Abstract
Mitochondrial preproteins that are imported via the translocase of the mitochondrial outer membrane (Tom)70 receptor are complexed with cytosolic chaperones before targeting to the mitochondrial outer membrane. The adenine nucleotide transporter (ANT) follows this pathway, and its purified mature form is identical to the preprotein. Purified ANT was reconstituted with chaperones in reticulocyte lysate, and bound proteins were identified by mass spectrometry. In addition to 70-kDa heat-shock cognate protein (Hsc70) and 90-kDa heat-shock protein (Hsp90), a specific subset of cochaperones were found, but no mitochondria-specific targeting factors were found. Interestingly, three different Hsp40-related J-domain proteins were identified: DJA1, DJA2, and DJA4. The DJAs bound preproteins to different extents through their C-terminal regions. DJA dominant-negative mutants lacking the N-terminal J-domains impaired mitochondrial import. The mutants blocked the binding of Hsc70 to preprotein, but with varying efficiency. The DJAs also showed significant differences in activation of the Hsc70 ATPase and Hsc70-dependent protein refolding. In HeLa cells, the DJAs increased new protein folding and mitochondrial import, although to different extents. No single DJA was superior to the others in all aspects, but each had a profile of partial specialization. The Hsp90 cochaperones p23 and Aha1 also regulated Hsp90–preprotein interactions. We suggest that multiple cochaperones with similar yet partially specialized properties cooperate in optimal chaperone–preprotein complexes.
INTRODUCTION
The majority of mitochondrial proteins are encoded in the nucleus, translated on cytosolic ribosomes, and subsequently imported into mitochondria (Reichert and Neupert, 2004). The translocase of the mitochondrial outer membrane (TOM) is responsible for polypeptide translocation across the outer membrane; and inside the organelle, various mechanisms sort these polypeptides to their appropriate compartment—matrix, inner membrane or intermembrane space. The TOM complex contains integral membrane import receptors, Tom20 and Tom70, which mediate targeting of mitochondrial preproteins, and a general import pore that provides an aqueous channel for translocation. Tom20 is thought to be responsible for the import of preproteins bearing classical matrix-directed N-terminal signal sequences, or leader peptides. Tom70, the alternate receptor, is thought to mediate the import of preproteins that contain targeting information within their mature sequences (Koehler, 2004; Rehling et al., 2004). Although much of our knowledge of import derives from work in Saccharomyces cerevisiae and Neurospora crassa, the biological functions of the human Tom70 and Tom20 receptors seem to be largely conserved (Iwahashi et al., 1997; Schleiff et al., 1997; Yano et al., 1997; Suzuki et al., 2002).
The Tom70 receptor seems to be most critical for the import of particularly hydrophobic preproteins, such as members of the inner membrane metabolite carrier family (Sollner et al., 1990; de Marcos-Lousa et al., 2006). Before import, these preproteins typically depend on cytosolic chaperones to maintain their solubility. The ATP-dependent chaperones 70-kDa heat-shock cognate protein (Hsc70) and 90-kDa heat-shock protein (Hsp90) have key roles in this mechanism (Young et al., 2004). In addition to maintaining the solubility of the bound preprotein, Hsc70 and Hsp90 can both dock onto to a specific binding site in Tom70, an essential first step in preprotein targeting. The chaperone docking site lies in the central region of Tom70 next to the single N-terminal transmembrane domain (Young et al., 2003). At this stage, preprotein is thought to contact Tom70 directly in a C-terminal region separate from the chaperone docking site (Brix et al., 2000; Wu and Sha, 2006). ATP-dependent cycling by Hsp90 and most probably Hsc70 then assists the translocation of preprotein via the outer membrane TOM machinery (Fan et al., 2006).
In their other cellular functions, Hsc70 and Hsp90 interact with a wide range of cochaperone proteins, which connect additional specific chaperoning functions or biochemical activities to chaperone complexes. In some cases, the type of substrate polypeptide bound by a chaperone complex determines which specific cochaperone is recruited to Hsp90. For example, the immunophilin FKBP52 usually associates with, and assists in the maturation of, steroid hormone receptors, whereas Cdc37 has a comparable role for kinases (Pratt and Toft, 2003). Such specificity is not absolute: both Cdc37 and the immunophilin FKBP8 have been found in chaperone complexes with a folding-deficient mutant of the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel (Wang et al., 2006). Many cochaperones contain functionally conserved chaperone–interaction domains. Some interactions with Hsp90 and Hsc70 are mediated by structurally conserved tetratricopeptide repeat (TPR) clamp domains that recognize the C-terminal motifs of the chaperones. The TPR-domains in FKBP52 and the related cyclophilin Cyp40 connect to Hsp90. Those in the cochaperone Hop recognize Hsc70 and Hsp90 separately, allowing Hsc70–substrate complexes to recruit Hsp90 (Pratt and Toft, 2003). Also, a conserved TPR clamp domain in Tom70 forms its chaperone docking site (Young et al., 2003). J-domains, found in various cochaperones including the Hsp40-related proteins, stimulate ATP hydrolysis and substrate binding by Hsc70. J-domains do not form stable contacts with Hsc70, but they are essential for all known functions of Hsc70 (Walsh et al., 2004; Qiu et al., 2006). Several nucleotide exchange factors including Bag-1 transiently promote ATP rebinding and Hsc70 dissociation from substrate (Mayer and Bukau, 2005). Cochaperones that do not belong to these structural families include Cdc37, p23, and Aha1. p23 stabilizes the ATP- and substrate-binding state of Hsp90, whereas the recently discovered Aha1 stimulates the Hsp90 ATPase (Panaretou et al., 2002; Sullivan et al., 2002; Lotz et al., 2003; Morishima et al., 2003; Pearl and Prodromou, 2006).
It is not known what proteins in addition to Hsc70 and Hsp90 are complexed with Tom70-dependent preproteins before import, or what proteins are involved in complex formation. It seems certain that cochaperone proteins participate in these complexes. The preproteins are not related to structural families of proteins known to prefer specific cochaperones, and the involvement of many cochaperones cannot be predicted. The preproteins seem to be in high-molecular-weight complexes of ∼600 kDa, similar to other Hsp90-bound complexes, but with some heterogeneity (Young et al., 2003; Fan et al., 2006). The identity and function of bound cochaperones would lead to significant insight. First, are the cochaperones present specifically bound? Second, are there components unique to chaperone–preprotein complexes, and what role might they have? Third, do the cochaperones bound have implications for the function of Hsc70 and Hsp90?
