Summary
The Krüppel-like factors (KLFs) comprise a family of evolutionarily conserved zinc finger transcription factors that regulate numerous biological processes including proliferation, differentiation, development and apoptosis. KLF4 and KLF5 are two closely related members of this family and are both highly expressed in epithelial tissues. In the intestinal epithelium, KLF4 is expressed in terminally differentiated epithelial cells at the villus borders of the mucosa and inhibits cell growth, while KLF5 is expressed in proliferating epithelial cells at the base of the intestinal crypts and promotes cell growth. KLF4 and KLF5 respond to a myriad of external stress stimuli and are likely involved in restoring cellular homeostasis following exposure to stressors. Confirming their importance in maintaining tissue integrity, KLF4 and KLF5 are both dysregulated in various types of cancer. Here we review the recent advances in defining the physiological and pathobiological roles of KLF4 and KLF5, focusing on their functions in the intestinal epithelium.
Introduction
The Krüppel-like factors (KLFs) belong to the family of zinc-finger-containing transcription factors that regulate a diverse array of cellular processes, including development, differentiation, proliferation and apoptosis. This family of proteins shares homology with the founding member, Krüppel, which was first identified in Drosophila melanogaster by deletion of a gene that causes a “crippled” phenotype.(1–3) Phylogenetic analysis indicates that the KLF proteins are closely related to the Sp1 and Krox zinc finger protein families, but the KLF family is distinct in that the proteins contain three highly conserved zinc fingers with additional conserved residues between each finger.(4–7) KLFs recognize and bind to very similar “GT-box” or “CACCC element” consensus sequences; thus, the specificity of their activities is determined by differing amino termini and/or by tissue-specific expression.(4) Two members of the KLF family, Krüppel-like factor 4 (KLF4; also known as gut-enriched Krüppel-like factor or GKLF)(8,9) and Krüppel-like factor 5 (KLF5; also known as intestine-enriched Krüppel-like factor or IKLF)(10) are highly expressed in epithelial tissues including the intestinal epithelium. KLF4 is expressed in terminally differentiated epithelial cells at the lumenal surface of the intestinal mucosa, whereas KLF5 is expressed in actively dividing cells at the base of the intestinal crypts. This contrast between the two proteins carries over into their transcriptional activities, as KLF4 and KLF5 often exhibit opposing effects on shared transcriptional targets and carry out distinctive biological activities.(11) In light of the traditional roles of KLF proteins as well as the abundant expression of KLF4 and KLF5 in epithelial cells of the gut, these two proteins are likely to play key roles in maintaining cellular homeostasis in the intestinal epithelium. Indeed, altered expression of both KLF4 and KLF5 has been reported in colorectal cancer as well as in numerous other types of solid tumors.(12–26) The pathobiology of KLF4 and KLF5 is a complex question, as both proteins have been reported to have tumor suppressor activity as well as to promote tumorigenesis, depending on the cellular, tissue and genetic context that is being studied. This review will focus on advances in understanding the physiological and pathobiological roles of KLF4 and KLF5, highlighting their functions in the intestinal epithelium.
Expression and localization of KLF4 and KLF5 in the intestinal epithelium
Expression of the genes encoding KLF4 and KLF5 is developmentally regulated, with higher expression occurring toward the later stage of fetal development.(15,27) In the adult intestine, KLF4 and KLF5 play distinct and often opposing roles as evidenced in part by their tissue distribution. In situ hybridization and immunohistochemical staining show distinct tissue and cellular localization of KLF4 and KLF5 in the small and large bowel. Whereas KLF4 is expressed in the postmitotic, terminally differentiated epithelial cells in the upper regions of the villi and colonic crypts, KLF5 expression is found in actively proliferating epithelial cells in the bottom two-thirds of the crypts (Fig. 1). In concordance with this mutually exclusive expression pattern, KLF5 has been shown to negatively regulate the promoter of the KLF4 gene, while KLF4 activates the promoter of its own gene.(28,29) KLF4 and KLF5 regulate the KLF4 promoter by competing directly for binding to their cognate DNA sequence.(29)
Figure 1.
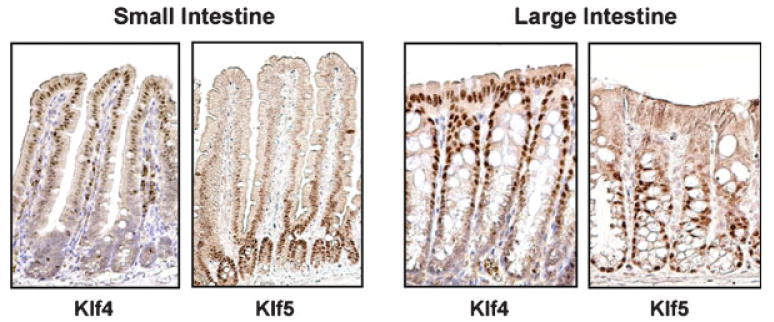
Localization of Klf4 and Klf5 in mouse small and large intestine. Immunostaining of Klf4 and Klf5 shows that Klf4 is present in the nuclei of terminally differentiated epithelial cells in the villi and upper crypts of the small and large intestine, respectively. In contrast, Klf5 exhibits nuclear expression in proliferating epithelial cells at the base of the crypts in both tissues.
