Abstract
To determine the influence of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T cells on the development of drug resistance mutations in the HIV-1 protease, we analyzed protease sequences from viruses from a human leukocyte antigen class I (HLA class I)-typed cohort of 94 HIV-1-positive individuals. In univariate statistical analyses (Fisher's exact test), minor and major drug resistance mutations as well as drug-associated polymorphisms showed associations with HLA class I alleles. All correlations with P values of 0.05 or less were considered to be relevant without corrections for multiple tests. A subset of these observed correlations was experimentally validated by enzyme-linked immunospot assays, allowing the definition of 10 new epitopes recognized by CD8+ T cells from patients with the appropriate HLA class I type. Several drug resistance-associated mutations in the protease acted as escape mutations; however, cells from many patients were still able to generate CD8+ T cells targeting the escape mutants. This result presumably indicates the usage of different T-cell receptors by CD8+ T cells targeting these epitopes in these patients. Our results support a fundamental role for HLA class I-restricted immune responses in shaping the sequence of the HIV-1 protease in vivo. This role may have important clinical implications both for the understanding of drug resistance pathways and for the design of therapeutic vaccines targeting drug-resistant HIV-1.
The development of drug resistance is one of the biggest obstacles in human immunodeficiency virus type 1 (HIV-1) therapy, at least in countries where antiretroviral therapy is available. Being one of the major drug targets, the HIV-1 protease (PR) is subjected to the acquisition of resistance mutations. The criteria of the Drug Resistance Mutation Group of the International AIDS Society (15) discriminate between three categories of resistance-associated mutations: major and minor drug mutations and drug-associated polymorphisms. The evolutionary pathways for most of these drug-associated amino acid substitutions are complex (5, 10, 27, 43) and difficult to predict. Considering the various mutational pathways for the development of drug resistance in patients with similar or even identical therapeutic regimens, it seems likely that immunological host factors may affect the emergence of resistance mutations. Over the past years, several studies have provided evidence that immunological pressure exerted by HLA class I-specific cytotoxic T lymphocytes (CTL) shapes the sequence of HIV-1 through the selection of CTL escape mutations (18, 21, 22, 28). To assess the influence of HIV-1-specific CD8+ T cells on the development and patterns of drug resistance, we analyzed HIV-1 full-length PR sequences from 94 patients in the context of patients' HLA class I alleles. We could demonstrate that there are positive and negative correlations between drug-associated amino acid substitutions and certain HLA class I alleles in the population, suggesting important interactions between the T-cell-mediated immune response selection and the development of resistance (16, 23, 28). Biological assays could verify that the observed associations between amino acid substitutions and HLA class I alleles predict epitopes recognized by CD8+ T cells, even if univariate statistical analyses are used.
MATERIALS AND METHODS
Samples and patients.
Full-length PR sequences from plasma samples from 94 HIV-1-seropositive subjects presenting at the Immunodeficiency Unit of the Department of Medicine III of the University Hospital Erlangen were analyzed. The study was approved by the ethics committee of the medical faculty of the University of Erlangen-Nuremberg, and the patients gave informed consent. For all patients, genotypic resistance testing was performed and the most recent HIV-1 PR sequence from each individual was included in the analysis. Up to the time of resistance testing, 82% of the patients had been treated with at least one of the following protease inhibitors (PIs): saquinavir (37%), indinavir (29%), nelfinavir (33%), ritonavir (24%), lopinavir (19%), amprenavir (18%), atazanavir (6%), and tipranavir (3%). All patients were HLA typed, and viral loads were determined using the Versant HIV-1 RNA assay (Bayer Diagnostics, Leverkusen, Germany) with a detection limit of 50 copies/ml of plasma (version 3.0). The median viral load of the patients included in the statistical analysis was 26,000 copies/ml (range, 150 to 1,800,000); the median CD4 cell count was 197 × 106 cells/liter (range, 2 × 106 to 896 × 106). Enzyme-linked immunospot (ELISPOT) assays were conducted using peripheral blood mononuclear cells (PBMC) not only from patients included in the statistical analysis but also from additional HLA-typed HIV-1-infected patients without available PR sequence data. HIV-1 subtype analyses, which were conducted with the genotyping tool from NCBI (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi), showed that 83 of our 94 patients (88.3%) harbored subtype B viruses. Of the remaining patients, three were infected with subtype C, four with the circulating recombinant form 01_AE (CRF01_AE), one with CRF03_AB, one with CRF15_01B, and two with subtype D. The epidemiological and phylogenetic relatedness of the viral sequences was analyzed by the neighbor-joining method (Wisconsin package; Genetics Computer Group) (7).
HLA typing.
HLA-A and -B typing was performed using standard serological techniques (Biotest AG, Dreieich, Germany) or genotypic analyses (enzyme-linked probe hybridization assay; Biotest AG) according to the manufacturers's guidelines. HLA-A and -B alleles in 56% of the patients were identified by genotyping, and those in 44% of the patients were identified by serological analysis. For all patients, HLA-C genotyping was performed using sequence-specific primer amplification (AllSet SSP; Dynal Biotech, Karlsruhe, Germany).
Cells and culture media.
PBMC were obtained by Ficoll-Hypaque (Biotest AG) density gradient centrifugation. PBMC and CD8+-T-cell lines were cultured in RPMI 1640 medium containing 10% fetal bovine serum, glutamine, streptomycin, and penicillin supplemented with 10 U/ml recombinant interleukin-2 (Proleukin, Chiron, CA). ELISPOT assays were conducted using RPMI 1640 medium supplemented with 5% human AB serum (Sigma-Aldrich, Steinheim, Germany).
Peptides.
Synthetic HIV-1 peptides (Table 1) were synthesized as C-terminal carboxamides by EMC Microcollections GmbH (Tübingen, Germany). Peptides used to confirm the predicted epitopes were synthesized according to the sequence from the reference virus HXB2 (GenBank accession number K03455) (32) and according to sequences from the patients' viral strains. Peptides with amino acid substitutions associated with specific HLA class I alleles in Fisher's exact tests are listed in Table 1. Lyophilized peptides were reconstituted at 2 mg/ml in sterile distilled water with 10% dimethyl sulfoxide and 1 mmol/liter dithiothreitol (Sigma-Aldrich).
TABLE 1.
Peptides used for ELISPOT and restriction analyses
| Peptide name | Amino acid sequencea | Position in proteinb | Corresponding restricting HLA allele(s)c |
|---|---|---|---|
| EW9 | EEMNLPGRW | 34-42 | B44, B40, B18 |
| EW9 M/I | EEINLPGRW | 34-42 | |
| EW9 N/D | EEMDLPGRW | 34-42 | |
| EW9 E/D | EDMNLPGRW | 34-42 | |
| EW9 DI | EDINLPGRW | 34-42 | |
| EW9 R/K | EEMNLPGKW | 34-42 | |
| EW9 P/S | EEMNLSGRW | 34-42 | |
| KI10 | KMIGGIGGFI | 45-54 | A2, B62 |
| KI10-I | KIIGGIGGFI | 45-54 | |
| KI10-V | KIIGGIGGFV | 45-54 | |
| KF9 | KMIGGIGGF | 45-53 | B62 |
| IG10 | IEICGHKAIG | 64-73 | B44, B40, B18 |
| IG10-V | IEICGHKVIG | 64-73 | |
| IK9 | ILIEICGHK | 62-70 | A3 |
| IR9 | ILIEICGHR | 62-70 | |
| LL10 | LDTGADDTVL | 24-33 | A2 |
| LL11 | LLDTGADDTVL | 23-33 | |
| LF11-L | LLDTGADDTVF | 23-33 | |
| LF11-I | LIDTGADDTVF | 23-33 | |
| KL9 | KVRQYDQIL | 55-63 | A2, Cw6 |
| KL9 I/V | KVRQYDQVL | 55-63 | |
| QG10 | QRPLVTVKIG | 7-16 | B51 |
| QG10-I | QRPIVTVKIG | 7-16 | |
| QG10-R | QRPLVTVRIG | 7-16 | |
| TF9 | TQIGCTLNF | 91-99 | B62, Cw3 |
| TF9-L | TQLGCTLNF | 91-99 | |
| VI11 | VRQYDQIPIEI | 56-66 | B13 |
| VI11-V | VRQYDQVPIEI | 56-66 | |
| RI10 | RQYDQIPIEI | 57-66 | |
| QI9 | QYDQIPIEI | 58-66 | |
| VE10 | VRQYDQIPIE | 56-65 | |
| YC9 | YDQIPIEIC | 59-67 | |
| WI9 | WKPKMIGGI | 42-50 | Cw3 |
| LI10 | LPGRWKPKMI | 38-47 | Cw3 |
Substitutions are indicated in bold italics.