To address these questions, we reconstituted chaperone complexes in reticulocyte lysate (RL) as a model cytosol. This method was used to originally identify most of the cochaperones involved in steroid receptor and kinase maturation (Pratt and Toft, 2003). RL is uniquely suited for such experiments. Although it contains essentially no organellar contamination, it is the basis for all experiments reconstituting cell-free mitochondrial import; therefore, it contains all necessary factors. A mitochondrial preprotein, the adenine nucleotide transporter (ANT) is uniquely purifiable in biochemical amounts (Pebay-Peyroula et al., 2003), and it was used as the starting point for the reconstitution of complexes.
MATERIALS AND METHODS
Chemicals and Reagents
Unless stated otherwise, all chemical reagents were from Sigma Diagnostics Canada or BioShop Canada (Mississauga, ON, Canada). Restriction enzymes and other recombinant DNA reagents were from New England Biolabs (Ipswich, MA), Invitrogen (Carlsbad, CA), and Stratagene (San Diego, CA). Geldanamycin (GA) was from LC Laboratories (Boston, MA). Untreated rabbit RL was from Green Hectares (Oregon, WI). Antibodies against Hsp90, Hsc70, Hop, FKBP52, and Hsp40 were from Assay Designs (Ann Arbor, MI); those against Cdc37, Cyp40, and p23 were from Affinity BioReagents (Golden, CO); that against DJA1 was from Labvision/NeoMarkers (Fremont, CA); and that against luciferase was from Sigma Diagnostics Canada. The antibody against Tpr2 was a gift from Dr. W.M.J. Obermann (Houston, TX); those against DJA2 and DJA4 were gifts from Dr. M. Mori (Kumamoto, Japan), and that against Hsp60 was a gift from Dr. F. U. Hartl (Martinsried, Germany). Additional antibodies specific for DJA1, DJA2, and DJA4 were raised in rabbits against the synthetic peptides LVDFDPNQER, PEVPNIIGET, and PNEQNWRQHR, respectively, and they were confirmed with immunoblots against the purified DJA proteins, and HeLa cells were transfected with each antibody. Antibodies against the myc (9E10) and hemagglutinin (HA.11) epitope tags were from Covance/BAbCO (Richmond, CA).
Plasmids
The sequences encoding bovine phosphate carrier (PiC) and N. crassa Rieske iron-sulfur protein (ISP) were in pGEM-3 (Promega, Madison, WI) (Sollner et al., 1989; Zara et al., 1992). The sequence encoding murine ANT2 (NM_007451) was amplified by polymerase chain reaction (PCR) from a cDNA library and inserted into pGEM-11Z (Promega) (Fan et al., 2006). The sequences encoding nontagged human Hsp90α and rat Hsc70 were in pET15b and pET11a (Novagen, San Diego, CA), respectively, and the sequences encoding residues 566-732 of human Hsp90α (C-90) and residues 151-263 of human Bag-1 (C-Bag) were in pProExHTa (Clontech, Mountain View, CA) (Young and Hartl, 2000; Sondermann et al., 2001; Young et al., 2003). Recombinant nontagged human p23 was in pET28 (Novagen) (Young and Hartl, 2000), and the sequence encoding human DJA1 (NM_001539) also in pET28a was a gift from C.H.I. Ramos (Laboratório Nacional de Luz Síncrotron, Campinas SP, Brazil) (Borges et al., 2005). The sequences encoding human Hsc70 (NM_006597), DJA2 (NM_005880), and DJA4 (NM_018602) as well as the sequences encoding amino acids 98-397 of human DJA1 (C-A1), amino acids 100-412 of human DJA2 (C-A2), and amino acids 99-397 of human DJA4 (C-A4) were amplified from a cDNA library and inserted into pProExHTa (Clontech). Sequences of DJA1, DJA2, and DJA4 were inserted into pGEM-11Z (Promega) without tags, and into pcDNA3.1 myc-His C (Invitrogen). Human Aha1 in pProExHTa was a gift from W.M.J. Obermann (Houston, TX) (Lotz et al., 2003). The sequence encoding firefly luciferase was amplified by PCR from pGL3 (Promega) and inserted into pcDNA3.1 (Invitrogen) without a tag; the pSV40-β-galactosidase vector was from Promega. The vectors pCAGGS-PiC-3HA, pGR-ΔLBD, and pMTV-luciferase (glucocorticoid responsive element [GRE]-luciferase) were as reported previously (Hollenberg et al., 1987; Hollenberg and Evans, 1988; Brychzy et al., 2003; Young et al., 2003).
Protein Purification
The expression vectors for full-length His-tagged DJA1, DJA2, and DJA4 were grown in Rosetta 2 Escherichia coli cells (Novagen), and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside at 30°C for 1 h, at 37°C for 30 min, and at 30°C for 3 h, respectively. The cells were harvested and resuspended in buffer containing 750 mM NaCl, 60 mM imidazole and 20 mM KH2PO4, pH 7.5, with Complete protease inhibitors (Roche Diagnostics, Indianapolis, IN). Cells were lysed by cavitation in a French press, and the cell debris was removed by centrifugation. The supernatant was loaded onto a 5-ml nickel-Sepharose high-performance column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and proteins were eluted with buffer containing 1 M imidazole, 500 mM NaCl, and 20 mM KH2PO4, pH 7.5. Peak fractions were loaded on a Superdex 200 Hi-Load 16/60 column (GE Healthcare) and eluted with buffer HS (500 mM NaCl, 20 mM HEPES-KOH, pH 7.5, and 5 mM MgOAc2). Nonaggregated peak fractions were collected, and the yield was determined by absorbance at 280 nm. The DJA proteins lacking the N-terminal J-domain, C-A1, C-A2, and C-A4, were purified identically to the full-length proteins. Hsc70 was expressed for 4 h at 30°C and purified on nickel-Sepharose high-performance equlibrated in buffer containing 500 mM NaCl, 20 mM imidazole, and 20 mM KH2PO4, pH 7.5, and eluted in buffer containing 300 mM imidazole and 20 mM KH2PO4, pH 7.5. Hsc70 was further purified by ion exchange on a Mono Q 5/50 GL column and gel filtration on a Superdex 200 Hi-Load 16/60 column (GE Healthcare) equilibrated in buffer CG (100 mM KOAc, 20 mM HEPES-KOH, pH 7.5, and 5 mM MgOAc2). Bovine ANT was purified as described previously (Klingenberg et al., 1979; Pebay-Peyroula et al., 2003), with the following modifications. Mitochondria were prepared from bovine heart and treated with atractyloside, and then they were solubilized in buffer AB (500 mM NaCl, 10 mM HEPES-KOH, pH 7.5, and 1 mM EDTA) containing 2% Triton X-100. Insoluble material was removed by ultracentrifugation at 140,000 × g for 30 min, and the supernatant was loaded onto a hydroxyapatite HTP-gel column (Bio-Rad, Hercules, CA) equilibrated in buffer AB containing 0.1% Triton X-100, and peak flow-through fractions containing ANT were collected. Recombinant C-90, C-Bag, Aha1, p23, and firefly luciferase were purified as described previously (Young and Hartl, 2000; Brychzy et al., 2003; Lotz et al., 2003; Young et al., 2003).