In the intestine, Wnt signaling has been shown to play a critical role in driving the proliferation of epithelial cells.(30,31) The expression pattern of KLF5 is similar to several Wnt signaling pathway components that are localized to the proliferating zones of the crypt.(32) Indeed, KLF5 has been reported to be a downstream target of canonical Wnt signaling(33) and may be an important component of cell proliferation regulated by Wnt. In addition, KLF5 staining patterns in the colonic epithelium correlate with those of the proliferation marker, Ki67 (McConnell and Yang, unpublished observations), again suggesting a role for KLF5 in mediating proliferation of gut epithelial cells. In contrast to KLF5, which localizes with pro-proliferative factors of the Wnt pathway, KLF4 exhibits an expression pattern in the intestinal epithelium that is similar to APC, a negative regulator of Wnt signaling. Like KLF4, APC expression increases as intestinal epithelial cells progress from the base of the crypt to the villus,(34) and KLF4 has been shown to be induced upon activation of the APC gene in HT29 colon cancer cells.(19) KLF4 is also involved in the Wnt signaling pathway through cross talk between KLF4 and the Wnt-responsive target, β-catenin.(35) KLF4 has been shown to interact directly with β-catenin and to repress β-catenin-mediated gene expression, as well as to inhibit the tumorigenic activity of colon cancer cells that contain APC or β-catenin mutations.(35) Thus, both KLF4 and KLF5 are responsive to signaling pathways that are critical for regulation of proliferation in the intestinal epithelium.
Roles of KLF4 and KLF5 in cell cycle regulation and proliferation
Progression through the cell cycle is driven by cyclins and their respective cyclin- dependent kinases (Cdks) (Fig. 2). Conversely, inhibition of cell cycle progression is regulated by inhibitors of the Cdks, which include p16ink4a, p21Cip1/Waf1, p27Kip1 and p57Kip2.(36–38) Evidence points to a role for both KLF4 and KLF5 in regulating expression of several of these cell cycle machinery components. Since KLF4 is highly expressed in terminally differentiated, postmitotic intestinal epithelial cells, suggesting a link to growth arrest, Shields et al. examined the role of KLF4 in cell cycle progression in vitro. In actively proliferating NIH3T3 cells, little or no KLF4 is expressed; however, when growth of NIH3T3 cells becomes arrested, either by serum starvation or contact inhibition, KLF4 levels are significantly induced.(9) Furthermore, ectopic expression of KLF4 in NIH3T3 cells results in inhibition of DNA synthesis.(9) Expression profiling of KLF4 through microarray analysis confirms that KLF4 activates numerous genes that encode negative regulators of the cell cycle as well as suppresses expression of genes that promote cell cycle progression.(39) In addition, transcriptional profiling further reveals a global inhibitory function for KLF4 in regulating the expression of groups of genes involved in macromolecular synthesis, including transcription, protein and cholesterol biosynthesis.(40) Taken together, these observations suggest that KLF4 is a growth-arrest-associated gene and is itself an inhibitor of cell proliferation.
Figure 2.
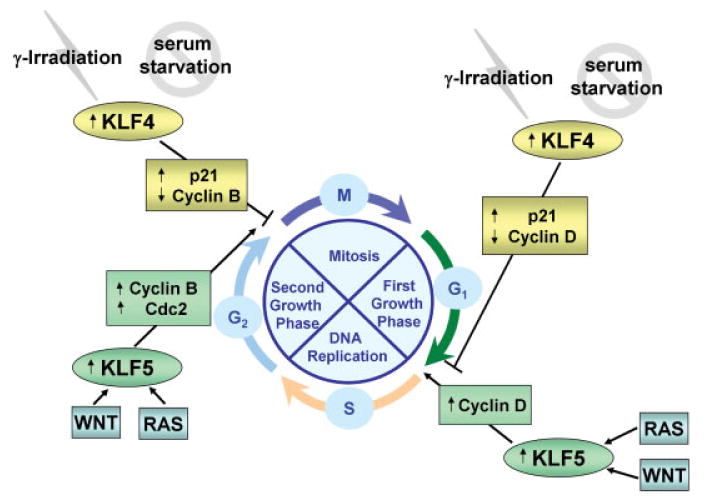
Participation of KLF4 and KLF5 in cell cycle regulation. KLF4 is induced by DNA damage or serum starvation and has a growth inhibitory role by regulating expression of keycell cycle genes, including the cell cycle inhibitor, p21Cip1/Kip1. In contrast, KLF5 plays a growth-promoting role in response to mitogenic signals through its activation of cyclin D, cyclin B, and the cyclin B kinase, Cdc2.
