, the underlying molecular mechanisms regulating sporulation, encystment, and germination of oomycete zoospores are almost unknown. It is not even clear to what extent transcripts found in the zoospores encode proteins that were synthesized when the spores were formed in the zoosporangium, proteins that are being synthesized in the zoospores, or proteins that will be made at the onset of germination. Using in vitro translation, Gwynne and Brandhorst (12) showed that great changes in gene expression take place upon zoospore formation in the water mold Achlya ambisexualis, and expressed sequence tags have been obtained for zoospores and mycelium of Phytophthora sojae (23). Except for these studies, there is little knowledge about gene expression in the different life stages of oomycetes.
Recognizing that zoospore encystment and differentiation are important steps in establishing an infection, we are interested in learning more about the molecular changes taking place during these events. In a pilot study we have compared gene expression during different stages of the asexual life cycle of the water mold S. parasitica, strain Spt. The zoospores of this strain can be easily manipulated to encyst, germinate, and undergo repeated zoospore emergence with almost perfect synchrony (8), which is difficult with most oomycete isolates, and it is thus a good model for studying changes in gene expression during the asexual spore cycle and for identifying stage-specific transcripts. In this study we have employed differential-display reverse transcription-PCR (ddRT-PCR) in order to obtain markers for the spore and mycelium life stages and have used some of the spore-specific transcripts (sst transcripts) as markers for identifying different phases of the asexual life cycle.
MATERIALS AND METHODS
Microorganism and growth conditions.
S. parasitica strain Spt (26) was taken from our stock cultures. It was grown in a peptone-glucose medium (PG1) and sporulated by being washed in lake water (8). After it was confirmed that spontaneous encystment and germination were not taking place, cysts were produced by vortexing as described by Diéguez-Uribeondo et al. (8). Germinating cysts were produced by adding culture medium to a final concentration of 10% followed by vortexing and incubation at 20°C for 20 to 240 min. Cysts were collected by centrifugation at 2,700 × g for 10 min at 10°C, frozen in liquid nitrogen, and stored at −80°C. Mycelia were collected after 3 days at 20°C, quick-frozen in liquid nitrogen, and stored at −80°C. Sporulating mycelia were incubated at 20°C for 0 to 8 h and frozen in liquid nitrogen.
RNA isolation.
Total RNA and mRNA were isolated from encysted spores using a Trizol kit (Life Technologies) and a QuickPrep Micro mRNA isolation kit (Pharmacia), respectively. The frozen cysts were thawed and resuspended in the extraction buffer. The resuspended cysts were studied with an inverted microscope to confirm that more than 95% of the cysts were at the same stage. The cysts were disrupted by vortexing with 425- to 600-μm glass beads (Sigma) in the extraction buffer or by sonication. Successful disruption of cysts was confirmed by microscopy, and RNA isolation was performed according to the manufacturer’s instructions. Total RNA from growing and sporulating mycelium was isolated with Trizol (Life Technologies) according to the manufacturer’s instructions.
ddRT-PCR.
Gene expression in spores, germinating cysts, and mycelium was compared by using ddRT-PCR. For this, mRNA was isolated as described above, and cDNA synthesis and RT-PCR were performed as described by Bauer et al. (2). Differentially expressed bands were reamplified under the same PCR conditions and cloned into pBluescript KS(+) by T-vector cloning (17). The primer T12MC was used in combination with five different arbitrary primers (10-IS, CAGGCTATCA; 10-PP, GGGGAAACTT; BMR-10, GGTACTAAGG; DD-4, AACCGGGTTC; and AP-1, AGCCAGCGAA). The subclone puf1 was isolated with the anchor primer T12MC and the arbitrary primer 10-IS.
Cloning of puf1.
A cDNA library was constructed from mRNA isolated from encysted zoospores. cDNA was synthesized with a Zap-cDNA kit (Stratagene) and ligated into Uni-Zap XR (Stratagene). The library was screened with a probe from clone puf1, and positive clones were in vivo excised according to the protocol for Uni-Zap XR (Stratagene).
DNA sequencing and sequence analysis.
The clones were sequenced on both strands with an ABI-Prism 377 DNA sequencer (Perkin-Elmer) using vector- and gene-specific primers. The sequences were analyzed using Macvector 4.1.4 (Eastman Kodak Company) and Assemblylign 1.0.2 (International Biotechnologies). Related proteins were identified using BLAST 2.0 (1). The deduced amino acid sequence was aligned with sequences of other proteins belonging to the Puf family, using ClustalW 1.7 multiple-sequence alignment (30). Sequences used in alignments and in sequence comparisons are as follows: Pumilio (Drosophila melanogaster) (15), accession number A46221; F16P2.43 (Arabidopsis thaliana), accession number ACC95216; PufA (Dictyostelium discoideum) (28), accession number AF128626; Puf3 (YLL013c) (Saccharomyces cerevisiae) (20), accession number Z73118; Puf5 (MPT5) (S. cerevisiae) (29), accession number D26184; and FBF2 (Caenorhabditis elegans) (38), accession number U23176.
Preparation of probes.