Chaperone–ANT Complexes
RL was desalted into buffer CG on NAP-10 columns or a Hi-Prep 26/10 fast desalting column (GE Healthcare). Purified ANT was chemically biotinylated on cysteine side chains with maleimide PEO2-biotin (Pierce Chemical, Rockford, IL) for 3 h at room temperature, and excess biotin was removed with NAP-10 columns equilibrated in buffer MT (100 mM NaCl, 10 mM HEPES-KOH, 1 mM EDTA and 0.1% Triton X-100). The biotinylated ANT (ANT-B) was bound to immobilized streptavidin-agarose (Pierce Chemical) for 1 h. The ANT-B was washed twice with buffer CG to remove stabilizing detergent, and it was reconstituted by incubation with 50% RL in buffer CG, supplemented with 2 mM ATP. After 15 min at room temperature, the reactions were terminated with 0.1 U/μl apyrase, grade VII. The reconstituted ANT-B complexes were reisolated, and the protein components were eluted with Laemmli loading buffer. After separation by Laemmli SDS-polyacrylamide gel electrophoresis (PAGE) and total protein staining with Coomassie Blue, bands were excised and subjected to in-gel tryptic digestion as described previously (Shevchenko et al., 1996). Peptides were analyzed by C18 reverse phase separation followed by ion trap liquid chromatography-tandem mass spectrometry on a Daltonics Esquire HCT+ instrument (Bruker, Newark, DE). Spectra were formatted and searched against mammalian sequences in the National Center for Biotechnology Information database by using the Mascot search engine (Matrix Science, Boston, MA). Data were validated manually.
For gel filtration analysis, ANT-B was bound to monomeric avidin agarose (Pierce Chemical) for 1 h, washed with buffer CG, and eluted with 1% SDS and 8 M urea. The eluted ANT-B was diluted 1:400 into 50% RL in buffer CG, supplemented with 2 mM ATP. After 15 min at room temperature, the reaction was terminated with apyrase as described above and loaded onto a Superose 6 10/300 GL column (GE Healthcare) equilibrated in buffer CG. Fractions were collected and separated by SDS-PAGE, transferred onto nitrocellulose, detected with horseradish peroxidase-conjugated streptavidin (Pierce Chemical), and visualized on chemiluminescent-sensitive film. In control reactions, cell-free translation reactions of ANT were performed using the TNT coupled RL system with SP6 polymerase (Promega), and they were labeled with [35S]methionine (GE Healthcare). Reactions were diluted 1:4 into buffer CG and analyzed by gel filtration as described above. Fractions were analyzed by SDS-PAGE and autoradiography. Phosphorimager quantitation was performed with a FujiFilm BAS-1800II analyzer (FujiFilm, Stamford, CT) and ImageGauge software (Fuji, Tokyo, Japan).
Mitochondrial Import Assays
Rat liver mitochondria were isolated and import reactions were performed as described previously (Lingelbach et al., 1986; Young et al., 2003; Fan et al., 2006). For cleaner mitochondrial preparations by gradient centrifugation (Vijayasarathy et al., 1989), mitochondria were layered over 12-ml steps of 42, 45, and 60% sucrose in 10 mM HEPES-KOH, pH 7.5, and 1 mM EDTA, in SW28 tubes (Beckman Coulter, Fullerton, CA). After centrifugation at 100,000 × g for 1 h, mitochondria were harvested from the 45–60% interface, diluted in the same buffer without sucrose, collected by centrifugation, and resuspended at 10 mg/ml in buffer MC (250 mM sucrose, 80 mM KOAc, 20 mM HEPES-KOH, pH 7.5, and 5 mM MgOAc2) supplemented with 10 mM Na-succinate, 1 mM dithiothreitol, 2 mM ATP, and 0.4 mM ADP. Cell-free translations of the preproteins were performed with the TNT-coupled RL system with SP6 or T7 polymerase supplemented with [35S]methionine, and then reactions were terminated with 1 mM methionine and adjusted to 250 mM sucrose. Typical import reactions contained 25% reticulocyte lysate and 5 mg/ml mitochondria at 30°C for 30 min, and negative control reactions were inhibited with 1 μM valinomycin. Mitochondria were reisolated and digested with 250 μg/ml proteinase K (PK) at 4°C for 10 min, followed by 1 mM phenylmethylsulfonyl fluoride to stop the digestion. Undigested and digested samples were analyzed by SDS-PAGE and autoradiography or phosphorimager analysis.
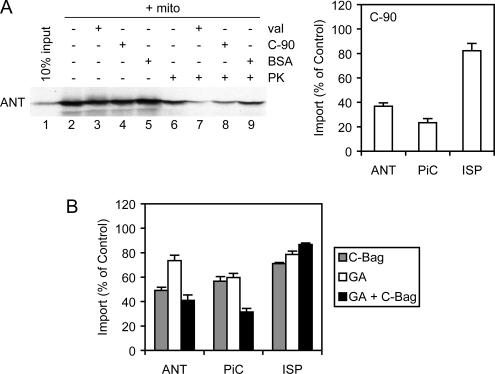
Inhibition of import by C-90, C-Bag, and geldanamycin (GA) was assayed as reported previously (Young et al., 2003; Fan et al., 2006). Purified C-90 and C-Bag were added to reactions at a final concentration of 20 and 5 μM, respectively. Then, 18 μM GA or the equivalent volume of vehicle control dimethyl sulfoxide (1%) was added to the translation reactions, and excess GA was removed before import reactions by using MicroBioSpin 6 columns (Bio-Rad) pre-equilibrated in buffer MC containing 0.4 mM EGTA, centrifuged at 16°C with 2 mM ATP added to the collection tube. Inhibition of import by C-A1, C-A2, and C-A4 was assayed as for C-90, with each mutant added at 20 μM, and reactions adjusted to a final concentration of 90 mM NaCl in buffer MC to maintain solubility of the mutants. Control reactions adjusted to 90 mM NaCl with no additional protein showed that the added ionic strength had no significant effect on import.