The primary mechanism by which KLF4 participates in cell cycle regulation was elucidated from studies examining the role of KLF4 in DNA-damage-induced cell-cycle arrest. Inducible expression of KLF4 in the RKO colon cancer cell line blocks G1/S cell cycle progression and correlates with increased mRNA levels of the Cdk inhibitor, p21Cip1/Waf1.(41) In addition, treatment of cultured cells with the DNA-damaging agent methanesulfonate(42) or γ-irradiation(43) increases the levels of KLF4 mRNA in a p53-dependent manner and likewise correlates with increased expression of p21Cip1/Waf1.(42) It was further determined that KLF4 transactivates the p21Cip1/Waf1 promoter by binding to a specific Sp1-like cis-element in the proximal region of the promoter.(42) Upon binding, KLF4 recruits p53 to the p21Cip1/Waf1 promoter, allowing p53 to drive transcription of the p21Cip1/Waf1 gene. Activation of p21Cip1/Waf1 expression following DNA damage causes cell cycle arrest at both the G1–S and G2–M transition points. KLF4 was shown to be necessary and sufficient in mediating this p53-dependent checkpoint function at both of these transition points.(43,44) In addition to its effects on p21Cip1/Waf1 expression, KLF4 has been reported to inhibit expression of cyclin D1(45) and cyclin B,(44) which promote progression through the G1–S and G2–M boundaries, respectively. In response to DNA damage, KLF4 has been shown to suppress transcription of the cyclin E gene to prevent chromosomal amplification.(46) KLF4 has also been shown to block mRNA expression of ornithine decarboxylase, a biosynthesis regulatory enzyme that promotes cell proliferation and is implicated in the progression of colon cancer.(47) Thus, KLF4 plays a critical role in activating cell cycle checkpoints to prevent improper progression through the cell cycle and to maintain DNA integrity.
In distinct contrast to KLF4, KLF5 has been shown to have a positive effect on cell cycle progression and proliferation. The level of KLF5 transcripts is low in serum-deprived NIH3T3 cells, but is acutely induced upon addition of serum or phorbol ester.(48) Ectopic expression of KLF5 in either NIH3T3 cells(48) or IEC6 intestinal epithelial cells(49) results in an increased rate of cellular proliferation and leads to anchorage-independent growth. KLF5 expression has been shown to be induced by the mitogen-activated protein kinases (MAPKs), MAPK kinase (MEK)/extracellular signal-regulated kinase (ERK), through activation of the transcription factor, Egr1.(50,51) As a downstream target of this pathway, KLF5 has been shown to be a mediator of the transforming effect of oncogenic H-Ras via MAPK signaling.(52,53) Upon activation of MAPK by H-Ras, KLF5 expression is induced, and KLF5 activates the transcription of several cell cycle promoting gene products, including cyclin D1, cyclin B1 and Cdk1/Cdc2.(52,53) Additional evidence for the pro-proliferative effect of KLF5 is provided by a recent study examining the role of KLF5 in proliferation of primary squamous esophageal epithelial cells.(54) Ectopic expression of KLF5 in mouse primary esophageal keratinocytes induces increased proliferation. KLF5 also activates transcription of the epidermal growth factor receptor (EGFR) gene. EGFR signaling, in turn, activates MEK/ERK which further induces KLF5 expression, creating a positive feedback loop. Conversely, suppression of KLF5 in these cells results in a marked reduction in proliferation. The authors conclude that KLF5 promotes proliferation of basal cells in squamous esophageal epithelia by amplifying EGFR signaling via induction of EGFR expression.
Yet another example of the importance of KLF5 in mediating cell proliferation is the finding that the inhibitory effect of all-trans retinoic acid (ATRA) on proliferation of intestinal epithelial cells is mediated by inhibition of KLF5 expression.(49) This inhibitory effect is seen in both non-transformed IEC6 cells and colon cancer cell lines. Thus, in various types of epithelial and fibroblast cells, induction of KLF5 has a pro-proliferative effect. In vivo evidence for a growth-promoting role for KLF5 in the intestine is provided by the finding that mice with a heterozygous disruption of the Klf5 gene have shorter intestinal crypts and villi.(55)
Enhancement of the pro-proliferative effect of KLF5 has recently been reported through direct interaction of KLF5 with protein inhibitor of activated STAT1 (PIAS1), a small ubiquitin-like modifier (SUMO) ligase.(56) Interaction of these two proteins was identified through a yeast two-hybrid screen using KLF5 as bait and was confirmed by co-immunoprecipitation of the proteins as well as through co-localization by immunofluorescence. Co-expression of KLF5 with PIAS1 was shown to synergistically increase the transcriptional activity of KLF5 in activating the cyclin D1 and Cdc2 promoters and to increase the proportion of cells actively synthesizing DNA, as compared to effects seen with ectopic expression of KLF5 alone.(56)
Regulation of apoptosis by KLF4 and KLF5
Both KLF4 and KLF5 have been implicated in the regulation of apoptosis. The function of KLF4 in modulating apoptosis is apparent during DNA damage, when cells must decide either to activate cell cycle checkpoint and repair machinery or to undergo apoptosis to eliminate damaged cells. Using an inducible expression system for KLF4 in RKO colon cancer cells, it was shown that induction of KLF4 significantly reduces the percentage of apoptotic cells following γ-irradiation.(57) Related to its regulation of p21Cip1/Waf1, KLF4 tips the balance towards growth arrest following DNA damage through its induction of p21Cip1/Waf1 expression.(42) Importantly, upregulation of KLF4 also inhibits expression of the gene encoding the pro-apoptotic protein Bax following DNA damage.(57) This activity is reported to be due to the ability of KLF4 to inhibit transactivation of the Bax promoter by p53.(57) Another report studying breast cancer cells indicates that KLF4 modulates apoptosis through regulating expression of p53 itself. In this study, KLF4 binds directly to the p53 promoter in MDA-MB-134 breast cancer cells and suppresses transcription of p53.(21) Furthermore, inhibition of KLF4 expression in MDA-MB-134 cells using KLF4-specific siRNA molecules results in restoration of p53 levels and an abrupt induction of apoptosis.(58) A contrasting role for KLF4 has been reported in experiments conducted in the esophageal cancer cell line, TE2, which does not express KLF4 or KLF5.(23) Ectopic expression of either KLF4 or KLF5 in these cells is reported to enhance detachment-induced apoptosis (anoikis). In addition, ectopic expression of KLF5 in TE2 cells reduces cell viability, reportedly through the activation of Bax expression to promote apoptosis. Disparate roles for KLF4 and KLF5 may be accounted for by various genetic backgrounds in the cancer cell lines and/or by tissue-specific differences. However, as transcriptional regulators, both KLF4 and KLF5 appear to influence activation of apoptotic pathways through the activation or repression of key genes whose products are involved in cell fate decisions.