Probes for Northern and Southern blots were cleaved with the appropriate enzymes and gel purified. Probes puf1, sst2, sst15, and mst3 are the clones originally identified by ddRT-PCR. Two probes were prepared from the library clone puf1f. puf1f-sma corresponds to residues 1 to 912 of the submitted sequence. Probe puf1f-1213 contains the 3′ end of clone puf1f, from position 1213, and includes the complete conserved domain. A probe was also prepared against tef1, coding for elongation factor 1α, from the oomycete Phytophthora infestans (32). The probe corresponds to positions 175 to 700 of PPI116 and was a kind gift from Francine Govers, Wageningen, The Netherlands. All probes were labeled with [α-32P]dCTP using a Megaprime labeling kit (Amersham) and were purified on a Nick column (Pharmacia).
Northern and Southern blots.
Northern blot analysis was performed with 10 μg of total RNA or 1 μg of mRNA as described by Sambrook et al. (24). A 0.28- to 6.58-kb size marker was obtained from Promega. The gel was blotted to a Hybond N+ membrane (Amersham) and UV cross-linked. Ribosomal bands were visualized under UV light before and after blotting. Genomic DNA for Southern blots was isolated with a Puregene kit (Gentra Systems) and cleaved with BamHI, EcoRI, KpnI, SalI, and SmaI. Southern blotting was performed as described by Sambrook et al. (24) using a Hybond N+ transfer membrane (Amersham). Hybridization was performed according to the manufacturer’s instructions. The membranes were exposed to a BAS 2000 II image plate (Fujix), and images were analyzed with TINA 2:0 (Raytest Isotopenmessgeräte GmbH).
Gene-specific RT-PCR.
Zoospores and cysts were collected by centrifugation for 1 min at 400 × g or 13,000 × g, respectively. RNA was isolated with a GenElute mammalian total RNA kit (Sigma), and after DNase treatment, cDNA was synthesized from 250 ng of RNA using 1 μM T30V or gene-specific primer. The cDNA was purified with a Qiaquick PCR purification kit and eluted in water. Each 12-μl PCR mixture contained cDNA corresponding to 25 ng of RNA, 1.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.4 mM concentration of each primer, and 1 U of Taq polymerase (Gibco) mixed with Platinum Antitaq antibodies (Gibco). Replicates of each PCR were run for 25 to 40 cycles. Sequences and annealing temperatures for the different primers were as follows: puf1 specific, TCGAGCTCGCGCTGCAAATGTA and GTTGGCGTACTGGTCCTTCATCAT, 60°C; sst2 specific, CAGGCAACGACTTTTTCAA and TGATGACCTGGGCTTCTTG, 54°C; sst15 specific, CGACGGTAAGCGGTATGC and AAGCGGACAACCACTGGA, 54°C; and actin specific, TTACTCGTTCACCACCAC and CCGCCCGACAACACAATG, 53°C. A second set of puf1 primers was designed against regions conserved between Puf1 and other Puf proteins. The sequences of these primers, referred to as universal Puf primers, were GAICAITAIGGIAAYTAYGTIATHCARCAYG and ACIACITAITTIGCRTAYTGRTCYTTCATCAT, and they were used at 54°C. The PCR products were separated on agarose gels and stained with ethidium bromide (24). The universal Puf and sst15-specific primer sets readily produced bands after 25 cycles of replication, while the sst2- and puf1-specific primer sets required 30 cycles. The two different sets of puf1 primers give the same results in all experiments, and all results have been repeatedly obtained in independent experiments.
Nucleotide sequence accession numbers.
The accession numbers of the transcripts described in this paper are as follows: puf1, AJ245441; sst2, AJ413213; sst15, AJ413214; and mst3, AJ413215.
RESULTS
Identification of stage-specific transcripts by ddRT-PCR.
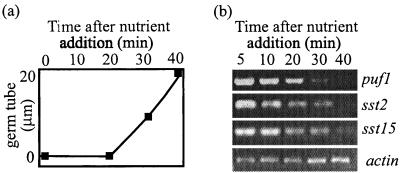
In order to obtain stage-specific markers for studying spore differentiation, we compared gene expression in mycelium, asexual spores, and two germling stages by ddRT-PCR. With five primer combinations, a total of 220 relatively discrete bands could be seen on the differential display gels. Approximately 45 of these bands were specific for the spore stage and 40 were specific for the mycelium stage, while only a few bands appeared to be specific for the germling stages. Of the 14 bands which were subcloned, only two could be unambiguously identified from their sequences, one (puf1) showing a high similarity to D. melanogaster maternal Pumilio protein (score of 10−99) and another coding for a 40S ribosomal protein (score of 10−20). Other subclones showed similarity to, for example, various glucanases (sst2) (best score of 9 × 10−5), acyl-coenzyme A binding proteins (best score of 5 × 10−16), and NADH dehydrogenase (score of 5 × 10−32). Five of the subcloned bands, four spore specific and one mycelium specific, were tested by Northern blotting. Four of these bands were confirmed as being stage specific (Fig. 1), and the fifth band was upregulated in spores.