Purified ANT was radiolabeled with Na-[125I] (GE Healthcare) by using Iodo-Beads (Pierce Chemical) according to the manufacturer's instructions. 125I-ANT was desalted using a NAP-10 column in buffer MT to remove excess radiolabel, concentrated by trichloroacetic acid precipitation, and resuspended in 0.1% SDS. Import was initiated by 1:100 dilution into reactions containing 4 mg/ml mitochondria and 40% RL in buffer MC, supplemented with 2 mM succinate, 0.2 mM dithiothreitol, 2 mM ATP, and 0.4 mM EGTA. To assay import of ANT-B, the biotinylated protein was recovered with monomeric avidin agarose as described for gel filtration analysis. The eluted ANT-B was diluted 1:10 into distilled water and then a further 1:100 dilution into reactions containing RL and mitochondria as described for 125I-ANT. Control reactions using cell-free translated ANT showed that the final concentration of 0.001% SDS did not interfere with mitochondrial import in any way.
Coprecipitation Experiments
To assay binding of preproteins to the DJA proteins, an assay established for Tom70 (Young et al., 2003; Fan et al., 2006) was adapted. DJA1, DJA2, and DJA4 or the truncation mutants C-A1, C-A2, and C-A4 were prebound on nickel-Sepharose in buffer HS for 30 min at 4°C. Cell-free translations of ANT and PiC were performed as described above with SP6 polymerase, diluted 1:20 into buffer CG containing 20 mM imidazole, 0.1% Triton X-100, and 2 mg/ml ovalbumin, and added to the immobilized DJA proteins. The final reactions contained 5 μM wild-type DJA protein or 10 μM truncation mutant, and 5% translation mixture. After 5 min at room temperature, the binding reactions were terminated by the addition of 0.1 U/μl apyrase. The protein complexes were recovered at 4°C for 30 min, and then they were washed with buffer CG containing 20 mM imidazole and 0.1% Triton X-100. Protein complexes were eluted with Laemmli loading buffer, and they were analyzed by SDS-PAGE and autoradiography or phosphorimager quantitation. To form DJA heterocomplexes, ANT-B was diluted into 50% RL as described for the gel filtration analysis, but binding reactions were supplemented 1:20 with cell-free translations of each nontagged DJA. The reactions were then coprecipitated with different His-tagged DJAs as described above.
To assay formation and dissociation of Hsc70, Hsp90, and DJA complexes with ANT, methods established for the ligand binding domain of the glucocorticoid receptor (Young and Hartl, 2000; Sondermann et al., 2001; Brychzy et al., 2003) were adapted. To assay binding, ANT-B was prebound to streptavidin-agarose and washed with buffer CG as described above. Cell-free translation reactions of Hsc70, Hsp90, or each DJA were performed as described above with T7 or SP6 polymerase, diluted 1:10 or 1:14 into 40% RL in buffer CG containing 2 mM ATP, and added to the immobilized ANT-B. Aliquots were taken at the indicated time points, and complexes were reisolated and washed with buffer CG containing 0.1% Triton X-100. Protein complexes were eluted and analyzed as described above. Where indicated, binding reactions were performed in the presence of 20 μM C-A1, C-A2, C-A4 or C90, or 4 μM Aha1. To assay dissociation of complexes, cell-free translation reactions of Hsc70 and Hsp90 were complexed with ANT-B as described above for 15 min at room temperature, and complexes were reisolated and washed with buffer CG. Dissociation reactions consisting of buffer CG with or without 2 mM ATP, 4 μM Aha1, 5 μM C-Bag, 5 μM p23, or 20 μM C-90 were incubated with the complexes for 10 min at room temperature. Released and remaining immobilized fractions were separated and analyzed by SDS-PAGE and autoradiography or phosphorimager quantitation.
Hsc70 Activity
ATPase activities were measured as described previously (Young and Hartl, 2000; Brychzy et al., 2003). Briefly, reactions contained 4 μM Hsc70, 20 μM C-Bag, and the indicated concentration of each DJA protein, in buffer CG supplemented with 78 mM NaCl to maintain solubility of the DJA proteins, 4 mM ATP, and 5 μCi/ml α-[32P]ATP. Reactions were assembled on ice and initiated by incubation at 30°C. At various times, aliquots were removed and terminated by adjusting to 25 mM EDTA. Samples were separated by thin layer chromatography (TLC) on polyethylene-imine cellulose (Mallinckrodt Baker, St. Louis, MO) developed in 0.5 M LiCl and 0.5 M formic acid. ADP produced was quantified by phosphorimager analysis and linear rates calculated.
To assay refolding of firefly luciferase, the protein was denatured in buffer CG containing 6 M guanidinium-HCl and 1 mM dithiothreitol for 15 min at room temperature. Refolding reactions were preassembled on ice, containing 4 μM Hsc70, 4 μM DJA protein, and 0.5 μM C-Bag, either with 1 mM ATP or treated with 0.1 U/μl apyrase, in buffer CG supplemented with 39 mM NaCl, and warmed to 30°C immediately before use. Luciferase was diluted 1:100 into reactions to a final concentration of 5.4 nM and incubated at 30°C. Control reactions contained 50% RL in buffer CG supplemented with 39 mM NaCl and 1 mM ATP. At various times, aliquots were diluted 1:45 into luciferase assay reagent (Promega), and luciferase activity was measured in a Lumat LB 9507 luminometer (Berthold Technologies, Bad Wildbad, Germany).
Cell Culture
HeLa cells were maintained in DMEM containing 4.5 g/l glucose, 36 mg/l pyruvate, 2 mM glutamine, and 10% fetal bovine serum (Invitrogen). Cells were transfected using Lipofectamine and PLUS Reagent (Invitrogen) in six-well plates with 4 μg of myc-His-tagged DJA plasmid or vector alone, 0.5–1.5 μg of pSV40-β-galactosidase, and 0.5 μg of either pcDNA3.1-luciferase, or pCAGGS-PiC-3HA, or pMTV-luciferase with 0.5 μg of pGR-ΔLBD. Two days after transfection, cells were harvested and lysed by freeze-thaw in β-galactosidase Reporter lysis buffer (Promega). Lysates were cleared by centrifugation at 20,000 × g for 10 min and assayed for β-galactosidase and luciferase activities with the appropriate kits (Promega). Equal amounts of lysate were analyzed by immunoblots. The immunoblots were quantified with a FujiFilm LAS-1000 CH luminescent image analyzer and the ImageGauge software.
RESULTS
Mitochondrial Import of ANT
Mammalian ANT is an inner membrane metabolite carrier protein, and it was expected to follow the chaperone–Tom70 mitochondrial import pathway (de Marcos-Lousa et al., 2006), as has been established for PiC. Like many such carriers, ANT lacks a signal sequence, and it contains targeting information within its mature sequence, consistent with Tom70 dependence. ANT is composed almost entirely of six transmembrane helices, and its hydrophobicity suggested reliance on chaperones. Our previous work also showed that the Hsp90 inhibitor novobiocin blocked import of ANT, consistent with strong chaperone involvement (Fan et al., 2006). However, the relative importance of Hsc70 and Hsp90 to ANT import remained unknown, and this was addressed using previously established assays.