Involvement of KLF4 and KLF5 in differentiation and development
Most of the evidence for roles of KLF4 and KFL5 in differentiation and development has been derived from knockout or overexpression transgenic mouse studies. The predominant phenotype of the Klf4−/− knockout mouse is a defect in the barrier function of the skin, which results in rapid loss of body fluids and postnatal lethality.(59) KLF4 is normally highly expressed in the differentiating layers of epidermis, and the barrier defect in the Klf4−/− mice is caused by perturbations in late-stage epidermal differentiation structures. Conversely, in transgenic mice induced to ectopically express KLF4 in the basal layer of the epidermis, KLF4 has been shown to accelerate terminal differentiation and to promote early formation of the epidermal permeability barrier.(60) The ability of KLF4 to affect barrier function is likely due to its ability to regulate gene clusters of the Sprr(61) and keratin families,(39) which are key components in maintaining epithelial barrier integrity. In examining the role of KLF5 in epidermal tissues, it has been reported that ectopic expression of KLF5 in the epidermis of transgenic mice results in disruption of epithelial–mesenchymal interactions that are required for proper skin formation and craniofacial development.(62) In addition, ectopic expression of KLF5 results in a loss of regenerative potential of keratinocytes, suggesting that KLF5 is a modulator for regenerative potential of the epidermis.(62)
In addition to functions in epidermal tissues, KLF4 appears to play a critical role in regulating differentiation, and possibly lineage specification, in the intestinal epithelium. Transcriptional targets of KLF4 that suggest a function in differentiation of intestinal tissues include Lama1, which encodes the basement membrane component, Laminin-1, and the gene encoding intestinal alkaline phosphatase, an enterocyte differentiation marker.(63,64) In examining in vivo functions of KLF4, analysis of Klf4−/− mice shows a 90% depletion of goblet cells in the colon as determined by morphology and ultrastructural analysis.(65) Interestingly, the goblet cell phenotype in Klf4−/− mice is minimal in the small intestine, which may be attributable to KLF4 being more highly expressed in colon than in small intestine.(9) Differences in goblet cell numbers are proposed to be due to the lack of goblet cell maturation in the Klf4−/− mice.(65) Colonic goblet cells in these mice do not show the normal goblet cell morphology, and the goblet cell marker MUC2 exhibits patchy expression throughout the colonic epithelium and is not confined to cells with “goblet-like” morphology. Thus, it is concluded that KLF4 is required for proper goblet cell maturation, and that goblet cells in the Klf4−/− mice represent immature goblet cells of the goblet cell lineage. The involvement of KLF4 in goblet cell maturation as well as its role in skin differentiation suggests that KLF4 functions in the switch from proliferation to differentiation.(58) Further substantiating this concept is a recent finding that KLF4 is one of four factors necessary for induction of pluripotent stem cells from mouse embryonic or adult fibroblasts.(66)
While a definitive role for KLF5 in intestinal epithelial cell differentiation has not been reported, it has been identified as a key regulator of adipocyte differentiation.(67) Klf4−/− mice are embryonic lethal, but experiments using Klf4−/− mice show that neonatal mice carry much less white adipose tissue than wild-type mice, suggesting a role for KLF5 in adipogenesis.(67) In these studies, constitutive overexpression of dominant-negative KLF5 in 3T3-L1 preadipocytes inhibits adipocyte differentiation, whereas overexpression of wild-type KLF5 induces differentiation in the absence of hormonal stimulation. This physiologic effect was reported to be through the interaction of KLF5 with C/EBPα/δ to activate the PPARγ2 promoter.
Potential roles for KLF5 in response to external stressors
In addition to roles for KLF5 in proliferation and differentiation, a number of studies point to a function for KLF5 as a mediator of external stress responses following tissue injury. KLF5 is induced by activation of the MAPK pathway in vascular smooth muscle cells and is a direct target of the early response gene, Egr-1.(68) Experiments in vascular tissues reveal that KLF5 is induced in response to cardiac tissue injury and is involved in the activation and proliferation of smooth muscle cells and fibroblasts within vascular lesions.(69) Further evidence of KLF5 mediating stress responses has been reported in experiments using Klf4−/− mice to determine responses to injury in vascular tissues.(55) This study reports that activation and proliferation of smooth muscle cells (SMCs) and fibroblasts following vascular injury is impaired in Klf4−/− mice, as are inflammatory responses and angiogenesis. Important for the effect of KLF5 on the modulation and proliferation of SMCs is the interaction of KLF5 with the retinoic acid receptor/retinoic X receptor (RAR/RXR) heterodimer to drive transcription of PDGF-A.(70) Thus, KLF5 appears to be a crucial determinant of the cellular response to cardiovascular injury, and may play similar roles in other tissues.