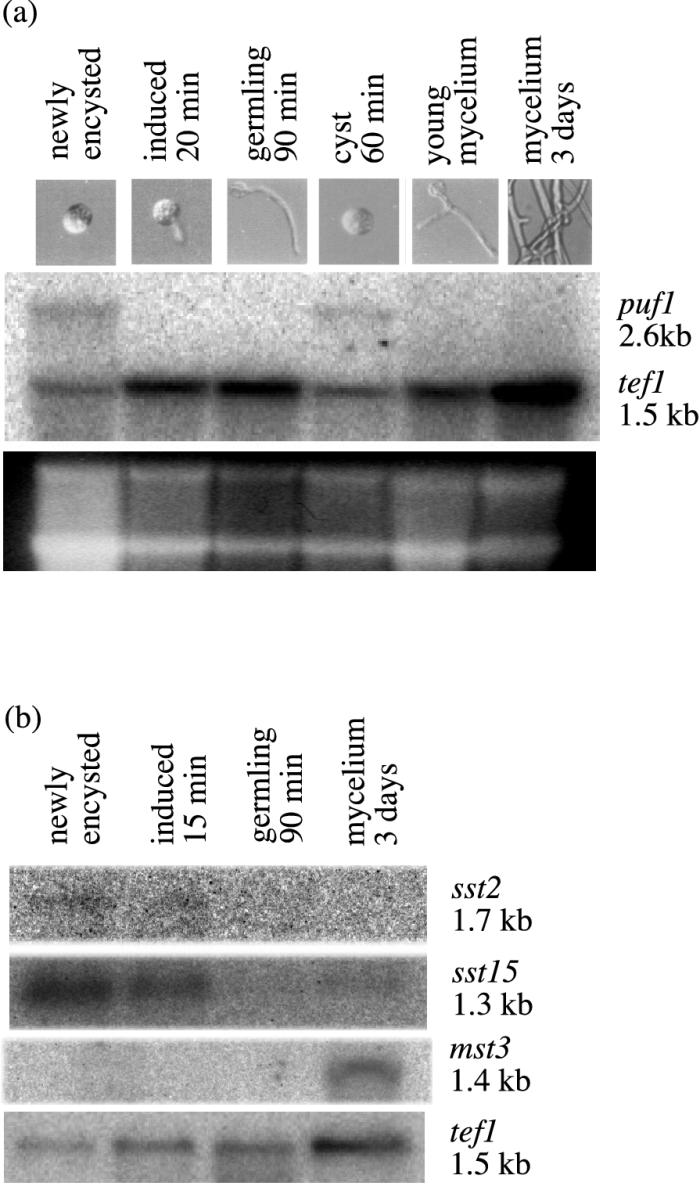
FIG. 1.
Confirmation of stage-specific transcripts by Northern blotting of total RNA. (a) Expression of puf1 in spores, cysts induced to germinate (20 min after induction), germlings (90 min after induction), nongerminating cysts collected 60 min after encystment, young (4-h) mycelium from germinating cysts, and 3-day-old mycelium. The blot was hybridized simultaneously with probes puf1f-sma (upper blot) and tef1 (lower blot). Ribosomal bands were stained with ethidium bromide. (b) Expression of sst2, sst15, and mst3 in zoospores, cysts induced to germinate (15 min after induction), germlings (90 min after induction), and 3-day-old mycelium. The blots were reprobed with tef1.
Cloning of a putative RNA binding protein, Puf1.
One of the spore-specific clones showed homology to a family of RNA binding proteins named the Puf family (38) which comprises many proteins that play important roles in regulation of development, making it an interesting object for further studies. To obtain the full sequence of the transcript designated puf1, we screened a cDNA library made from mRNA from asexual spores, using the subcloned 213-nucleotide (nt) PCR product as a probe. The lengths of the longest clones, puf1f [2,412 nt plus poly(A)] and puf1b [2,359 nt plus poly(A)], agree with the size estimated from Northern blots, 2.6 kb. Southern blot analysis with puf1 or a probe from the 5′ part of the transcript (puf1f-sma) showed a simple pattern suggesting that the gene is present in one copy in the genome (results not shown). Similar results were obtained when Southern blot analysis was performed with the probe containing the conserved 3′ region of the puf1 transcript (puf1f-1213) under medium- or low-stringency conditions (results not shown). The cDNA sequence contains an open reading frame (ORF) corresponding to 773 amino acids. A stop codon, TGA, is present 6 nt upstream of the putative start methionine, and there are two stop codons downstream from the end of the putative ORF, suggesting that this putative ORF represents the complete coding region.
Puf1 contains a complete putative RNA binding domain.
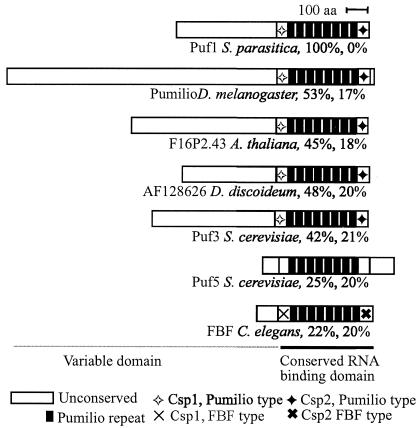
To identify conserved regions, we aligned the putative Puf1 protein to other members of the Puf family. A schematic view of this family, with percent similarities, is shown in Fig. 2. The proteins shown are some for which a function has been reported or which show high similarity to Puf1. In addition to the members shown, the Puf family contains many proteins which are more or less distantly related (37, 38).
FIG. 2.
A selection of members of the Puf family. Characteristic of this family is an RNA binding domain consisting of eight repeats and two flanking sequences, Csp1 and Csp2. The eight repeats are well conserved within the Puf family, while the Csp motifs vary between different subclasses. The N termini of the proteins consist of a nonconserved domain of unknown function. Percent identities and similarities between the RNA binding domains of the different proteins and that of Puf1 are indicated.