Both Hsc70 and Hsp90 dock onto a juxtamembrane TPR clamp domain of Tom70, a key step in the targeting of chaperone-bound preproteins. The C-terminal 16-kDa fragment of Hsp90 (C-90), containing the TPR clamp domain recognition motif, competes with Hsc70 and Hsp90 for binding to Tom70, thereby blocking Tom70-dependent import (Young et al., 2003). The import of ANT was therefore tested in the presence of excess C-90 competitor. Radiolabeled ANT was generated by cell-free translation in RL, and it was imported into isolated rat liver mitochondria. ANT is not proteolytically processed upon import, so the extent of import was assessed by resistance to externally added PK. Imported ANT was resistant to digestion (Figure 1A, lanes 2 and 6), but when import was abolished by valinomycin disruption of the inner membrane potential, PK resistance was reduced to a low background level (Figure 1A, lanes 3 and 7). Addition of the C-90 competitor also reduced the amount of imported ANT, although not to the valinomycin background, whereas the addition of the control protein serum albumin had no effect (Figure 1A, lanes 8 and 9). Quantitation revealed that C-90 impaired ANT import to below 40% of control reactions (Figure 1A). This effect was similar to that observed for Tom70-dependent PiC (Young et al., 2003), whereas import of the Tom20-dependent Rieske ISP was only marginally affected. ANT thus seems to follow the Tom70 import route.
Figure 1.
Mitochondrial import of ANT. (A) ANT (input, lane 1) was radiolabeled by cell-free translation and imported into isolated rat liver mitochondria (+ mito, lanes 2–9). As a negative control, import was abolished with valinomycin (val, lanes 3 and 7). The true extent of import was determined by resistance to added PK (lanes 6–9). Import was assayed in the presence of 20 μM C-90 fragment, a competitor of chaperone-Tom70 interactions (C-90, lanes 4 and 8), or the same mass concentration of bovine serum albumin as a negative control (BSA, lanes 5 and 9). Reactions were analyzed by SDS-PAGE and autoradiography. Right, phosphorimager quantitation of import of ANT, PiC, and ISP are shown relative to control reactions. Standard deviations here, and in all figures, were determined from at least three independent experiments. (B) Import of ANT, PiC, and ISP was assayed upon inhibition with 5 μM C-Bag, treatment with 18 μM GA, or combined inhibition with both reagents, and the quantitation relative to control reactions is shown.
The individual contributions of Hsc70 and Hsp90 to ANT import were tested. Hsc70 can be dysregulated by an excess of its nucleotide exchange factor, the C-terminal domain of Bag-1 (C-Bag), which promotes the release of polypeptide substrate from Hsc70 (Sondermann et al., 2001; Mayer and Bukau, 2005). C-Bag inhibited the import of ANT to ∼50% of control reactions (Figure 1B), and as established previously (Young et al., 2003), PiC import was inhibited to ∼60%. The specific Hsp90 inhibitor GA (Pearl and Prodromou, 2006) was used to block Hsp90 function during import, and it partially diminished ANT import to ∼72% of the control (Figure 1B). As reported previously (Young et al., 2003), GA inhibition of PiC import was also partial, to ∼60% of the control (Figure 1B). Most importantly, combined inhibition of Hsc70 and Hsp90 by C-Bag and GA further reduced ANT import to ∼40% of the control, similar to the established level of ∼30% observed for PiC. In contrast, combined C-Bag and GA treatment had no inhibitory effect on ISP import (Figure 1B). In PiC, the combined effect of Hsc70 and Hsp90 inhibition indicated that the chaperones substitute for each other when one chaperone is inhibited (Young et al., 2003). For ANT, combined chaperone inhibition had an effect closer to that of Hsc70 inhibition alone, suggesting that ANT is more dependent on Hsc70 relative to Hsp90 for its import. Combined chaperone inhibition blocked the import of ANT to the same level as C-90 competition for Tom70 (Figure 1, A and B), as expected from the chaperone docking mechanism.
Reconstitution of Chaperone–ANT Complexes
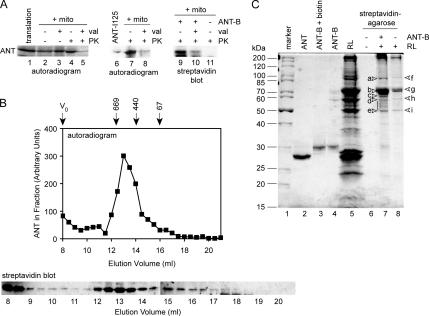
ANT can be purified in biochemical amounts from bovine heart (Pebay-Peyroula et al., 2003). Mass spectrometry analysis determined that the purified protein was identical in mass to that predicted by the cDNA sequence (data not shown), indicating that mature ANT should be reverted to its preprotein state by denaturation. Dilution of denatured ANT into RL should then reconstitute the chaperone–preprotein complexes involved in targeting. We confirmed that purified ANT could indeed be reimported into isolated mitochondria. First, purified ANT was radiolabeled by tyrosine iodination (Figure 2A, lane 6) and diluted into import reactions containing RL and mitochondria. After PK digestion, PK-resistant ANT was observed significantly above that in reactions treated with valinomycin (Figure 2A, lanes 7 and 8). The PK-resistant material in the valinomycin control reactions was similar to that normally observed in reactions using cell-free translated ANT (Figure 2A, lanes 1–5). In a second approach, purified ANT was chemically biotinylated on its cysteine side chains (Figure 2C, lanes 2–4, Coomassie stain). Import of ANT-B after dilution into reactions containing RL and mitochondria was detected by streptavidin blots (Figure 2A, lanes 9–11). A number of high-molecular-weight background bands were observed due to endogenous biotinylated mitochondrial proteins (data not shown). Nevertheless, imported ANT-B was detected above the valinomycin background after PK digestion (Figure 2A, lanes 9 and 10), and mock reactions lacking ANT-B showed no corresponding band (Figure 2A, lane 11). Thus, the purified ANT can be reimported into mitochondria, even after chemical modification.
Figure 2.