Recent evidence from our laboratory indicates that exposure of rat intestinal epithelial cells (IEC6) to the bacterial component, lipopolysaccharide (LPS), results in induction of KLF5 expression through the MAPK pathway.(71) Induction of KLF5 correlates with induction of the NF-κB subunits, p50 and p65. Suppression of KLF5 by small interfering RNA (siRNA) inhibits NF-κB activity in response to LPS. Importantly, suppression of KLF5 leads to abrogation of many downstream targets of NF-κB due to LPS stimulation, including interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α).(71) Previous studies have demonstrated a synergistic effect of KLF5 and the NF-κB subunits, p50 and p65, on transcriptional activation of target genes through physical interaction of KLF5 with p50 or p65.(51,72) KLF5 expression is likewise induced in vitro in IEC6 cells and in vivo in mouse colonic epithelial cells by the bacterial pathogen, Citrobacter rodentium (McConnell and Yang, unpublished observations). These results indicate that KLF5 is an important mediator of the proinflammatory response to LPS and is likely involved in immune responses to bacterial infection in intestinal tissues. This mechanism may represent a relatively “late” response as compared to the classical mechanism of NF-κB activation through post-translational modification.
Pathobiology of KLF4
Because KLF4 plays critical roles in differentiation and maintenance of cell cycle checkpoint functions, it is logical to assume that KLF4 would function as a tumor suppressor. Indeed, numerous studies have reported loss of KLF4 expression in human tumors, including colorectal, stomach, esophageal and bladder cancers.(12-19,73–76) Mutational analysis indicates that the KLF4 gene is subject to deletion, mutation and methylation silencing in a significant proportion of colon and gastric cancers.(17,18) In support of a tumor suppressor role for KLF4, overexpression of KLF4 in the human colon cancer cell line RKO reduces colony formation, cell migration and invasion, and in vivo tumorigenecity.(77) In mouse models, KLF4 mRNA levels are reduced in the ApcMin/+ mice,(12) and recent evidence from our laboratory indicates that ApcMin/+Klf4+/− mice have increased intestinal tumor burden as compared to ApcMin/+Klf4+/+ mice (Ghaleb and Yang, unpublished observations). In studies using a conditional Klf4-knockout mouse specific for the gastric epithelium, loss of Klf4 results in increased proliferation and altered differentiation in the stomach, culminating in precancerous changes.(16)
In light of the evidence supporting a tumor suppressor role for KLF4, it is surprising that several reports derived from genetic screens identify KLF4 as a putative oncogene rather than a tumor suppressor.(20–22) In agreement with these studies, elevated levels of KLF4 have been reported in up to 70% of mammary carcinomas,(78) and KLF4 is frequently overexpressed in squamous-cell carcinomas of the oropharynx.(20) Furthermore, ectopic expression of KLF4 in basal keratinocytes of transgenic mice results in dysplastic lesions that resemble squamous cell carcinoma in situ.(79,80) Recent evidence suggests that the role KLF4 plays may depend on the genetic context in which it is functioning.(21,58) A potential oncogenic activity of KLF4 was discovered in examining the mechanism by which KLF4 allows bypass of oncogenic RASv12-induced senescence. In this study, KLF4 suppresses the expression of p53 by acting directly on its promoter, thus allowing for RASv12-mediated transformation and providing resistance to DNA-damage-induced apoptosis. This mechanism appears to be at work in breast cancer, where p53 expression is often suppressed at the level of transcription. Indeed, depletion of KLF4 from breast cancer cells was shown to restore p53 expression, resulting in p53-dependent apoptosis.(21) Thus, the oncogenic potential of KLF4 appears to lie in its ability to inhibit apoptosis. In addition to p53 suppression, another mechanism by which KLF4 has been reported to suppress apoptosis is through direct downregulation of the Bax promoter.(57) Genetic contexts in which KLF4 could act as an oncogene, therefore, would be those where KLF4 is induced, but the tumor suppressor activity of KLF4 is disrupted, in other words, where p21Cip1/Waf1 is functionally inactive. Activation of the Rasv12-cyclin D pathway provides such a scenario, as overexpression of cyclin D overrides the growth-inhibitory effects of p21Cip1/Waf1 in KLF4-induced cell cycle arrest.(21) Fig. 3A presents a model for the “two faces” of KLF4 in tumor suppression and transformation.
Figure 3.