While little or no sequence similarity could be seen between the N-terminal region of Puf1 and other Puf proteins, the alignments showed that the ORF encodes a complete RNA binding domain of approximately 323 amino acids consisting of eight repeats of approximately 36 amino acids, which are characteristic of the Puf family (38). The repeats are flanked by two short sequence motifs (36, 38) named Csp1 and Csp2 (38), which together with the repeats form an extended structure of tandem helical repeats (9) that covers the entire RNA binding domain. The sequences of the Csp motifs are not conserved among all Puf proteins, and those of Puf1 resemble the Csp motifs of Pumilio, while they show little or no homology to their counterparts in FBF (Fig. 2). The alignments also show that the RNA binding domain of Puf1 is highly conserved, sharing practically all residues that are conserved between other Puf proteins (results not shown).
puf1 is specifically expressed in asexual spores.
To confirm the stage-specific nature of the transcripts identified by ddRT-PCR, their expression was examined by Northern blotting at different stages of the asexual life cycle. puf1 was detected in nongerminating encysted zoospores collected 15 min or 1 h after encystment, while in cysts harvested approximately 25 min after being triggered to germinate, when the germ tube had emerged, the transcript had disappeared (Fig. 1a). The puf1 transcript could be detected neither in 90-min germlings nor in young or old mycelia (Fig. 1a). The transcripts sst2 and sst15 were detected in cysts not triggered to germinate and in cysts collected 20 min after being triggered to germinate, while they were absent in older germlings and vegetative mycelium (Fig. 1b). We also confirmed one of the mycelium-specific transcripts, mst3, which was detected only in the 3-day-old mycelium (Fig. 1b). The data from the Northern blots thus confirm the stage-specific expression of the transcripts studied. As a control for gene expression in these experiments, we used tef1. The expression pattern of this transcript in S. parasitica, with low expression levels in spores and higher levels in the mycelium, has been confirmed in multiple experiments.
puf1 is transcribed in the late stages of sporulation.
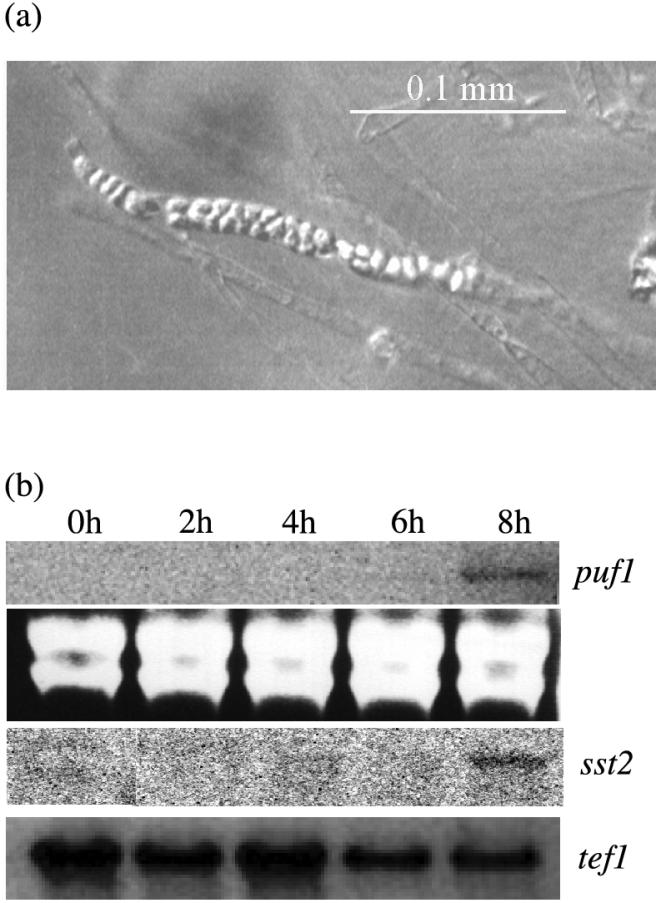
Having shown its spore-specific nature, we were interested to know if puf1 was expressed during sporulation. Sporangia of Saprolegnia are formed directly at the end of the hyphal tips and cannot be separated from the mycelium. Instead, mycelia were harvested at different time points after induction of sporulation. The first immature sporangia were seen 4 h after induction of sporulation. After 6 h most hyphal tips were converted into sporangia, most of which were immature but a few of which had visible spores. After 8 h most sporangia had matured, and many of them contained visible spores waiting to be released (Fig. 3a). A very faint band could be seen on Northern blots after 6 h, and after 8 h the signal was evident (Fig. 3b). Similar results were obtained for another of the spore-specific transcripts, sst2 (Fig. 3b), while the expression of tef1 did not change significantly during sporulation (Fig. 3b). These results indicate that the transcripts puf1 and sst2 are expressed when spores are being formed in the sporangia.
FIG. 3.
Expression of puf1 in sporulating mycelium harvested 0, 2, 4, 6, and 8 h after washing. (a) Interference contrast microscopy of mature zoosporangia at 8 h after induction of sporulation, with visible spores ready to be released. (b) Northern blot of total RNA hybridized with probe puf1f-sma (upper panel). The next three panels, from top to bottom, show ribosomal bands stained with ethidium bromide and Northern blots hybridized with sst2 and tef1, respectively.
puf1 is present in cysts but not in zoospores.