Reconstitution of chaperone–ANT complexes. (A) Left, a representative reaction of cell-free translated ANT (lane 1) imported into mitochondria (+ mito, lanes 2–5) as in Figure 1 is shown. Middle, purified ANT was radioiodinated (ANT-I125, lane 6) and diluted into reactions containing RL, ATP, and mitochondria (lanes 7 and 8). Right, purified ANT was biotinylated (ANT-B) and diluted into reactions containing RL, ATP, and mitochondria (lanes 9 and 10), whereas mock reactions contained no ANT-B (lane 11). Negative control reactions were treated with valinomycin (val) to abolish import (lanes 3, 5, 8, and 10). The extent of import was assessed by resistance to PK (lanes 4, 5, 7, 8, and 9–11). (B) Top, ANT was radiolabeled by cell-free translation and resolved on a Superose 6 10/300 GL column. The amount of ANT in each fraction was determined by SDS-PAGE and phosphorimager quantitation, and arbitrary phosphorimager units are represented. The column void volume (V0) and elution volumes of molecular size standards (indicated in kilodaltons) are marked. Bottom, ANT-B was diluted into reactions containing RL and ATP and resolved on the same gel filtration column. The ANT-B in each fraction was detected by SDS-PAGE and streptavidin blots. The elution volume corresponding to the fractions is marked (indicated in milliliters). (C) Molecular weight markers (indicated in kilodaltons) are shown (lane 1) and purified ANT (lane 2), ANT after the biotinylation reaction (ANT-B, lane 3) and removal of excess biotin by desalting (lane 4). ANT-B bound to streptavidin-agarose was incubated in reactions containing RL (lane 5) and ATP. Complexes were recovered and eluted with Laemmli loading buffer (lane 7). Negative control reactions contained no ANT-B or RL (lane 6) or no ANT-B (lane 8). Discrete bands labeled a, b, c, and the range between d and e, were analyzed by mass spectrometry. Bands in the negative control reactions labeled f, g, h, and i also were analyzed.
Chaperone complexes with Tom70-dependent preproteins in RL have a characteristic profile on gel filtration chromatography. As reported previously (Young et al., 2003; Fan et al., 2006), cell-free translated ANT eluted in a broad peak at ∼13-ml elution volume on a 24-ml Superose 6 column, with an apparent molecular size of ∼600 kDa (Figure 2B). Some ANT was also observed in the void volume of the column (8-ml elution volume), most likely representing aggregated material (exclusion limit ∼5 MDa). In parallel, ANT-B was diluted into chaperone binding reactions containing RL and ATP for 15 min at room temperature (Young and Hartl, 2000; Brychzy et al., 2003), and then it was analyzed on the same gel filtration column. Streptavidin blots detected a similar peak eluting around 13 ml (Figure 2B), in addition to void volume material. The reconstituted ANT-B complexes, therefore, reproduce those normally studied by cell-free translation as preimport intermediates, in molecular size as well as import competence.
To identify the components of chaperone–ANT complexes, ANT-B (Figure 2C, lane 4) was incubated with RL (Figure 2C, lane 5) as described above but in larger reactions. ANT-B complexes were then isolated with streptavidin-agarose and analyzed by total protein staining (Figure 2C, lane 7). Several bands were visible in the 40- to 100-kDa range. Very little ANT-B was observed, because the streptavidin–biotin interaction was mostly resistant to the SDS-containing buffer used to elute the beads. The protein pattern was clearly distinct from that of the RL (Figure 2C, lane 5). Mock reactions lacking ANT-B returned far fewer bands (Figure 2C, lane 8), and no bands were detected with streptavidin-agarose alone (Figure 2C, lane 6). Visible bands were subjected to tryptic digestion followed by mass spectrometry identification of peptides. Relevant proteins identified are summarized in Table 1. Although the exact rabbit sequences for many of the proteins are not known, the very high degree of conservation between chaperones and cochaperones within mammals (>97% identity) allowed unambiguous identification based on human, rat, mouse or pig sequences. Accession numbers are for human sequences, unless otherwise indicated.
Table 1.
Proteins identified by mass spectrometry from reconstituted chaperone—ANT complexes (Figure 2C, lanes 7 and 8)
| Name | Gene | Protein accession no. | Molecular weight (kDa) | Peptides identified | Sequence coverage (%) |
|---|---|---|---|---|---|
| Hsp90α | HSPCA | NP_005339 | 84.6 | 24 | 21 |
| Hsp90β | HSPCB | NP_031381 | 83.2 | 17 | 22 |
| Hsc70 | HSPA8 | NP_006588 | 70.8 | 63 | 40 |
| Hop | STIP1 | NP_006810 | 62.6 | 12 | 17 |
| Tpr2 | DNAJC7 | NP_003306 | 55.4 | 5 | 7 |
| DJA2 | DNAJA2 | NP_005871 | 45.7 | 6 | 16 |
| DJA4 | DNAJA4 | NP_061072 | 44.8 | 14 | 22 |
| DJA1 | DNAJA1 | NP_001530 | 44.8 | 3 | 14 |
| Hip | ST13 | NP_598487 | 41.6 | 11 | 14 |
Accession numbers are for human sequences, except Hip for which the mouse sequence was matched. Predicted molecular weights shown are usually lower than apparent molecular weights by SDS-PAGE.
The band at ∼90 kDa (Figure 2C, lane 7, labeled a) was identified as a mixture of Hsp90α and Hsp90β, the two highly similar cytosolic forms of the chaperone. The heavy band ∼70 kDa (labeled b) was found to be Hsc70. The greater amount of Hsc70 compared with Hsp90 is in agreement with higher dependence on Hsc70 for import (Figure 1B). The bands below 70 kDa (labeled c, and a range between d and e) contained a number of cochaperones of Hsc70 and Hsp90. Samples from regions of the gels below the 40-kDa molecular weight did not return reliably identifiable peptides except for ANT itself. The major visible bands in negative control reactions (Figure 2C, lane 8, labeled f, g, h, and i) were also analyzed, and they were found to be exclusively keratins and hemoglobin multimers (keratin 1, NP_006112; keratin 10, NP_000412; keratin 9, NP_000217; and hemoglobin β, NP_000509). These were unavoidable background proteins and ruled out as chaperone complex components. Importantly, no peptides from the chaperone-related components were identified in the negative controls.
Components of the Chaperone–ANT Complexes.