Numbers represent amino acid positions.
HLA alleles found to be restricting alleles after statistical and biological evaluations are listed. The B44 supertype allele B40 was tested due to the similarity of B44 and B40 binding motifs. Samples from additional randomly chosen patients with various HLA alleles were tested with each peptide. In comparison to the HXB2 reference sequence, the sequences of the peptides EW9 and VI11 and those of the corresponding variant peptides include the S37N or L63P substitution, at least one of which was detected in samples from most of the patients.
Gamma interferon (IFN-γ) ELISPOT assays.
Specific CD8+-T-cell lines were generated by peptide stimulation of PBMC, and ELISPOT analyses were performed as described previously (38). To control for cross presentation, on average samples from 25 randomly chosen HIV-1-positive HLA-mismatched patients were tested for the ability to recognize the epitopes in peptide stimulation assays.
Genotypic analysis.
Viral RNA was isolated from plasma by using the QIAamp viral RNA kit (QIAGEN, Hilden, Germany). Reverse transcription and amplification of the 1.5-kb pol fragment (encoding HIV-1 full-length PR and amino acids 1 to 300 of the reverse transcriptase) were performed as reported previously (44). The resulting PCR products were directly sequenced on an ABI 3100 automated DNA sequencer with DyeDeoxy chain terminators. Sequencing was performed by using either the ViroSeq HIV-1 genotyping kit from Applied Biosystems or the method described by Schmidt et al. (37). To determine mutations and polymorphisms in the patients' samples, strain HXB2 was used as reference virus. To ensure that HXB2 was an adequate reference strain for our analysis, a consensus sequence for the patients' viruses included in the study was determined. This consensus sequence differed from the HXB2 sequence at only three amino acid positions (3, 37, and 63). All mutations, including those present as mixtures in virus samples from single patients, were included in the analyses. The estimated detection limit for minority species was 25%.
Statistics and bioinformatics.
For statistical evaluations, substitutions were defined as amino acid exchanges in comparison to the amino acid sequence from HXB2. Data were analyzed using SPSS version 14 (SPSS, Chicago, IL). Statistical analyses were limited to HLA alleles and mutations and polymorphisms which occurred with a frequency of at least 5% in the cohort, leaving 38 HLA types and 41 amino acid positions to examine. For the distributions of HLA alleles and mutations and polymorphisms, see Table S1 in the supplemental material. Correlations between polymorphisms and mutations in HIV-1 and HLA alleles were evaluated by Fisher's exact test (two-tailed); data were calculated for 1,558 tests, and no correction for multiple testing was applied. In all analyses, P values of <0.05 were considered relevant. Given that we intended to verify the observed associations between HLA class I alleles and amino acid substitutions in biological assays, a rather high type I error was accepted in favor of a low type II error. Additionally, we report odds ratios (OR) as a measure of the effect size. Odds ratios were calculated using the website http://medweb.uni-muenster.de/institute/imib/lehre/skripte/biomathe/bio/vierf.html.
Analyses were performed for broad and split HLA alleles (information concerning the definition of broad and split alleles can be found at the website of the Anthony Nolan Research Institute, http://www.anthonynolan.org.uk/HIG/lists/broad.html) but, due to feasibility, not for genotypic subtypes. In a first analysis, we tested for identity or difference in comparison to HXB2. If more than one mutation was detected at a specific amino acid position, more detailed analyses, investigating the various occurring substitutions separately, were performed. To control for influences of PI therapy, we analyzed potential associations between the usage of any specific PIs ever taken by the patients and the occurrence of polymorphisms and mutations or the presence of HLA alleles (Fisher's exact test, two-tailed). Additionally, Fisher's exact tests (two-tailed) have been conducted to exclude the possibility that correlations between HLA alleles and amino acid substitutions were driven solely by linkage disequilibrium between HLA alleles. HIV-1 subtype-specific polymorphisms were excluded from the analysis in order to eliminate false-positive correlations. Following the statistical evaluation, potential epitopes were predicted by the SYFPEITHI prediction system (http://www.syfpeithi.de) (31), the epitope location finder (http://hiv-web.lanl.gov/content/hiv-db/ELF/epitope_analyzer.html) (19), or sequence comparison with known peptide binding motifs. Proteasomal cleavage of proteins was predicted by using the major histocompatibility complex class I antigenic peptide processing prediction tool (http://www.mpiib-berlin.mpg.de/MAPPP/index.html) (13). The consensus sequence for HIV-1 in our cohort was calculated by using the Wisconsin package from the Genetics Computer Group (7) and included in the phylogenetic tree (Fig. S1 in the supplemental material).
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank. The accession numbers are EF158477 to EF158570.
RESULTS
Associations between HLA class I alleles and sequence variation in HIV-1 protease.
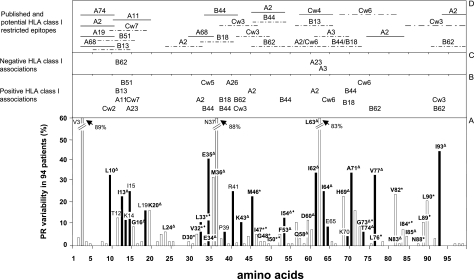
In order to determine the influence of HLA class I genes on the frequencies and patterns of mutations emerging upon antiretroviral therapy, HIV-1 PR sequence data from 94 HLA-typed patients were analyzed. By applying the criteria of the International AIDS Society Drug Resistance Mutation Group (15), no major drug mutations in viruses from the 17 PI-naïve patients were found by genotypic resistance testing. Common polymorphisms were detected in all pretreatment samples. In samples from patients receiving treatment, the following major drug resistance mutations were found: D30N (2.2% of the samples), L33I/F (10.6%), M46I/L (22.3%), I47V (1%), G48V (2.2%), V82A/F/L/T (23.4%), I84V (7.4%), and L90M (20.2%). Additionally, minor drug resistance mutations and drug-associated polymorphisms (L10I/F, I13V, G16E, K20R, L24I, V32I, E34Q, E35D, M36I, K43T, F53L, I54M/V, D60E, I62V, I64L/M/V, L63P, H69K, A71V/T, G73S/T, T74P, V77I, N83D, I85V, and I93L) were detected in patient samples with frequencies of 1 to 89% (see Fig. 1). Despite the risk of false-positive correlations due to multiple testing, all statistical analyses to discover associations between HLA class I alleles and mutations and polymorphisms emerging in HIV-1 PR over the course of infection were performed using Fisher's exact tests. Instead of applying a mathematical correction like Bonferroni's correction, which has been recommended for biomathematical analyses of large data sets to enhance the specificity and to reduce the probability of false-positive associations, we verified observed correlations in biological assays. All correlations between mutations and HLA class I alleles with P values of <0.05 were considered to be relevant candidates for further analysis. The mutations and the correlating HLA alleles as well as known and predicted epitopes were plotted on a map of HIV-1 PR (Fig. 1). Thirty associations with a P value of <0.05 were found between patients' HLA alleles and mutational patterns. According (to HIV-1 subtype analysis performed with the genotyping tool from NCBI (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi), three correlations turned out to be based on HIV-1 subtype-specific polymorphisms. HIV-1 subtypes C and D as well as CRF01_AE, CRF03_AB, and CRF15_01B differ in several amino acids (e.g., those at positions 19, 36, and 41) from HXB2 (32), which was used as a reference strain in this study. Accordingly, the correlations between B17 and M36, B17 and R41, and L19 and A23 (Table 2) were presumably a consequence of genetic lineage, not immune response escape. Hence, these correlations were eliminated from additional analyses. Additionally, the correlation between Cw5 and substitutions at position 35 is supposedly due to the known linkage disequilibrium between B44 and Cw5 (17, 29). Indeed, all Cw5-positive subjects (n = 9) in our cohort also were positive for HLA-B44. This left 26 associations between HLA alleles and mutations at 19 different amino acid positions for further investigation. Of these 26 correlations, 2 concerned major and 16 concerned minor drug resistance mutations, while 8 associations were observed between HLA alleles and sequence polymorphisms (Table 2). Epidemiological and phylogenetic relatedness among viral isolates was excluded based on the neighbor-joining method (35). Only one couple showed highly homologous viral strains due to recent heterosexual transmission (Fig. S1 in the supplemental material).