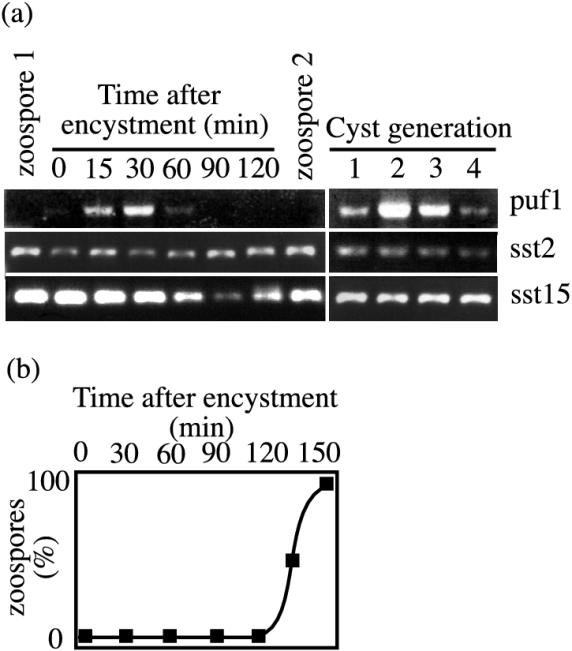
The asexual spores of Saprolegnia are released from sporangia in the form of motile zoospores. After encysting, the spores may either germinate in response to nutrient stimuli or release a second zoospore generation. The differences in gene expression between the different phases of the asexual spore stage have not been studied at all in oomycetes. The ipiO gene of P. infestans is found in both zoospores and cysts, but no transcripts whose expression changes at the transition between these stages are known. To see if we could detect any changes in the expression of the different spore-specific transcripts, we analyzed their expression in zoospores and nongerminating cysts collected at different time points from after encystment by RT-PCR. Swimming zoospores were triggered to encyst by vortexing, after which the cysts were collected at different time points from 0 to 120 min. At the same time points the number of released zoospores was counted. We also collected zoospores from the first generation as well as zoospores from the second generation released by the cysts that had been produced by encysting first-generation zoospores. We found that one of the transcripts, puf1, was absent in the zoospores while it was present in cysts collected 15 min after encystment (Fig. 4a). The transcript could still be detected 60 min after encystment, after which it disappeared. The second zoospore generation emerged from the cysts approximately 120 min after encystment (Fig. 4b). In the second zoospore generation also, no puf1 transcript could be detected (Fig. 4a), while it was present in all subsequent cyst generations (Fig. 4a). The other transcripts, sst2 and sst15, which were analyzed using the same poly(T)-primed cDNA, were present throughout the spore life stage, in both zoospores and cysts (Fig. 4a). Zoospores of S. parasitica are very rapidly encysted by mechanical stimuli, and the results from several experiments showed that it was crucial that zoospores were frozen within minutes after collection to avoid induction of puf1 during preparation. It thus seems that transcription of this cyst-specific transcript is initiated almost immediately after the signal to encyst is received and that it is subsequently lost when the cyst is getting ready to release a new zoospore.
FIG. 4.
puf1 is induced transiently upon encystment. (a) RT-PCR with gene-specific primers for puf1 (upper panels), sst2 (middle panels), and sst15 (lower panels) on total RNA from the first zoospore generation; from cysts harvested 0, 15, 30, 60, 90, and 120 min after vortexing; and from the second zoospore generation. RT-PCR was also performed with the same primers on total RNAs from the first, second, third, and fourth cyst generations. (b) Percentages of cysts that have released a second zoospore generation at different time points after encystment.
Induction of germination triggers disappearance of puf1.
The Northern blots showed that the spore-specific transcripts were degraded upon germination. We analyzed the time point of disappearance of the spore-specific transcripts by RT-PCR. Zoospores were triggered to germinate by vortexing in presence of a nutrient trigger. After 20 min the first cysts showed visible signs of germination, and 10 min later all cysts had produced a germ tube (Fig. 5a). We found that the level of puf1 dropped below the detection level at between 30 and 40 min after induction of germination (Fig. 5b) and thus much faster than in a cyst undergoing repeated zoospore emergence. The levels of the other spore-specific transcripts, sst2 and sst15, also decreased upon germination, although sst15 could still be detected at 40 min (Fig. 5b). In contrast, the levels of a control transcript, encoding actin, did not change during the experiment (Fig. 5b).
FIG. 5.
Disappearance of spore-specific transcripts upon germination. (a) Length of germ tubes in cysts 0 to 40 min after induction of germination by vortexing together with a nutrient trigger. (b) RT-PCR with primers for puf1, sst2, sst15, and actin on total RNA from cysts harvested 5 to 40 min after induction of germination.