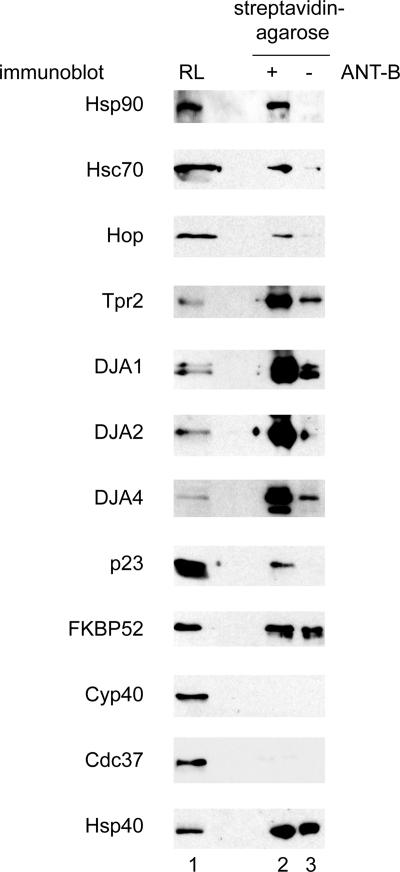
In addition to the Hsc70 and Hsp90 chaperones, several cochaperone proteins were found (Table 1). Hop recognizes Hsc70 and Hsp90 through two separate TPR domains. It is thought to coordinate the transfer of substrates from Hsc70 to Hsp90, which is the major pathway for many Hsp90-dependent substrates (Chen and Smith, 1998; Scheufler et al., 2000; Pratt and Toft, 2003; Young et al., 2004). Hop is found in many Hsp90–substrate complexes, and its presence in complexes with ANT was not unexpected. Tpr2 binds both Hsc70 and Hsp90 through its two TPR domains, and it activates the Hsc70 ATPase through its J-domain. It has been proposed to return substrates from Hsp90 to Hsc70, and its expression level in cells optimized the maturation of the Hsc70- and Hsp90-dependent glucocorticoid receptor (Brychzy et al., 2003; Young et al., 2004). The presence of Tpr2 in ANT complexes suggests that it has a similar role to optimize the balance between Hsc70 and Hsp90, in this case favoring Hsc70. Furthermore, three distinct Hsp40-related J-domain cochaperones were found. DJA1 (Hdj2, HSDJ) and DJA2 (Hdj3, HIRIP4) have been reported to be the constitutively expressed members of the Hsp40 family (Terada and Mori, 2000). DJA4 (Hdj4) has been only recently discovered, and it has been hypothesized to be functionally specialized (Hafizur et al., 2004). Mouse DJA4 is highly expressed in testis, but we have also identified it in other tissues (data not shown). The presence of these proteins in the chaperone–preprotein complexes was tested directly by immunoblots (Figure 3). The main components described above were detected in the reconstituted ANT-B complexes: Hsp90 and Hsc70 as well as Hop, Tpr2, DJA1, DJA2, and DJA4, confirming the mass spectrometry data.
Figure 3.
Components of chaperone–ANT complexes. Complexes associated with ANT-B were reconstituted in RL and coprecipitated as in Figure 2C, and then they were analyzed by immunoblots with antibodies against the indicated proteins. Samples shown are total RL (lane 1), ANT-B complexes (lane 2), and negative control reactions lacking ANT-B (lane 3).
Importantly, many other known cochaperones of Hsp90 and Hsc70 were not detected by mass spectrometry. These included the immunophilin FKBP52, the cyclophilin Cyp40 and the kinase-binding Cdc37, all of which have been reported to have general chaperone activity, in the sense of unfolded polypeptide binding (Bose et al., 1996; Freeman et al., 1996; Kimura et al., 1997). Also, the Hsp90 cochaperone p23 (Pearl and Prodromou, 2006), which stabilizes the substrate-binding ATP-bound state of Hsp90, was not detected. p23 is common to many Hsp90 complexes, and it has intrinsic chaperone activity. The presence or absence of these proteins was tested independently by immunoblots. First, immunoblots unambiguously detected p23 in chaperone–ANT complexes but not in negative control reactions (Figure 3). It is possible that the small size (predicted molecular weight 18 kDa) and strong acidic charge of p23 limited the identifiable tryptic peptides derived from the protein or that the poorly staining protein was simply missed in the excision from gels. Next, immunoblots detected FKBP52 at similar low levels in both the ANT complexes and the negative control reactions (Figure 3), suggesting it was nonspecifically bound and therefore not a major component of the ANT complex. Cyp40 and Cdc37 were not detected at all in the ANT-B complexes, supporting the mass spectrometry data. Hsp40 (DJB1, Hdj1), the stress-inducible J-domain cochaperone that is structurally divergent from the DJAs (Cintron and Toft, 2006; Qiu et al., 2006) also seemed to be nonspecifically bound.
The only cochaperone positively identified by mass spectrometry that could not be validated by immunoblots was Hip, possibly due to the quality of the antibody (data not shown). Hip stabilizes the ADP-bound form of Hsc70, but it is mainly thought to counterbalance the Hsc70 nucleotide exchange factors (Hohfeld et al., 1995; Mayer and Bukau, 2005). A number of subunits of the TCP-1 chaperonin (TRiC, CCT) were matched to low numbers of peptides in the mass spectrometry analysis, and their presence could not be validated by immunoblots (data not shown). Several proteasome subunits were similarly identified, but they likely represent clearance of nonproductively targeted preprotein and they were not pursued. In summary, we propose that the proteins in Table 1, with the addition of p23 and exception of Hip, comprise the major chaperones and cochaperones involved in Tom70-dependent import. Furthermore, the exclusion of certain proteins from the chaperone–preprotein complexes suggests a substantial degree of specificity in complex formation.
DJA Interaction with Preprotein
We were struck by the presence of three related DJA proteins in the chaperone–ANT complexes. Although all three were present, it was possible that one in particular might be more effective than the others. For example, the higher number of peptides derived from DJA4 might suggest a greater involvement of that cochaperone with preprotein complexes. Alternatively, there might be subtler differences between them, such that they function together in import, and perhaps other biological processes. In the yeast S. cerevisiae, the two cytosolic Hsp40-related proteins Ydj1 and Sis1 have distinct biological roles (Johnson and Craig, 2001; Fan et al., 2004); however, Ydj1 is orthologous to the three human DJA proteins, whereas Sis1 is related to the divergent Hsp40/DJB1. The homology between the human DJAs (70–85% similarity) suggests they have a closer functional relationship. The DJA proteins contain the J-domain, zinc finger domain, and C-terminal dimerization domain (Figure 4C, diagram) conserved from the eponymous DnaJ of E. coli (Terada et al., 1997; Terada and Mori, 2000; Borges et al., 2005; Qiu et al., 2006). As expected, their N-terminal J-domains were found to activate ATP hydrolysis by Hsc70 (Terada and Mori, 2000; Hafizur et al., 2004). Based on experiments with the E. coli and S. cerevisiae orthologues, the binding site for unfolded polypeptides should reside in the central- to C-terminal regions of the DJA proteins (Walsh et al., 2004; Mayer and Bukau, 2005; Cintron and Toft, 2006; Qiu et al., 2006). Like most J-domains, those in the DJA proteins are well conserved, and the greatest divergence in sequence is within the expected substrate binding region. This suggested that one difference between the DJAs might be the range of substrates bound, or the strength of binding. However, sequences outside the J-domains of some proteins have been implicated in Hsc70 interactions, so there might also be differences between the DJAs in the activation of Hsc70. We therefore addressed these possibilities systematically.