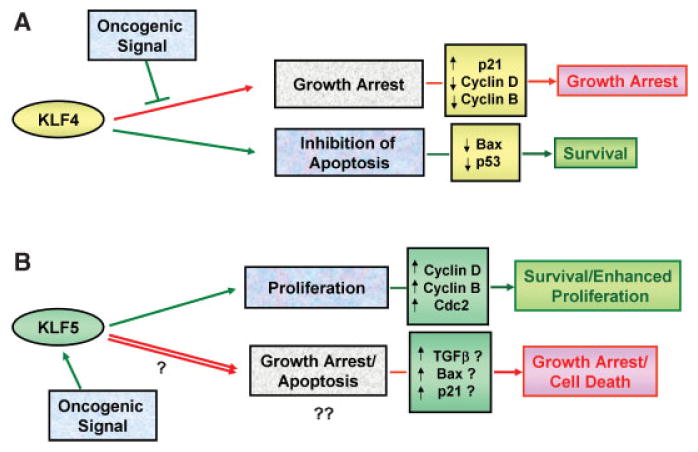
Models for the dual roles of KLF4 and KLF5 in tumor suppression and oncogenesis. A: KLF4 normally acts as a tumor suppressor through p21Cip1/Kip1-dependent cell cycle arrest. However, in the presence of an oncogenic signal (such as Rasv12), the p21Cip1/Kip1 pathway is inactivated, and the anti-apoptotic activity of KLF4 provides a survival advantage, allowing for cellular transformation. B: The pro-proliferative activity of KLF5 contributes to transformation in response to oncogenic signals, such as H- or K-Ras. However, in certain contexts, KLF5 may also possess growth inhibitory activities in the presence of an oncogenic signal that allow it to act as a tumor suppressor, overriding its growth-promoting effects.
In summary, KLF4 has crucial cell-cycle-checkpoint functions in response to DNA damage that are, in large part, carried out through its induction of p21Cip1/Waf1 expression. These functions contribute to its tumor suppressor activity, particularly in the gastrointestinal tract. However, KLF4 also exhibits anti-apoptotic activity, which, in certain genetic contexts, can drive cellular transformation in cooperation with other tumor-promoting events, such as the activation of Rasv12.
Pathobiology of KLF5
Because of the pro-proliferative function of KLF5 in the intestinal epithelium, KLF5 is a likely suspect in mediating oncogenic events in this tissue. An excellent example of this potential is the ability of KLF5 to mediate the transforming activity of oncogenic H-Ras in vitro.(52) In H-Ras transformed NIH 3T3 cells, KLF5 is upregulated via H-Ras activation of MAPK. Subsequently, increased expression of KLF5 is responsible for inducing expression of cyclin D1, cyclin B and Cdc2 at the level of transcription.(52,53) As another example, overexpression of KLF5 in the human bladder cancer cell line, TSU-Pr1, accelerated the cell cycle progression of these cells and was shown to stimulate their tumorigenic potential when injected into SCID mice.(81) Furthermore, KLF5 expression has been reported to be a prognostic factor for overall survival in patients with sporadic breast cancer.(7) Higher expression of KLF5 correlates with shorter overall survival of breast cancer patients, whereas lower KLF5 expression correlates with better survival. In contrast to these reports, a number of studies examining KLF5 status in various types of tumors indicate that KLF5 is downregulated by several mechanisms, including deletion, gene silencing and protein degradation.(26,82,83) Because of this selective loss, KLF5 has been implicated as a tumor suppressor in esophageal, breast, prostate and intestinal cancers.(23–26,82,84) As with KLF4 having “hidden” oncogenic potential, it is possible that KLF5 possesses a yet unidentified tumor suppressor activity. Thus, while KLF5 has been shown to promote proliferation and mediate transforming events, KLF5 may also, in certain genetic contexts, activate expression of growth-suppressive or pro-apoptotic factors that can “override” KLF5's growth-promoting activity. A model of this theory is presented in Fig. 3B. A downstream target of KLF5 that is a potential candidate for mediating this tumor suppressor effect is TGF-β.(55) TGF-β is a well-documented negative regulator of epithelial cell growth, whose tumor suppressor activity is activated early in tumorigenesis.(85) Likewise, inactivation of KLF5 is reported to be an early event in intestinal cancers.(24) Others have reported that KLF5 potentially mediates pro-apoptotic or anti-proliferative effects through the induction of Bax and p21Cip1/Waf1.(23)
To summarize, KLF5 has pro-proliferative functions in intestinal epithelial cells through its activation of key cell-cycle-promoting genes. In addition to its growth-promoting role, KLF5 is implicated in injury response pathways as well as the activation of inflammatory pathways following exposure to bacterial components or pathogens. The growth-promoting activity of KLF5 can be manipulated to mediate transforming events, such as those activated by oncogenic H-Ras. However, a number of studies report the loss of KLF5 expression in certain cancers, suggesting a putative tumor suppressor role. From these reports, it is clear that the role of KLF5 in cancer is a complex issue, and its function in tumorigenesis, like KLF4, is likely influenced by its genetic context.
Conclusion
KLF4 and KLF5 are two members of the Krüppel-like family of transcription factors that exert important biological effects on cellular proliferation and differentiation in the intestinal epithelium. Both proteins are activated in response to stressful stimuli, such as DNA damage or tissue injury, and are integral components of pathways necessary for restoring tissue homeostasis. KLF4 is a critical mediator of the p53–p21Cip1/Waf1 axis following DNA damage, inducing growth arrest to allow for DNA repair. KLF5 is activated in response to bacterial pathogens in the colon and is likely important for the pro-proliferative and pro-inflammatory responses to infection. Because of the functions of these proteins in maintaining intestinal tissue homeostasis, KLF4 and KLF5 are often altered in gastrointestinal and other cancers and play significant roles in the process of tumorigenesis. The physiological roles and the pathobiology of KLF4 and KLF5 in the intestinal epithelium will continue to be unveiled as the involvement of these proteins in cancer and inflammation is explored further.