FIG. 1.
Association of HLA class I alleles and sequence variation in HIV-1 PR. (A) Bars indicate the percentages of subjects with viruses carrying mutations at each individual amino acid position within HIV-1 PR. Mutations and polymorphisms were defined as variations from the sequence of HIV-1 HXB2 PR (32). Amino acid substitutions showing correlations with defined HLA alleles are indicated by black bars. Drug-associated mutations are given in bold; major drug mutations are marked by an asterisk, and minor drug mutations are marked by a triangle. Newly defined drug mutations presumably involved in darunavir resistance (for preliminary data, see Johnson et al. [15]) are marked by a diamond. (B and C) HLA alleles associated positively (B) or negatively (C) with mutations at the indicated amino acid positions. (D) Previously defined CTL epitopes (19) are indicated by solid lines; dashed lines indicate predicted epitopes.
TABLE 2.
Statistically relevant correlations between HLA class I alleles and amino acid substitutions in the PR and defined epitopes
| Position of mutation | HLA type | P value | OR | nd | Epitope sequencee
|
Position of epitopef | ELISPOT reultsg | |
|---|---|---|---|---|---|---|---|---|
| Known | Newly defined | |||||||
| Major mutations | ||||||||
| L33a,b,c | A2 | 0.005 | 12.6 | 9 | LLDTGADDTVL | 23-33 | 29/76 | |
| M46a,b | A2 | 0.049 | 2.9 | 14 | KMIGGIGGFI | 45-54 | 15/42 | |
| Minor mutations | ||||||||
| L10a,b | Cw2 | 0.032 | 4.0 | 9 | ||||
| I13a | A11 | 0.012 | 5.6 | 6 | VTIKIGGQLK | 11-20 | ||
| I13a | B62 | 0.018 | 0.0082 | 0 | ||||
| I13a | B13 | 0.020 | 8.6 | 4 | ||||
| E35a | B44 | 1.2 × 109 | 34.4 | 23 | EEMNLPGRW | KMIGGIGGF | 34-42 | 34/36 |
| K43a | B62 | 0.012 | 6.6 | 5 | WKPKMIGGI | 45-53 | 19/24 | |
| K43a | Cw3 | 0.000073 | 21.2 | 8 | LPGRWKPKMI | 42-50 | 6/12 | |
| I54a,b,c | B44 | 0.017 | 4.6 | 7 | 38-47 | 4/15 | ||
| I62b | A2 | 0.027 | 3.0 | 20 | KVRQYDQIL | 55-63 | 25/76 | |
| I62b | A23 | 0.028 | 0.009 | 0 | ||||
| I64b | A3 | 0.030 | 0.2 | 2 | ILIEICGHK | 62-70 | 17/33 | |
| A71b | B44 | 0.013 | 3.5 | 14 | IEICGHKAIG | 64-73 | 12/23 | |
| T74a | Cw6 | 0.028 | 8.6 | 3 | ||||
| V77 | B62 | 0.037 | 3.5 | 9 | ||||
| I93b | B62 | 2.0 × 106 | 31.8 | 15 | TQIGCTLNF | 91-99 | 6/9 | |
| I93b | Cw3 | 0.00038 | 8.9 | 17 | TQIGCTLNF | 91-99 | 5/11 | |
| Polymorphisms | ||||||||
| K14 | B51 | 0.001 | 13.0 | 6 | QRPLVTVKIG | 7-16 | 17/20 | |
| I15 | A23 | 0.012 | 7.5 | 6 | ||||
| I15 | Cw7 | 0.041 | 0.32 | 10 | ||||
| P39 | B44 | 0.048 | 6.1 | 4 | EEMNLPGRW | 34-42 | 34/36 | |
| P39 | B18 | 0.046 | 10.6 | 2 | EEMNLPGRW | 34-42 | 3/5 | |
| R41 | A26 | 0.032 | 9.1 | 3 | ||||
| E65 | Cw6 | 0.045 | 6.3 | 2 | KVRQYDQIL | 55-63 | 11/26 | |
| K70 | B18 | 0.019 | 21.5 | 2 | IEICGHKAIG | 64-73 | 4/10 | |
| Mutations showing associations that were lost after correction for HIV-1 subtypes | ||||||||
| L19 | A23 | 0.034 | 5.4 | 4 | ||||
| M36a,b | B17 | 0.030 | 10.2 | 4 | ||||
| R41 | B17 | 0.012 | 14.7 | 4 | ||||
The mutation is included in the tipranavir score (more than four tipranavir-associated mutations lead to ≥3-fold resistance and more than seven tipranavir-associated mutations lead to ≥10-fold resistance compared to that of the wild-type virus) (3, 15).
The mutation is included in the atazanavir score (three or more atazanavir-associated mutations lead to a reduced virological response) (15).
The mutation contributes to darunavir resistance (for preliminary data, see Johnson et al. [15]).
n, number of patients with the indicated HLA type harboring the viral strain with the indicated mutation.
Residues affected by the substitutions are shown in bold italics. In comparison to the HXB2 reference sequence, the sequences of the peptides EW9 and VI11 and those of the corresponding variant peptides include the S37N or L63P substitution, at least one of which was detected in samples from most of the patients.
Numbers represent amino acid positions.
Recognition of peptides by CD8+ T cells from patients with the appropriate HLA allele. Values indicate the number of cell samples responding to the wild-type or a respective mutant peptide versus the number of cell samples tested. To exclude cross presentation, on average samples from 25 randomly chosen HIV-1-positive patients with various HLA alleles were additionally tested. No cross presentation was observed.
Of the remaining 26 correlations, 22 indicated a positive linkage between the appearance of a mutation and a given HLA type, while only 4 correlations denoted a negative association. These negative associations were found between amino acid substitutions at the following positions and the following HLA types: position 13 and B62 (P = 0.018; OR = 0.0027), position 15 and Cw7 (P = 0.019; OR = 0.27), position 62 and A23 (P = 0.028; OR = 0.0017), and position 64 and A3 (P = 0.030; OR = 0.2) (Table 2). Detailed analyses of these mutations revealed that B62 is negatively associated with the development of I13V and Cw7 with the development of I15V while A23 is negatively associated with the development of I62V and A3 with the development of I64V. This finding indicates that these substitutions rarely occur in B62-, A23-, A3-, or Cw7-positive patients.
HLA correlations with mutations in previously described epitopes and confirmation of published report of correlations.
Our approach of using Fisher's exact test to screen the PR for new epitopes recognized by CTL was supported by the detection of correlations between amino acid substitutions in previously described epitopes and the appropriate restricting HLA allele. The amino acid mutations E35D and P39S are located in the previously described B44-restricted epitope EEMSLPGRW (EW9; PR amino acids 34 to 42) (33). Whereas the correlation between B44 and the E35D mutation was highly significant (P = 1.2 × 109), B44 correlated only weakly with P39S (P = 0.048) (Table 2). Another association was identified between M46L/I (P = 0.049), a major drug mutation (15) located in the previously described A2-restricted epitope KMIGGIGGFI (KI10; PR amino acids 45 to 54) (30), and HLA-A2. HLA-A11 was associated with I13V (P = 0.012), a minor tipranavir resistance mutation (15) located in the previously described A11-restricted epitope VTIKIGGQLK (amino acids 11 to 20) (24) (Table 2). Furthermore, we confirmed the recently published report of correlations between K14R and HLA-B5 and K43R and HLA-B15 as well as V77I and HLA-B15 (14). In addition, we were able to map these correlations to the split alleles B51(5) (P = 0.001) and B62(15) (P = 0.012 and 0.037), respectively.