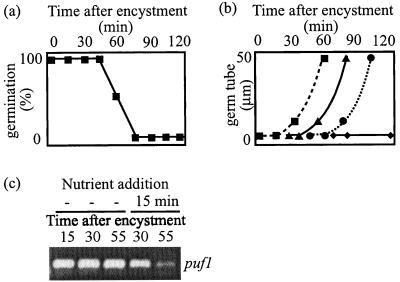
The disappearance of puf1 in nongerminating cysts occurs 30 to 60 min before the cysts release new zoospores (Fig 4). This made us ask whether the disappearance represents an early step on the path toward repeated zoospore emergence and whether it correlates with physiological changes in the cyst. We thus tested the ability of the cysts to germinate in response to a nutrient trigger, PG1, at different time points after encystment. Zoospores were encysted by vortexing, and the nutrient trigger was added after 0 to 120 min. After the addition of the trigger, the cysts were monitored for an additional 1 h, and the number of cysts producing a germ tube or releasing new zoospores was counted. In this experiment 100% of the cysts germinated when the nutrient trigger was added within the first 45 min after encystment. At 60 min the germination frequency was reduced to approximately 50%, and when the nutrient trigger was added at later time points, no cysts germinated (Fig. 6a). Once a cyst was triggered to germinate, a germ tube emerged within 30 min and reached a length of 50 μm within about 1 h (Fig. 6b). The cysts that failed to germinate instead released swimming zoospores approximately 120 min after encystment. The conclusion of this experiment is that a cyst which has not been triggered to germinate within the first hour, when puf1 is expressed, becomes determined to undergo repeated zoospore emergence.
FIG. 6.
(a) Percentage of cysts germinating in response to a nutrient trigger added at different time points after encystment by vortexing. (b) Germ tube length in cysts triggered to germinate by addition of a nutrient trigger (PG1) at different time points after encystment by vortexing: ▪, PG1 added at 0 min; ▴, PG1 added at 25 min; •, PG1 added at 50 min; ⧫, PG1 added at 75 min. (c) Disappearance of puf1 upon delayed induction of germination. RT-PCR with universal Puf primers was done with nongerminating cysts collected at 15, 30, or 55 min after encystment by vortexing or with cysts triggered to germinate by addition of PG1 15 min after vortexing followed by an additional 15 or 40 min of incubation.
In the previous experiments the trigger to germinate was given at the same time as the encystment trigger and thus before the appearance of puf1. We wanted to test whether the rate of puf1 disappearance can also be enhanced by a germination trigger when given at a later time point. A nutrient trigger was added to zoospores 15 min after encystment had been induced by vortexing, when puf1 was already being transcribed at its maximum level. In this experiment the transcript still remained for an additional 15 min after the addition of PG1, while after 40 min the transcript had disappeared (Fig. 6c) and the cysts had started to produce a germ tube. At all time points up to 60 min, the control cysts not exposed to PG1 expressed puf1 at a high level (Fig. 6c). This means that the disappearance of puf1 takes place between 20 and 40 min after the addition of the germination trigger, irrespective of whether it is added directly upon encystment, (Fig. 5a) or at a later time point (Fig. 6c), and that the disappearance correlates with the time of germ tube emergence. Taken together our results show that puf1 is present exclusively in undetermined cysts and that it is lost when the cysts either are triggered to germinate or become determined to release new zoospores.
Induction of puf1 is due to de novo synthesis of mRNA.
Cytoplasmic deadenylation may shorten the poly(A) tail of a transcript to a length at which oligo(dT) selection and poly(T)-primed cDNA synthesis may be hampered (20). To test whether the transient appearance of puf1 in cysts could be due to cytoplasmic polyadenylation and subsequent deadenylation, we synthesized cDNA with a gene-specific primer in parallel with the poly(T) primer. The transcript was detected by PCR in 15-min-old cysts but not in zoospores irrespective of the primer used for the cDNA synthesis, while the control transcript sst15 was detected in both stages (Fig. 7). This shows that the observed induction of puf1 upon encystment results from de novo synthesis of the transcript and not from modifications of the poly(A) tail.
FIG. 7.
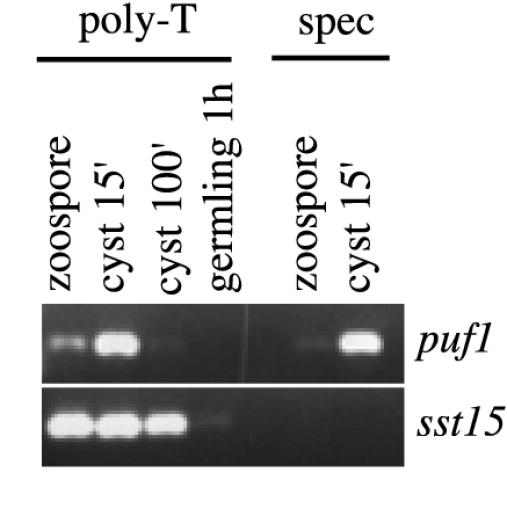
Induction of puf1 results from de novo synthesis. RT-PCR was done on cDNA primed with primer T30MC (left) or the universal Puf primer (right). PCR with universal Puf primers (upper panel) and sst15-specific (spec) primers (lower left panel) was done with zoospores, cysts 15 min after vortexing, cysts 100 min after vortexing, or germlings 1 h after addition of a nutrient trigger.
DISCUSSION
puf1 is a marker of undetermined cysts.
The zoospores of oomycetes generally exhibit low transcriptional activity, which is rapidly increased upon germination (22, 25). Expression during the asexual life cycle has been studied for only a few transcripts. One such transcript is ipiO, which is expressed in zoospores, in cysts, and also in young germlings and invading hyphae (33). In this work we report, for the first time in an oomycete, the finding of a transcript, puf1, which is expressed exclusively in one phase of the asexual spore stage.
While it has previously been shown that germination of oomycete asexual spores is dependent on de novo synthesis of mRNA (16, 22, 25), the question of whether transcription also occurs in nongerminating spores has not been thoroughly assessed. The transient expression of puf1 in cysts undergoing repeated zoospore emergence nevertheless provides evidence that the transcriptional machinery is also active in oomycete spores prior to the onset of germination.