Acknowledgments
Funding agency: This work was supported in part by grants from the National Institutes of Health; Grant number: (DK52230, DK64399, and CA84197).
Abbreviations
- ATRA
all-trans retinoic acid
- Cdk
cyclin-dependent kinase
- EGFR
epidermal growth factor receptor
- ERK
extracellular signal-regulated kinase
- GKLF
gut-enriched Krüppel-like factor
- IKLF
intestine-enriched Krüppel-like factor
- KLF
Krüppel-like factor
- LPS
Lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MEK
MAPK kinase
- RAR
retinoic acid receptor
- RXR
retinoic X receptor
- SMCs
smooth muscle cells
- SUMO
small ubiquitin-like modifier
References
- 1.Jackle H, Rosenberg UB, Preiss A, Seifert E, Knipple DC, et al. Molecular analysis of Kruppel, a segmentation gene of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:465–473. doi: 10.1101/sqb.1985.050.01.058. [DOI] [PubMed] [Google Scholar]
- 2.Preiss A, Rosenberg UB, Kienlin A, Seifert E, Jackle H. Molecular genetics of Krüppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- 3.Wieschaus E, Nusslein-Volhard C, Kluding H. Krüppel, a gene whose activity is required early in the zygotic genome for normal embryonic segmentation. Dev Biol. 1984;104:172–186. doi: 10.1016/0012-1606(84)90046-0. [DOI] [PubMed] [Google Scholar]
- 4.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 5.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, et al. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- 8.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 9.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conkright MD, Wani MA, Anderson KP, Lingrel JB. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27:1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, et al. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang DT, Bachman KE, Mahatan CS, Dang LH, Giardiello FM, et al. Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett. 2000;476:203–207. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, et al. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–256. doi: 10.1016/s0006-291x(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 14.Shie JL, Chen ZY, O'Brien MJ, Pestell RG, Lee ME, et al. Role of gut-enriched Kruppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G806–G814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 15.Ton-That H, Kaestner KH, Shields JM, Mahatanankoon CS, Yang VW. Expression of the gut-enriched Kruppel-like factor gene during development and intestinal tumorigenesis. FEBS Lett. 1997;419:239–243. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Wei D, Gong W, Kanai M, Schlunk C, Wang L, et al. Drastic downregulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 18.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, et al. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on C DX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster KW, Ren S, Louro ID, Lobo-Ruppert SM, McKie-Bell P, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 21.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 24.Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of K LF5. J Biol Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 26.Rozenblum E, Vahteristo P, Sandberg T, Bergthorsson JT, Syrjakoski K, et al. A genomic map of a 6-Mb region at 13q21-q22 implicated in cancer development: identification and characterization of candidate genes. Hum Genet. 2002;110:111–121. doi: 10.1007/s00439-001-0646-6. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi S, Laub F, Matsumoto N, Asaka M, Ramirez F, et al. Developmental expression of the mouse gene coding for the Krüppel-like transcription factor KLF5. Dev Dyn. 2000;217:421–429. doi: 10.1002/(SICI)1097-0177(200004)217:4<421::AID-DVDY9>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 28.Mahatan CS, Kaestner KH, Geiman DE, Yang VW. Characterization of the structure and regulation of the murine gene encoding gut-enriched Krüppel-like factor (Krüppel-like factor 4) Nucleic Acids Res. 1999;27:4562–4569. doi: 10.1093/nar/27.23.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Krüppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Krüppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 31.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregorieff A, Pinto D, Begthel H, Destree O, Kielman M, et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Ziemer LT, Pennica D, Levine AJ. Identification of a mouse homolog of the human BTEB2 transcription factor as a beta-catenin-independent Wnt-1-responsive gene. Mol Cell Biol. 2001;21:562–574. doi: 10.1128/MCB.21.2.562-574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KJ, Johnson KA, Bryan TM, Hill DE, Markowitz S, et al. The APC gene product in normal and tumor cells. Proc Natl Acad Sci USA. 1993;90:2846–2850. doi: 10.1073/pnas.90.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Chen X, Kato Y, Evans PM, Yuan S, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–2064. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 37.Vidal A, Koff A. Cell-cycle inhibitors: three families united by a common cause. Gene. 2000;247:1–15. doi: 10.1016/s0378-1119(00)00092-5. [DOI] [PubMed] [Google Scholar]
- 38.Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, et al. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitney EM, Ghaleb AM, Chen X, Yang VW. Transcriptional profiling of the cell cycle checkpoint gene kruppel-like factor 4 reveals a global inhibitory function in macromolecular biosynthesis. Gene Expr. 2006;13:85–96. doi: 10.3727/000000006783991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, et al. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, et al. The gut-enriched Krüppel-like factor (Krüppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon HS, Yang VW. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J Biol Chem. 2004;279:5035–5041. doi: 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, et al. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–4025. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen ZY, Shie JL, Tseng CC. Gut-enriched Krüppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 48.Sun R, Chen X, Yang VW. Intestinal-enriched Krüppel-like factor (Krüppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chanchevalap S, Nandan MO, Merlin D, Yang VW. All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Krüppel-like factor 5. FEBS Lett. 2004;578:99–105. doi: 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagai R, Kowase K, Kurabayashi M. Transcriptional regulation of smooth muscle phenotypic modulation. Ann N Y Acad Sci. 2000;902:214–222. doi: 10.1111/j.1749-6632.2000.tb06316.x. discussion 222–213. [DOI] [PubMed] [Google Scholar]