Variations in the B44-restricted EW9 epitope can influence recognition by CD8+ T cells.
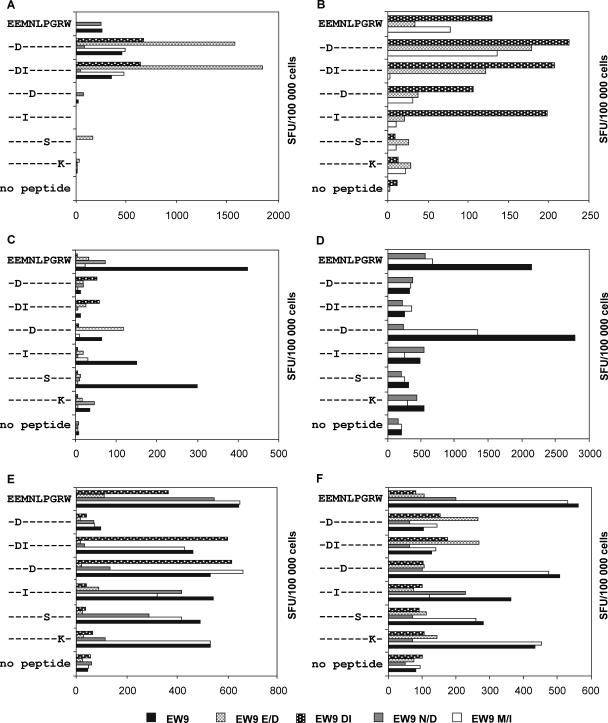
The HLA-B44-restricted epitope EEMSLPGRW (EW9) (33) is a variable epitope spanning amino acids 34 to 42 in the PR for which the Los Alamos Database reports various amino acid substitutions (20). In terms of drug resistance, these mutations are either minor mutations or (resistance-associated) polymorphisms. PBMC from HIV-1-infected, HLA-B44-positive patients were stimulated separately with five peptides corresponding to different EW9 variants (EW9, EW9 E/D, EW9 DI, EW9 M/I, and EW9 N/D) (Table 1). Outgrowing cell lines, displaying a strong CD8+-T-cell response against at least one of the EW9 peptides, were analyzed by ELISPOT assay for cross-reactivity by using a panel of peptides comprising the most common polymorphisms and mutations within the EW9 epitope. ELISPOT analysis revealed that 94% of the tested cell samples from HLA-B44-positive patients displayed CD8+-T-cell responses against the wild-type peptide or against at least one mutant peptide. The E35D mutation induced a strong decrease in T-cell recognition in cell samples from 56% of the analyzed patients; however, there were patient samples that could mount specific CD8+-T-cell responses to peptides containing the E35D mutation (Fig. 2A; Table 3). Samples from only a few patients showed a narrow CD8+-T-cell response to EW9 and EW9 variants, whereas samples from the majority of the patients showed broad responses, recognizing at least three of the EW9 peptides (Fig. 2). According to the various recognition patterns, we postulate that most patients harbored various CD8+-T-cell clones with different T-cell receptor (TCR) specificities. Representative graphs are shown in Fig. 2.
FIG. 2.
Cross-reactivity of CD8+ T cells recognizing the HLA-B44-restricted CTL epitope EW9 (amino acids 34 to 42). PBMC from HLA-B44-positive patients were stimulated with EEMNLPGRW (EW9; wild-type) and the variant peptides EEINLPGRW (EW9 M/I), EEMDLPGRW (EW9 N/D), EDINLPGRW (EW9 DI), and EDMNLPGRW (EW9 E/D; mutations are indicated in bold italics) and subsequently tested for reactivity by IFN-γ ELISPOT analysis. After stimulation, outgrowing CD8+-T-cell lines were analyzed for cross-reactivity with the peptides EW9, EW9 E/D, EW9 DI, EW9 M/I, EW9 N/D, EW9 P/S, and EW9 R/K, which carry the most frequent drug resistance mutations and/or polymorphisms occurring in the epitope. Peptides tested for cross-reactivity are given on the left side of each graph. Dashes indicate consensus with the wild-type peptide sequence; amino acid substitutions are given in capital letters. The numbers of spot-forming units (SFU) triggered by wild-type EW9, corresponding variant peptides, and the no-peptide control in CD8+ T cells are shown by differently shaded bars.
TABLE 3.
Influence of PR mutations on T-cell recognition
| Wild-type sequencea | Mutant peptidea | Position of mutationb | HLA typec | No. of samples with impaired CD8+-T-cell activity/no. of samples testedd | Median decrease in recognition (%) (range) | No. of samples with improved CD8+-T-cell activity/no of samples testedd | Median increase in recognition (%) (range) |
|---|---|---|---|---|---|---|---|
| QRPLVTVKIG | QRPIVTVKIG | 10 | B51 | 8/8 | 36 (19-87) | 0/8 | |
| QRPLVTVKIG | QRPLVTVRIG | 14 | B51 | 6/8 | 24 (12-42) | 0/8 | |
| LLDTGADDTVF | LIDTGADDTVF | 24 | A2 | 4/12 | 40 (27-69) | 7/12 | 33 (8-65) |
| LLDTGADDTVL | LLDTGADDTVF | 33 | A2 | 2/8 | 23 (12-34) | 5/8 | 26 (16-31) |
| EEMNLPGRW | EDMNLPGRW | 35 | B44 | 13/23 | 86 (51-98) | 7/23 | 87 (21-98) |
| EEMNLPGRW | EEMNLSGRW | 39 | B44 | 21/23 | 83 (30-97) | 2/23 | 17 (10-24) |
| KMIGGIGGFI | KIIGGIGGFI | 43 | B62 | 6/6 | 72 (60-97) | 0/6 | |
| KMIGGIGGFI | KIIGGIGGFI | 46 | A2 | 6/15 | 25 (12-36) | 4/15 | 33 (18-60) |
| KMIGGIGGFI | KMIGGIGGFV | 54 | A2 | 2/15 | 40 (35-45) | 3/15 | 40 (10-48) |
| KVRQYDQIL | KVRQYDQVL | 62 | A2 | 1/3 | 15 | 1/3 | 16 |
| RQYDQIPIEI | RQYDQVPIEI | 62 | B13 | 6/8 | 35 (14-81) | 2/8 | 24 (11-36) |
| KVRQYDQIL | KVRQYDQVL | 62 | Cw6 | 2/4 | 72 (52-91) | 0/4 | |
| ILIEICGHK | ILIEICGHR | 70 | A3 | 12/12 | 64 (15-89) | 0/12 | |
| IEICGHKAIG | IEICGHKVIG | 70 | B18 | 0/4 | 0/4 | ||
| IEICGHKAIG | IEICGHKVIG | 71 | B44 | 3/11 | 37 (26-51) | 7/11 | 39 (27-56) |
| TQIGCTLNF | TQLGCTLNF | 93 | B62 | 3/6 | 46 (38-62) | 3/6 | 30 (16-42) |
| TQIGCTLNF | TQLGCTLNF | 93 | B62/Cw3 | 6/13 | 57 (15-75) | 5/13 | 60 (10-80) |
Residues affected by the substitutions are shown in bold italics. In comparison to the HXB2 reference sequence, the sequences of peptides EW9 and VI11 and those of the corresponding variant peptides include the S37N or L63P substitution, at least one of which was detected in samples from most of the patients.
The position of the mutated amino acid relative to the HXB2 reference sequence is indicated.
Restricting HLA allele.