We showed that the transcript was induced by de novo synthesis almost instantly following the trigger to encyst and that it was lost when the cysts became determined for either zoospore release or germination. It is thus a marker for an undetermined cyst stage, which is different from zoospores and cysts predestined to undergo repeated zoospore emergence or germination and which represents the first step in spore differentiation. These cysts have the ability, in response to environmental signals, to “decide” whether to germinate or detach from the surface by releasing a new zoospore. Since encystment appears to be a nonspecific process, the regulation of the alternative pathways may contribute significantly to substrate specificity. In the Saprolegniales, the cysts are also the structures from which infectious penetration pegs emerge (19). These cysts may thus harbor several transcripts which play important roles during the early events of establishing an infection, and comparing gene expression in the zoospore and cyst stages may help to identify some of those transcripts.
Puf1 belongs to a family of posttranscriptional regulators.
The predicted Puf1 protein was identified as a member of a conserved family of RNA binding proteins named the Puf family, after Pumilio and FBF, which were the first two proteins of this class to be functionally characterized (38). Pumilio (35, 36) and FBF (38) bind sequence specifically to short motifs, named NRE and PME, respectively, present in the 3′ untranslated region of a target mRNA. Our alignments show that Puf1 contains a complete RNA binding domain with highly conserved repeats, and since RNA binding activity is conserved within the Puf family (37), we expect that Puf1 also binds mRNA.
Several of the characterized Puf proteins are involved in posttranscriptional regulation by binding to specific transcripts and blocking their translation. In contrast, the targeted transcripts and thus the biological role differ between the systems. In the D. melanogaster embryo, Pumilio functions in determining polarity (18, 34) and maintaining the germ line stem cells (10, 21). In the germ line of C. elegans, FBF promotes the switch from sperm to oocyte production (31, 38), and in D. discoideum, PufA regulates the development of reproductive structures (28). Repression of the targeted transcript may also require the presence of other proteins, which in turn may be differentially expressed (14, 27). In S. cerevisiae other Puf proteins regulate mRNA turnover by promoting deadenylation and degradation of targeted transcripts (20, 29), affecting a large number of transcripts (20), including the HO endonuclease involved in regulation of the mating-type switch (29).
Possible roles of Puf1.
The transition from one life stage to another often affects the expression of a large number of transcripts; for example, 1,000 of the 6,200 protein-encoding genes in the budding yeast genome are reported to be either up- or downregulated during sporulation (6). The development of zoospores into mycelium is thus likely to involve major changes in gene expression, and the undetermined cysts represent a transition stage in this process. Upon becoming triggered to germinate, spore-specific mRNAs should be degraded, while mRNAs required for hyphal outgrowth and infection should be translated, correlating with the time point of puf1 disappearance. In cysts not triggered to germinate, disappearance of puf1 is instead an early step on the path toward repeated zoospore emergence, marking the transition from an undetermined cyst to a cyst producing a new zoospore.
Although precise predictions of the role of Puf1 cannot be made, it is easy to imagine roles for a posttranscriptional regulator such as a Puf protein in maintaining the undetermined cyst stage or in regulating mRNA turnover upon germination or zoospore release. An example of a possible mechanism is that one or more factors involved in germination are transcribed upon encystment but that their translation is blocked by Puf1 until the cyst receives a trigger to germinate. Another possibility is that translation of transcripts encoding zoospore-specific proteins is blocked in the undetermined cyst. These transcripts might then either become degraded upon germination or be translated again when the cyst is releasing a new zoospore.
The absence of puf1 in zoospores raises the question of why it is present in the sporulating mycelium. One possibility is that it is expressed in the nonflagellated primordial spores and subsequently lost before zoospores are released, in a manner similar to what is seen during repeated zoospore emergence. If that holds true, its role during sporulation may be similar to that in the cyst, perhaps preventing germination in the sporangium or premature zoospore release.
Acknowledgments
This study was supported by grants from Carl Tryggers Stiftelse to Lage Cerenius and from Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) to Kenneth Söderhäll.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, D., H. Muller, J. Reich, H. Riedel, V. Ahrenkiel, P. Wharton, and M. Strauss. 1993. Identification of differentially expressed mRNA species by an improved display technique (ddRT-PCR). Nucleic Acids Res. 21: 4272–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerenius, L., and K. Söderhäll. 1984. Repeated zoospore emergence from isolated spore cysts of Aphanomyces astaci. Exp. Mycol. 8: 370–377. [Google Scholar]
- 4.Cerenius, L., and K. Söderhäll. 1985. Repeated zoospore emergence as a possible adaptation to parasitism in Aphanomyces. Exp. Mycol. 9: 259–377. [Google Scholar]
- 5.Cerenius, L., and K. Söderhäll. 1986. Saprolegniaceae: zoospore formation, virulence and pathogenesis in animal hosts, p. 97–116. In R. Dayal (ed.), Advances in zoosporic fungi. M D Publications Pvd. Ltd., New Delhi, India.