- 51.Aizawa K, Suzuki T, Kada N, Ishihara A, Kawai-Kowase K, et al. Regulation of platelet-derived growth factor-A chain by Kruppel-like factor 5: new pathway of cooperative activation with nuclear factor-kappaB. J Biol Chem. 2004;279:70–76. doi: 10.1074/jbc.M306621200. [DOI] [PubMed] [Google Scholar]
- 52.Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, et al. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579:4757–4762. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, Goldstein BG, Nakagawa H, Katz JP. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J. 2007;21:543–550. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 55.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 56.Du JX, Yun CC, Bialkowska A, Yang VW. Protein inhibitor of activated STAT 1 interacts with and up-regulates activities of the pro-proliferative transcription factor Kruppel-like factor 5. J Biol Chem. 2007;282:448–479. doi: 10.1074/jbc.M603413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26:2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 59.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 60.Jaubert J, Cheng J, Segre JA. Ectopic expression of Krüppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 61.Patel S, Kartasova T, Segre JA. Mouse Sprr locus: a tandem array of coordinately regulated genes. Mamm Genome. 2003;14:140–148. doi: 10.1007/s00335-002-2205-4. [DOI] [PubMed] [Google Scholar]
- 62.Sur I, Rozell B, Jaks V, Bergstrom A, Toftgard R. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci. 2006;119:3593–3601. doi: 10.1242/jcs.03070. [DOI] [PubMed] [Google Scholar]
- 63.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, et al. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hinnebusch BF, Siddique A, Henderson JW, Malo MS, Zhang W, et al. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–G30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- 65.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 67.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ Res. 1999;85:787–795. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 69.Sakamoto H, Sakamaki T, Kanda T, Hoshino Y, Sawada Y, et al. Smooth muscle cell outgrowth from coronary atherectomy specimens in vitro is associated with less time to restenosis and expression of a key Transcription factor KLF5/BT EB2. Cardiology. 2003;100:80–85. doi: 10.1159/000073043. [DOI] [PubMed] [Google Scholar]
- 70.Fujiu K, Manabe I, Ishihara A, Oishi Y, Iwata H, et al. Synthetic retinoid Am80 suppresses smooth muscle phenotypic modulation and in-stent neointima formation by inhibiting K LF5. Circ Res. 2005;97:1132–1141. doi: 10.1161/01.RES.0000190613.22565.13. [DOI] [PubMed] [Google Scholar]
- 71.Chanchevalap S, Nandan MO, McConnell BB, Charrier L, Merlin D, et al. Krüppel-like factor 5 is an important mediator for lipopolysaccharide-induced proinflammatory response in intestinal epithelial cells. Nucleic Acids Res. 2006;34:1216–1223. doi: 10.1093/nar/gkl014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sur I, Unden AB, Toftgard R. Human Krüppel-like factor5/KL F5:synergy with NF-kappaB/Rel factors and expression in human skin and hair follicles. Eur J Cell Biol. 2002;81:323–334. doi: 10.1078/0171-9335-00257. [DOI] [PubMed] [Google Scholar]
- 73.Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN, et al. Downregulation of gut-enriched Kruppel-like factor expression in esophageal cancer. World J Gastroenterol. 2002;8:966–970. doi: 10.3748/wjg.v8.i6.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi BJ, Cho YG, Song JW, Kim CJ, Kim SY, et al. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. 2006;202:585–589. doi: 10.1016/j.prp.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 75.Kanai M, Wei D, Li Q, Jia Z, Ajani J, et al. Loss of Krüppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res. 2006;12:6395–6402. doi: 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- 76.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 77.Dang DT, Chen X, Feng J, Torbenson M, Dang LH, et al. Overexpression of Krüppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;22:3424–3430. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foster KW, Frost AR, McKie-Bell P, Lin CY, Engler JA, et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 79.Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang CC, Liu Z, Li X, Bailey SK, Nail CD, et al. KLF4 and PCNA identify stages of tumor initiation in a conditional model of cutaneous squamous epithelial neoplasia. Cancer Biol Ther. 2005;4:1401–1408. doi: 10.4161/cbt.4.12.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen C, Benjamin MS, Sun X, Otto KB, Guo P, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulationin the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- 82.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 83.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, et al. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 84.Dong JT. Prevalent mutations in prostate cancer. J Cell Biochem. 2006;97:433–447. doi: 10.1002/jcb.20696. [DOI] [PubMed] [Google Scholar]
- 85.Akhurst RJ, Balmain A. Genetic events and the role of TGF beta in epithelial tumour progression. J Pathol. 1999;187:82–90. doi: 10.1002/(SICI)1096-9896(199901)187:1<82::AID-PATH248>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]