Values indicate the proportions of patients with samples showing impaired or improved CD8+-T-cell recognition of peptides with mutations (mutant peptides) and without mutations (wild-type peptides) at the indicated positions. For the remaining samples, the recognition of wild-type and mutant peptides by CD8+ T cells was similar.
Interestingly, we also found that cells from patients with HLA-B18 and HLA-B40, both belonging to the B44 supertype (39, 40), recognized several EW9 variants (data not shown). A correlation between B18 and P39S was detected in the statistical analyses (Table 2). Due to the low frequency of HLA-B40 alleles (n = 2) in the cohort used for the statistical evaluation, HLA-B40 was excluded from the analysis.
Variations in the A2-restricted KI10 epitope can influence recognition by CD8+ T cells.
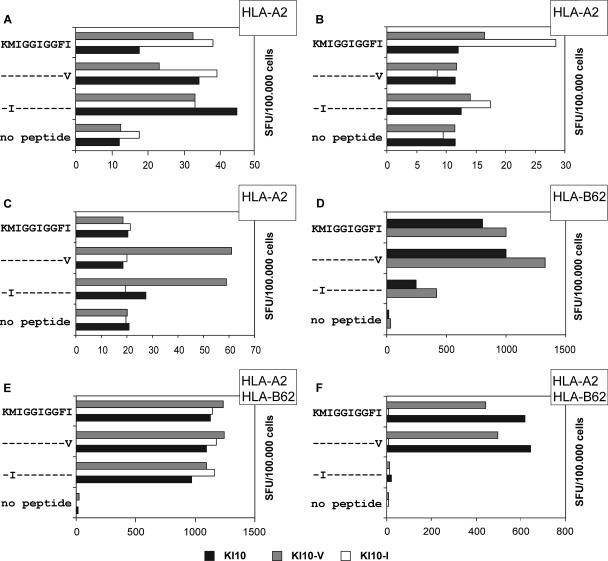
In contrast to EW9, the HLA-A2-restricted epitope KMIGGIGGFI (KI10; PR amino acids 45 to 54) (30), containing the drug resistance mutations M46I/L and I54V, is rather conserved. Cells from roughly 35% of the A2-positive patients showed CD8+-T-cell responses after stimulation with the KI10 variant peptides KIIGGIGGFI and KMIGGIGGFV (mutations are shown in bold). Representative graphs are shown in Fig. 3A to C. Cells from most patients displayed CD8+-T-cell responses against the wild-type peptide and the two variants. Some experiments provided evidence that these responses might be due to the generation of CD8+-T-cell clones with different recognition specificities in samples from individual patients (Fig. 3A). However, in samples from some A2-positive patients CD8+-T-cell recognition was abolished by the mutations M46I and I54V (Fig. 3B) whereas in samples from other A2-positive patients a CD8+-T-cell response was induced only by the mutant peptides and not by wild-type KI10 (Fig. 3C).
FIG. 3.
Cross-reactivity of CD8+ T cells recognizing the HLA-A2- and HLA-B62-restricted epitope KI10 (amino acids 45 to 54). PBMC from HLA-A2-positive patients (A to C), an HLA-B62-positive patient (D), and patients carrying both HLA-A2 and B62 (E and F) were stimulated with the peptide KMIGGIGGFI (wild type) and the variant peptides KIIGGIGGFI (KI10-I) and KMIGGIGGFV (KI10-V) comprising the drug mutations M46I and I54V, respectively. Outgrowing cell lines were evaluated for cross recognition of these peptides by IFN-γ ELISPOT analysis. Peptide sequences are given on the left side of each graph. Dashes indicate consensus with the HXB2 sequence; amino acid substitutions are given in capital letters. The numbers of spot-forming units (SFU) triggered by KI10, the corresponding variant peptides, and the negative control (no peptide) are shown by white, gray, and black bars. Restricting HLA alleles are indicated in the upper right corner of each graph.
Definition of new CD8+-T-cell epitopes in HIV-1 protease indicated by low and relevant P values.
An important goal of our study was the definition of new epitopes in HIV-1 PR. As already mentioned, we refrained from using mathematical corrections such as Bonferroni's correction due to the large type II error which is associated with such a strict correction. After using Bonferroni's correction in our analysis, only the associations between B44 and E35D and B62 and I93L would remain significant (P ≤ 0.00003) (Table 2). Eighteen potential epitopes were determined on the basis of observed correlations (P < 0.05) between HLA alleles and amino acid substitutions and the application of epitope prediction tools (http://www.syfpeithi.de [31] and http://hiv-web.lanl.gov/content/hiv-db/ELF/epitope_analyzer.html [19]) or sequence comparison with known CTL epitopes (Fig. 1). Ten epitopes, which were further investigated in biological assays, were chosen according to the following criteria: (i) stringent P values of <0.005, (ii) P values of <0.05, (iii) indication of proteasomal cleavage sites, and (iv) clustered correlations. Criteria i and ii were used in order to test if correlations with more stringent P values were more likely to be biologically verified than associations with higher P values. Criterion iii was supposed to provide evidence that selective pressure exerted by CD8+ T cells also acts on proteasomal cleavage sites in the PR and consequently plays an important role in peptide presentation (28), even though the mutated amino acid is located outside of the actual epitope. Criterion iv was adopted to test the hypothesis generated by Moore et al. (28) that clusters of HLA-associated polymorphisms are an indicator for HLA-restricted epitopes. In this context, even correlations with P values between 0.05 and 0.1 were analyzed. ELISPOT analyses showed that all peptides generated on the basis of the above-mentioned criteria were recognized by peptide-stimulated cells from patients with the appropriate HLA types (Table 2). To control for specificity and for cross presentation, all peptides were tested with samples from randomly chosen patients with mismatched HLA types.
The cross presentation of CD8+-T-cell epitopes by more than one HLA allele was observed only for HLA alleles that showed relevant correlations in the statistical analysis (Table 2). The sole exception was the IG10 epitope (IEICGHKAIG; PR amino acids 64 to 73), which was recognized not only by cells of B44- and B18-positive patients but also by peptide-stimulated cells of patients with HLA-B40 alleles belonging to the B44 supertype. A correlation between B40 and amino acid substitutions in this region was not found because B40 was excluded from the statistical evaluation due to its low frequency in the cohort (n = 2). In total, 3 out of the 10 newly defined epitopes could be presented by at least two different HLA alleles.
Mutations at proteasomal cleavage sites indicate CTL epitopes.
Earlier studies provided evidence that mutations outside of CTL epitopes can influence proteasomal cleavage and consequently can play an important role in peptide presentation (6, 26). The hypothesis that correlations between mutations at proteasomal cleavage sites and HLA alleles could be an indicator for HLA-restricted epitopes was verified using the peptides KVRQYDQIL (KL9; PR amino acids 55 to 63) and KMIGGIGGFI (KI10; PR amino acids 45 to 54). Due to associations between HLA-B62 and mutations at position 43 and between HLA-Cw6 and mutations at position 65, these peptides were tested for recognition by cells from B62- and Cw6-positive patients, respectively. Both peptides induced CD8+-T-cell responses in samples from patients with the respective HLA type (Fig. 3D; data not shown). HLA restriction was determined using peptide-pulsed target cells matched at only one major histocompatibility complex class I allele in ELISPOT assays (data not shown). KI10 and KMIGGIGGFV showed good recognition by B62-restricted CTL, whereas the drug resistance mutation M46I in the peptide KIIGGIGGFI (the mutation is indicated in bold) strongly impaired CTL recognition (Fig. 3D). Additional analyses showed that KMIGGIGGF (KF9; PR amino acids 45 to 53) is the optimal B62-restricted epitope.
Identification of a B13-restricted CTL epitope by analysis of clustered HLA correlations.