- 6.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- 7.Diéguez-Uribeondo, J., L. Cerenius, and K. Söderhäll. 1994. Saprolegnia parasitica and its virulence on three species of freshwater crayfish. Aquaculture 120: 219–228. [Google Scholar]
- 8.Diéguez-Uribeondo, J., L. Cerenius, and K. Söderhäll. 1994. Repeated zoospore emergence in Saprolegnia parasitica. Mycol. Res. 98: 810–815. [Google Scholar]
- 9.Edwards, T. A., S. E. Pyle, R. P. Wharton, and A. K. Aggarwal. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105: 281–289. [DOI] [PubMed] [Google Scholar]
- 10.Forbes, A., and R. Lehmann. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125: 679–690. [DOI] [PubMed] [Google Scholar]
- 11.Förster, H., M. D. Coffey, H. Elwood, and M. L. Sogin. 1990. Sequence-analysis of the small subunit ribosomal-rRNAs of 3 zoosporic fungi and implications for fungal evolution. Mycologia 82: 306–312. [Google Scholar]
- 12.Gwynne, D. I., and B. P. Brandhorst. 1982. Changes in gene expression during sporangium formation in Achlya ambisexualis. Dev. Biol. 91: 263–277. [DOI] [PubMed] [Google Scholar]
- 13.Judelson, H. S. 1997. The genetics and biology of Phytophthora infestans: modern approaches to a historical challenge. Fung. Genet. Biol. 22: 65–76. [DOI] [PubMed] [Google Scholar]
- 14.Kraemer, B., S. Crittenden, M. Gallegos, G. Moulder, R. Barstead, J. Kimble, and M. Wickens. 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9: 1009–1018. [DOI] [PubMed] [Google Scholar]
- 15.Macdonald, P. M. 1992. The Drosophila-pumilio gene—an unusually long transcription unit and an unusual protein. Development. 114: 221–232. [DOI] [PubMed] [Google Scholar]
- 16.MacLeod, H., and P. A. Horgen. 1979. Germination of the asexual spores of the aquatic fungus Achlya bisexualis. Exp. Mycol. 3: 70–82. [Google Scholar]
- 17.Marchuk, D., M Drumm, A. Saulino, and F. S. Collins. 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nuclei Acids Res. 19: 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murata, Y., and R. P. Wharton. 1995. Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80: 747–756. [DOI] [PubMed] [Google Scholar]
- 19.Nylén, L., and Unestam, T. 1975. Ultrastructure of the penetration of the crayfish integument by the fungal parasite Aphanomyces astaci (oomycete). J. Invertebr. Pathol. 26: 356–366. [Google Scholar]
- 20.Olivas, W., and R. Parker. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19: 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parisi, M., and H. F. Lin. 1999. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics 153: 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penington, C. J., J. R. Iser, B. R. Grant, and K. R. Gayler. 1989. Role of RNA and protein-synthesis in stimulated germination of zoospores of the pathogenic fungus Phytophthora palmivora. Exp. Mycol. 13: 158–168. [Google Scholar]
- 23.Qutob, D., P. T. Hraber, B. W. S. Sobral, and M. Gijzen. 2000. Comparative analysis of expressed sequences in Phytophthora sojae. Plant Physiol. 123: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Söderhäll, K., and L. Cerenius. 1983. Protein and nucleic acid synthesis during germination of the asexual spores of the aquatic fungus Aphanomyces astaci. Physiol. Plant 58: 13–17. [Google Scholar]
- 26.Söderhäll, K., M. W. Dick, G. Clark, M. Furst, and O. Constantinescu. 1991. Isolation of Saprolegnia parasitica from the crayfish Astacus leptodactylus. Aquaculture 92: 121–125. [Google Scholar]
- 27.Sonada, J., and R. P. Wharton. 2001. Drosophila brain tumor is a translational repressor. Genes Dev. 15: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Souza, G. M., A. M. da Silva, and A. Kuspa. 1999. Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development 126: 3263–3274. [DOI] [PubMed] [Google Scholar]
- 29.Tadauchi, Y., K. Matsumoto, I. Herskowitz, and K. Irie. 2001. Post-transcriptional regulation through the HO 3′-UTR by MPT5, a yeast homologue of Pumilio and FBF. EMBO J. 20: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tollervey, D., and J. F. Caceres. 2000. RNA processing marches on. Cell 103: 703–709. [DOI] [PubMed] [Google Scholar]
- 32.van’t Klooster, J. W., V.G. van den Berg, P. van West, and F. Govers. 2000. tef1, a Phytophthora infestans gene encoding translation elongation factor 1alpha. Gene 249: 145–151. [DOI] [PubMed] [Google Scholar]
- 33.van West, P., A. J. De Jong, H. S. Judelson, A. M. E. Emons, and F. Govers. 1998. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fung. Genet. Biol. 23: 126–138. [DOI] [PubMed] [Google Scholar]
- 34.Wharton, R. P., and G. Struhl. 1991. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67: 955–967. [DOI] [PubMed] [Google Scholar]
- 35.Wharton, R. P., J. Sonada, T. Lee, M. Patterson, and Y. Murata. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1: 863–872. [DOI] [PubMed] [Google Scholar]
- 36.Zamore, P. D., D. P. Bartel, R. Lehmann, and J. R. Williamsson. 1999. The Pumilio-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38: 596–604. [DOI] [PubMed] [Google Scholar]
- 37.Zamore, P. D., J. R. Williamson, and R. Lehmann. 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA binding proteins. RNA 3: 1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, B., M. Gallegos, A. Pouti, E. Durkin, S. Fields, J. Kimble, and M. P. Wickens. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 70–484. [DOI] [PubMed] [Google Scholar]