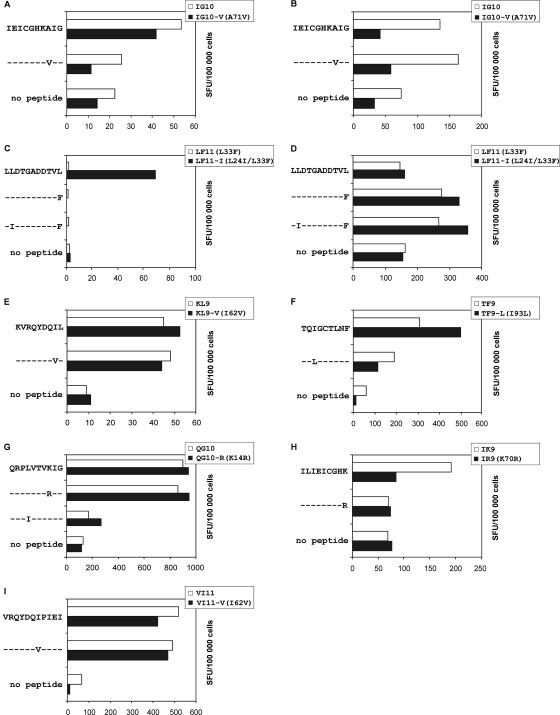
In an earlier study, Moore et al. (28) generated the hypothesis that clusters of associations between HLA and polymorphisms are an indicator for HLA-restricted epitopes. In our analysis, only one cluster of correlations with P values of <0.05 was detected. HLA-B44 was associated with amino acid substitutions at positions 35 (P = 1.2 × 109) and 39 (P = 0.048). Additional clusters were found if associations with higher P values (<0.1) were included in the analysis. For the epitopes LL11 (LLDTGADDTVL; PR amino acids 23 to 33), KI10 (KMIGGIGGFI; PR amino acids 45 to 54) and ILIEICGHK (IK9; PR amino acids 62 to 70), one correlation with a P value of <0.05 and an additional correlation with a P value of <0.1 were detected. Even though HLA-A3 correlated only weakly with K70R (P = 0.068), peptides including the mutation were tested in ELISPOT analyses. Given that mutations at position 70 affect the C-terminal anchor site of the epitope, the K70R substitution in the ILIEICGHR epitope (IR9; PR amino acids 62 to 70 [the mutation is shown in bold]) strongly reduced recognition in peptide-stimulated PBMC from HLA-A3-positive patients (Fig. 4H; Table 3). In order to conclusively prove that clustered correlations are strong indicators for HLA-restricted epitopes, an additional peptide, VRQYDQIPIEI (VI11; PR amino acids 56 to 66), was analyzed. The peptide and its variant VI11-V were generated based on two associations between HLA-B13 and the amino acid substitutions I62V and E65D, both showing relatively high P values (0.09 and 0.08). Despite the high P values, both peptides were recognized in ELISPOT analyses conducted with cells from B13-positive patients (Fig. 4I). Using a panel of truncations of the VI11 peptide and stimulated PBMC from HLA-B13-positive patients, we could define RQYDQIPIEI (RI10; PR amino acids 57 to 66) as the minimal epitope (data not shown). Additional restriction analyses proved that B13 is in fact the restricting HLA allele for RI10, VI11, and the mutant peptide VI11-V (data not shown). A summary of statistical and experimental results is given in Tables 2 and 3.
FIG. 4.
Influence of drug-induced mutations and polymorphisms on recognition by CD8+ T cells. PBMC from HLA-matched HIV-1-positive patients were stimulated with the indicated wild-type or mutant peptides and tested for recognition of the peptides by IFN-γ ELISPOT analyses. After peptide stimulation, outgrowing CD8+-T-cell lines were analyzed for cross recognition by using the peptides indicated on the left side of each graph. Dashes indicate consensus with the wild-type peptide sequence; amino acid substitutions are given in capital letters. The numbers of spot-forming units (SFU) triggered by wild-type peptides, corresponding variant peptides, and the negative (no-peptide) control are shown by white and black bars. The position(s) of the mutated amino acid(s) is indicated in parentheses in the upper right corner of each graph.
Influence of mutations in newly defined epitopes on recognition by CD8+ T cells.
To assess the influence of HLA-linked mutations and of additional resistance-associated mutations on T-cell recognition, PBMC from patients with the appropriate HLA alleles were stimulated with wild-type and variant peptides and outgrowing cells were tested for cross recognition in ELISPOT assays (Fig. 4 and Table 3). In principle, all analyzed mutations could impair recognition by CD8+ T cells. However, cells from subgroups of patients could recognize both the wild-type and the mutant peptides. In some cases, the mutant peptides showed even better recognition than the wild-type peptides. These differences regarding the recognition of the mutants indicate the usage of different TCRs by CD8+ T cells targeting these epitopes. Representative graphs are shown in Fig. 4. Table 3 gives a summary of the results obtained from ELISPOT assays.
DISCUSSION
Evasion of the immune response through CTL escape is an important factor in HIV-1 pathogenesis (11). Over the past years, some studies have provided evidence that selection pressure exerted by CTL can interfere with the development of drug resistance mutations (4, 16, 36, 38). Drug-targeted enzymes like the viral PR are exposed to both pharmacological selection pressure and immune response selection exerted by CD8+ T cells. This dual pressure may have important clinical implications, especially for the understanding of drug resistance pathways or the design of therapeutic vaccines. To determine the influence of HLA-specific CD8+-T-cell responses on the development of drug mutations or drug-associated polymorphisms in the PR, we analyzed PR sequences from 94 HLA class I-typed HIV-1-positive individuals. Twenty-six associations between patients' HLA class I alleles and mutations occurring in the autologous HIV-1 PR genes were detected. Five of the 26 correlations between patients' HLA alleles and the mutations, namely, the associations between K14R and B5, L19I and A9, E35D and B44(12), K43R and B15, and V77I and B15, were also reported in a recent study (14). Adding to the work presented in that study, we were able to map three of these associations to HLA split antigens, K14R to B51 (5) and K43R as well as V77I to B62 (15). Discrepancies between our findings and the findings from John et al. (14) may result from differences in the distributions of HLA class I alleles in ethnically diverse study populations, from the frequencies of HIV-1 subtypes in different populations, from differences in the use of PIs, and from the numbers of patients included in the studies. Interestingly, six of the correlations in our analysis were related to HLA-C alleles which were not considered in the study by John et al. (14). This demonstrates that HLA-C-linked immune response selection clearly contributes to the sequence variation in HIV-1.
For statistical analyses of many variables in large data sets, the application of statistical corrections such as Bonferroni's correction has been recommended. With the application of Bonferroni's correction in our study, P values of ≥0.00003 would be considered not statistically significant. Thus, only two of our correlations would remain significant. Applying such very strict limits, one has to take into account that the gain of specificity is achieved at the cost of sensitivity, resulting in the failure to detect biologically relevant correlations. To protect against a high type II error, we applied a less conservative limit (P < 0.05) and verified the associations in biological assays. Our approach of using Fisher's exact test to define new PR epitopes was confirmed by the detection of correlations between mutations in already described epitopes and the appropriate restricting HLA alleles. Correlations between resistance mutations and polymorphisms and the usage of specific PIs were not detected. This might be due to the fact that mutations clearly associated with resistance to only one or two specific PIs (e.g., D30N and G48V) were rarely found in our cohort whereas mutations which are induced by several PIs and which contribute to broad cross-resistance were frequently found. This study demonstrates that CD8+-T-cell epitopes can be predicted on the basis of associations between HLA alleles and amino acid substitutions which are detected in univariate statistical analyses. Additionally, we show that associations with P values of <0.05 and clustered associations with even higher P values (0.05 < P < 0.1) still can indicate biologically relevant selection pressure by CD8+ T cells. We could not see any differences in the patterns of recognition of epitopes predicted on the basis of correlations with P values of <0.005 and epitopes predicted on the basis of correlations with P values of <0.05. Instead, recognition frequencies and the magnitude of recognition were dependent on the restricting HLA allele and on the sequence variation within the epitope. Recent studies (2, 18) showed a dominant role for HLA-B in the HIV-1-specific CTL response in comparison to HLA-A and HLA-C. This finding was confirmed in our study by the detection of a stronger impact of HLA-B than of HLA-A and HLA-C on the protease sequence. Nevertheless, our results show that HLA-A and HLA-C also play an important role in HLA-linked immune response selection. Furthermore, we could confirm that selection pressure is also exerted on proteasomal cleavage sites and that correlations between HLA alleles and amino acids at proteasomal cleavage sites are good indicators for HLA-restricted epitopes (6, 26).
We demonstrated that peptides comprising the amino acid substitutions for which correlations were detected led to impaired peptide recognition by cells from subgroups of patients, indicating that these mutations can indeed act as escape mutations. However, samples from a significant number of patients were able to generate CD8+-T-cell responses against peptides containing these mutations. An analysis of recognition patterns showed that cells from several patients mounted oligoclonal CD8+-T-cell responses against certain epitopes, targeting a variety of viral variants. This result indicates that at least in some HIV-1-infected individuals the immune system can react to mutational escape by HIV-1 through the recruitment of CD8+ T cells with new TCR specificities. Longitudinal studies will have to deliver more-specific insights into the dynamics between drug-induced and HLA-induced selection pressure.
For the further assessment of the influence of CD8+ T cells on the development of resistance mutations, we focused on the B44-restricted epitope EW9 (EEMNLPGRW; PR amino acids 34 to 42) (33) and the A2-restricted epitope KI10 (KMIGGIGGFI; PR amino acids 45 to 54) (30). In our patient cohort, we observed a number of mutations in the EW9 epitope, indicating strong selection pressure by CD8+ T cells. Indeed, cells from most B44-positive patients showed CD8+-T-cell responses to this epitope, confirming that EW9 is highly immunogenic. The minor drug-associated mutation E35D is selected in the majority of B44-positive patients. According to other reports (33, 40), the glutamate residue at position 35 is likely to be the N-terminal anchoring amino acid in the B44-restricted peptide. Surprisingly, cells from most patients showed mutually exclusive recognition of peptides comprising either E35E (wild-type) or the E35D variant while those from only a few patients were able to generate different CD8+-T-cell lines recognizing both the E35E and the E35D variants. So far, we could not determine any factors, such as stage of disease, CD4 cell count, viral load, or therapy regimen, that could explain the distinct recognition patterns of the B44-positive patients. An analysis of B44 subtypes in our patients also failed to demonstrate an association between E35D recognition and certain B44 subtypes. One possible explanation for the different patterns of recognition of EW9 by cells from B44-positive patients might be that both N-terminal glutamates of the epitope can serve as the anchor amino acid and that in some patients a switch between E34 and E35 is possible. Whether B44-restricted CTL responses to EW9 influence disease progression is not clear yet. Results from earlier analyses investigating B44 as a protection or risk factor for HIV-1 infection have been ambiguous (8, 9, 34). A negative influence on drug efficacy has been demonstrated for both HLA-B44 (A. Saitoh, C. Powell, T. Fenton, C. Fletcher, and S. Spector, oral abstract 51d presented at the 12th Conference on Retroviruses and Opportunistic Infections, 2005) and the E35D substitution (1, 25). Our data show that HLA-B44 selects for the E35D mutation and thus establish a connection between these findings.
In contrast to EW9, the KI10 epitope (KMIGGIGGFI; PR amino acids 45 to 54) is a rather conserved epitope, comprising six drug resistance-associated mutations (M46I/L, I47A/V/L, G48V/M, I50L/V, F53L, and I54A/L/M/V). The observation that HLA-A2 favors the development of the resistance mutation M46I is supported by the finding that isoleucine at position 46 abolishes recognition by CD8+ T cells from some A2-positive patients. To explain why the recognition patterns for the M46 and the I46 variants are similar in other HLA-A2-positive patients, we hypothesize that both the methionine at position 46 and the isoleucine at position 47 could serve as N-terminal anchors and that the HLA-A2 molecule displays a preference for methionine. Replacement of the methionine by isoleucine at position 46 could lead to competition between the isoleucine at position 47 and the isoleucine at position 46 for binding to the N-terminal anchor pocket of the HLA-A2-molecule. A shift of anchor amino acids within the peptide could result in a significant conformational change of the TCR recognition site that possibly abolishes CD8+-T-cell recognition but might be compensated for by the generation of CD8+ T cells with other TCR specificities. This hypothesis is supported by the detection of two different T-cell clones with different recognition patterns for peptides including the M46 wild-type and M46I variant in samples from some patients.
Our analyses show that a number of novel epitopes overlap with or are embedded within previously described epitopes or other newly defined epitopes. For example, the Cw4-restricted QYDQIPIEI epitope (41) overlaps with the newly defined HLA-B13-restricted epitope RI10 (RQYDQIPIEI; PR amino acids 57 to 66). As an HLA binding motif for HLA-B13 has not been published yet, it is of interest that the RI10 epitope shows similarity at the putative anchor binding positions to the B13-restricted CTL epitope RQDILDLWI which we recently identified in HIV-1 Nef (12). Presentation of the same peptide by several HLA alleles was found for four peptides. In these cases, all restricting HLA alleles, with the exception of the infrequently occurring HLA-B40 allele, also showed relevant correlations with the respective amino acid substitutions in these epitopes.
In summary, the data presented here strongly suggest that escape from CD8+ T cells influences the emergence of polymorphisms as well as the development of minor and major drug resistance mutations in HIV-1-infected patients. Thus, this study provides an explanation for the findings of earlier studies showing that the development of resistance is not exclusively drug dependent but can also be influenced by host factors (38, 42). The finding that a variety of drug-related escape mutations in PR are immunogenic should be a further stimulus for the development of HIV-1 vaccines that can target drug-resistant HIV-1 strains.
Supplementary Material
Acknowledgments
This study was supported by the The German Competence Network on HIV/AIDS (HIVNET) (T. Harrer, project F2) and the DFG (T. Harrer, SFB 466: project B6).
We thank all patients participating in this study. We have no conflicting financial interests.
Footnotes
Published ahead of print on 3 January 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alexander, C. S., W. Dong, K. Chan, N. Jahnke, M. V. O'Shaughnessy, T. Mo, M. A. Piaseczny, J. S. Montaner, and P. R. Harrigan. 2001. HIV protease and reverse transcriptase variation and therapy outcome in antiretroviral-naive individuals from a large North American cohort. AIDS 15:601-607. [DOI] [PubMed] [Google Scholar]
- 2.Bihl, F., N. Frahm, L. Di Giammarino, J. Sidney, M. John, K. Yusim, T. Woodberry, K. Sango, H. S. Hewitt, L. Henry, C. H. Linde, J. V. Chisholm III, T. M. Zaman, E. Pae, S. Mallal, B. D. Walker, A. Sette, B. T. Korber, D. Heckerman, and C. Brander. 2006. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J. Immunol. 176:4094-4101. [DOI] [PubMed] [Google Scholar]
- 3.Boehringer Ingelheim Pharmaceuticals. 19April2005, posting date. Tipranavir Anti-Viral Drugs Advisory Committee (AVDAC) briefing document. Boehringer Ingelheim Pharmaceuticals, Ingelheim, Germany. http://www.fda.gov/ohrms/dockets/ac/05/briefing/2005-4139b1-02-boehringer.pdf.
- 4.Casazza, J. P., M. R. Betts, B. J. Hill, J. M. Brenchley, D. A. Price, D. C. Douek, and R. A. Koup. 2005. Immunologic pressure within class I-restricted cognate human immunodeficiency virus epitopes during highly active antiretroviral therapy. J. Virol. 79:3653-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Val, M., H. J. Schlicht, T. Ruppert, M. J. Reddehase, and U. H. Koszinowski. 1991. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 66:1145-1153. [DOI] [PubMed] [Google Scholar]
- 7.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabio, G., R. Scorza, A. Lazzarin, M. Marchini, M. Zarantonello, A. D'Arminio, P. Marchisio, A. Plebani, R. Luzzati, and P. Costigliola. 1992. HLA-associated susceptibility to HIV-1 infection. Clin. Exp. Immunol. 87:20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores-Villanueva, P. O., E. J. Yunis, J. C. Delgado, E. Vittinghoff, S. Buchbinder, J. Y. Leung, A. M. Uglialoro, O. P. Clavijo, E. S. Rosenberg, S. A. Kalams, J. D. Braun, S. L. Boswell, B. D. Walker, and A. E. Goldfeld. 17. April 2001. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. USA 98:5140-5145. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallant, J. E., P. Z. Gerondelis, M. A. Wainberg, N. S. Shulman, R. H. Haubrich, M. St. Clair, E. R. Lanier, N. S. Hellmann, and D. D. Richman. 2003. Nucleoside and nucleotide analogue reverse transcriptase inhibitors: a clinical review of antiretroviral resistance. Antivir. Ther. 8:489-506. [PubMed] [Google Scholar]
- 11.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630-640. [DOI] [PubMed] [Google Scholar]
- 12.Harrer, E. G., S. Bergmann, K. Eismann, M. Rittmaier, A. Goldwich, S. M. Muller, B. M. Spriewald, and T. Harrer. 2005. A conserved HLA B13-restricted cytotoxic T lymphocyte epitope in Nef is a dominant epitope in HLA B13-positive HIV-1-infected patients. AIDS 19:734-735. [DOI] [PubMed] [Google Scholar]
- 13.Holzhutter, H. G., C. Frommel, and P. M. Kloetzel. 1999. A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20 S proteasome. J. Mol. Biol. 286:1251-1265. [DOI] [PubMed] [Google Scholar]
- 14.John, M., C. B. Moore, I. R. James, and S. A. Mallal. 2005. Interactive selective pressures of HLA-restricted immune responses and antiretroviral drugs on HIV-1. Antivir. Ther. 10:551-555. [PubMed] [Google Scholar]
- 15.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 16.Karlsson, A. C., S. G. Deeks, J. D. Barbour, B. D. Heiken, S. R. Younger, R. Hoh, M. Lane, M. Sallberg, G. M. Ortiz, J. F. Demarest, T. Liegler, R. M. Grant, J. N. Martin, and D. F. Nixon. 2003. Dual pressure from antiretroviral therapy and cell-mediated immune response on the human immunodeficiency virus type 1 protease gene. J. Virol. 77:6743-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiefel, V., and A. Greinacher. 4 November 2006, posting date. Transfusionsmedizin und Immunhämatologie, p. 101-111. University of Rostock, Rostock, Germany. http://www-tmed.med.uni-rostock.de/tmed.pdf.
- 18.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 19.Korber, B. T. M., C. Brander, B. F. Haynes, R. Koup, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.). 2005. HIV molecular immunology 2005. LA-UR 06-0036. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM.
- 20.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.). 2005. HIV sequence compendium 2005. LA-UR 06-0680. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 21.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, S. Le Gall, G. Luzzi, A. Edwards, C. Brander, A. K. Sewell, S. Moore, J. Mullins, C. Moore, S. Mallal, N. Bhardwaj, K. Yusim, R. Phillips, P. Klenerman, B. Korber, P. Kiepiela, B. Walker, and P. Goulder. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 23.Mason, R. D., M. I. Bowmer, C. M. Howley, M. Gallant, J. C. Myers, and M. D. Grant. 2004. Antiretroviral drug resistance mutations sustain or enhance CTL recognition of common HIV-1 Pol epitopes. J. Immunol. 172:7212-7219. [DOI] [PubMed] [Google Scholar]
- 24.McKinney, D. M., R. Skvoretz, B. D. Livingston, C. C. Wilson, M. Anders, R. W. Chesnut, A. Sette, M. Essex, V. Novitsky, and M. J. Newman. 2004. Recognition of variant HIV-1 epitopes from diverse viral subtypes by vaccine-induced CTL. J. Immunol. 173:1941-1950. [DOI] [PubMed] [Google Scholar]
- 25.Meiselbach, H., A. H. Horn, T. Harrer, and H. Sticht. 2006. Insights into amprenavir resistance in E35D HIV-1 protease mutation from molecular dynamics and binding free-energy calculations. J. Mol. Model. 23:23. [DOI] [PubMed] [Google Scholar]
- 26.Milicic, A., D. A. Price, P. Zimbwa, B. L. Booth, H. L. Brown, P. J. Easterbrook, K. Olsen, N. Robinson, U. Gileadi, A. K. Sewell, V. Cerundolo, and R. E. Phillips. 2005. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J. Immunol. 175:4618-4626. [DOI] [PubMed] [Google Scholar]
- 27.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 28.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 29.Pedron, B., K. Yakouben, D. Adjaoud, A. Auvrignon, J. Landman, V. Guerin, G. Leverger, E. Vilmer, and G. Sterkers. 18. March 2005. Listing of common HLA alleles and haplotypes based on the study of 356 families residing in the Paris, France, area: implications for unrelated hematopoietic stem cell donor selection. Hum. Immunol. 66:721-731. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 30.Propato, A., E. Schiaffella, E. Vicenzi, V. Francavilla, L. Baloni, M. Paroli, L. Finocchi, N. Tanigaki, S. Ghezzi, R. Ferrara, R. Chesnut, B. Livingston, A. Sette, R. Paganelli, F. Aiuti, G. Poli, and V. Barnaba. 2001. Spreading of HIV-specific CD8+ T-cell repertoire in long-term nonprogressors and its role in the control of viral load and disease activity. Hum. Immunol. 62:561-576. [DOI] [PubMed] [Google Scholar]
- 31.Rammensee, H., J. Bachmann, N. P. Emmerich, O. A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213-219. [DOI] [PubMed] [Google Scholar]
- 32.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez, W. R., M. M. Addo, A. Rathod, C. A. Fitzpatrick, X. G. Yu, B. Perkins, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2004. CD8+ T lymphocyte responses target functionally important regions of protease and integrase in HIV-1 infected subjects. J. Transl. Med. 2:15. http://www.translational-medicine.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohowsky-Kochan, C., J. Skurnick, D. Molinaro, and D. Louria. 1998. HLA antigens associated with susceptibility/resistance to HIV-1 infection. Hum. Immunol. 59:802-815. [DOI] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Samri, A., G. Haas, J. Duntze, J. M. Bouley, V. Calvez, C. Katlama, and B. Autran. 2000. Immunogenicity of mutations induced by nucleoside reverse transcriptase inhibitors for human immunodeficiency virus type 1-specific cytotoxic T cells. J. Virol. 74:9306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt, B., H. Walter, B. Moschik, C. Paatz, K. van Vaerenbergh, A. M. Vandamme, M. Schmitt, T. Harrer, K. Uberla, and K. Korn. 2000. Simple algorithm derived from a geno-/phenotypic database to predict HIV-1 protease inhibitor resistance. AIDS 14:1731-1738. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, M., E. Harrer, A. Goldwich, M. Bauerle, I. Graedner, J. R. Kalden, and T. Harrer. 2000. Specific recognition of lamivudine-resistant HIV-1 by cytotoxic T lymphocytes. AIDS 14:653-658. [DOI] [PubMed] [Google Scholar]
- 39.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201-212. [DOI] [PubMed] [Google Scholar]
- 40.Sidney, J., S. Southwood, V. Pasquetto, and A. Sette. 2003. Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype. J. Immunol. 171:5964-5974. [DOI] [PubMed] [Google Scholar]
- 41.Stratov, I., C. J. Dale, S. Chea, J. McCluskey, and S. J. Kent. 2005. Induction of T-cell immunity to antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 79:7728-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang, J., and R. A. Kaslow. 2003. The impact of host genetics on HIV infection and disease progression in the era of highly active antiretroviral therapy. AIDS 17:S51-S60. [DOI] [PubMed] [Google Scholar]
- 43.Turner, D., J. M. Schapiro, B. G. Brenner, and M. A. Wainberg. 2004. The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients. Antivir. Ther. 9:301-314. [PubMed] [Google Scholar]
- 44.Walter, H., B. Schmidt, K. Korn, A. M. Vandamme, T. Harrer, and K. Uberla. 1999. Rapid, phenotypic HIV-1 drug sensitivity assay for protease and reverse transcriptase inhibitors. J. Clin. Virol. 13:71-80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.