Figure 4.
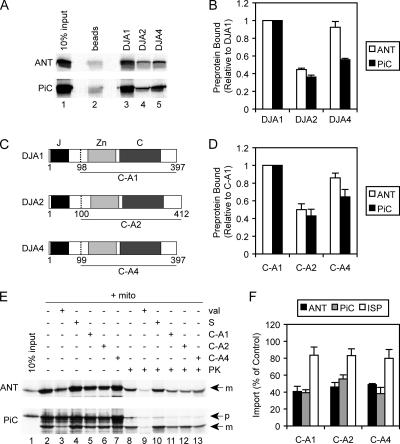
DJA interaction with preprotein. A, ANT, and PiC were radiolabeled by cell-free translation (lane 1) and coprecipitated with nickel-Sepharose alone (beads, lane 2), or with nickel-Sepharose and purified His-tagged DJA1, DJA2, or DJA4 at a final concentration 5 μM (lanes 3–5). Recovered material was analyzed by SDS-PAGE and autoradiography. (B) Phosphorimager quantitation is shown of the coprecipitation of ANT and PiC with DJA1, DJA2 and DJA4, as in described in A, relative to that with DJA1 set to 1. (C) Diagram of DJA1, DJA2, and DJA4 architecture. J-domain (J, black), zinc finger (Zn, light gray), and C-terminal domains (C, dark gray) are indicated. The start sites of C-A1, C-A2, and C-A4 deletion mutants are marked (dashed line, amino acid numbers indicated). (D) ANT and PiC were coprecipitated with purified His-tagged C-A1, C-A2, and C-A4 as described in A. Phosphorimager quantitation of the coprecipitation is shown, relative to that with C-A1 set to 1. (E) Cell-free translated ANT and PiC (lane 1) were imported into mitochondria (+ mito, lanes 2–13) as described in Figure 1. As a negative control, import was abolished with valinomycin (val, lanes 3 and 9). Reactions were supplemented with 90 mM NaCl alone (S, lanes 4 and 10) or with 20 μM C-A1, C-A2, or C-A4 with 90 mM NaCl (lanes 5–7 and 11–13). The extent of import was assessed by resistance to PK (lanes 8–13). The mature noncleaved form of ANT (m) is visible, as are the precursor (p) and proteolytically cleaved mature (m) forms of PiC. (F) Phosphorimager quantitation is shown of the import of ANT, PiC, and ISP in the presence of 20 μM C-A1, C-A2, or C-A4, as described in E, relative to control reactions with added NaCl.
The binding of the DJAs to the polypeptide substrate ANT was tested first. The polypeptide binding specificities of the human DJAs are so far unknown. For ease and accuracy of quantitation, we adapted a coprecipitation assay used to study preprotein interactions with Tom70 (Young et al., 2003; Fan et al., 2006). DJA1, DJA2, and DJA4 were purified as N-terminally His-tagged proteins. Radiolabeled ANT produced by cell-free translation was incubated with identical amounts (5 μM) of each DJA protein. The His-tagged proteins were reisolated with nickel-Sepharose, and the associated ANT was quantified. All three DJAs bound ANT at levels above the low background binding, but DJA2 bound noticeably less preprotein than DJA1 or DJA4 (Figure 4A, lanes 2–5). Quantitation showed that DJA2 binding of ANT was ∼40% of DJA1 binding, whereas DJA4 binding was not significantly different from that of DJA1 (Figure 4B). For comparison with another Tom70-dependent preprotein, the assay was applied to PiC (Figure 4, A and B). Again, DJA1 bound PiC well, and DJA2 seemed to have the poorest binding, at ∼40% of DJA1. However, DJA4 bound PiC relatively poorly, at <60% of DJA1 binding. These data suggested that the DJAs differ in their binding of polypeptides.
The full-length DJA proteins might bind polypeptides in a dynamic equilibrium, because their J-domains promote the transfer of polypeptides to Hsc70. The association of ANT and PiC with the DJAs might then depend on interactions with Hsc70 as well as the actual affinity of binding to the DJAs. To directly address the level of polypeptide binding, truncation mutants of the DJAs lacking the N-terminal J-domain were purified. The deletion mutants, C-A1 (residues 98-397 of DJA1), C-A2 (residues 100-412 of DJA2), and C-A4 (residues 99-397 of DJA4), were also N-terminally His-tagged, and they contained the complete zinc finger and C-terminal regions of the DJAs (Figure 4C). The binding of cell-free translated ANT and PiC to the mutants was tested as described above. C-A1 was found to bind strongly to both ANT and PiC, and C-A2 binding to both polypeptides was <50% of C-A1 binding (Figure 4D). C-A4 bound slightly less ANT than C-A1 and significantly less PiC, at ∼60% relative to C-A1. Overall, the interactions of the truncation mutants with polypeptides were similar to those of the full-length DJAs, suggesting that Hsc70 interactions did not greatly change the substrate binding profiles of full-length versus truncated DJAs. These results confirmed that the DJAs diverge in polypeptide binding, with DJA1 apparently binding the highest amount of preprotein. Also, as observed for DJA4, different polypeptides can be bound to varying degrees.
We wanted to evaluate the importance of DJA binding to preprotein for mitochondrial import. Immunodepletion of the DJAs from RL could not be achieved with the available antibodies (data not shown). So, we reasoned that the truncated DJAs should act as dominant-negative inhibitors of wild-type DJA function, by competing for the same binding sites in preprotein but disallowing transfer to Hsc70. Inhibition of Hsc70 binding should then impair the Tom70 docking step required for import. Alternatively, Hsp40 can activate substrate binding by Hsc70 without strongly interacting with substrate (Minami et al., 1996; Cintron and Toft, 2006). If Hsp40 or some mechanism other than the DJAs was sufficient for Hsc70 binding, the truncated DJAs should have less effect. Therefore, C-A1, C-A2, and C-A4 were added at 20 μM to mitochondrial import reactions of ANT and PiC. Note that PiC is proteolytically processed upon import to produce a faster migrating band on SDS-PAGE (Zara et al., 1992), but the extent of true import was quantified as described for ANT by resistance to externally added PK (Figure 4E). Indeed, all three mutants clearly inhibited import of ANT and PiC to between 40 and 60% of control reactions (Figure 4F). The partial but significant inhibition of import approached the level observed with the C-90 competitor or combined Hsc70 and Hsp90 inhibition in Figure 1. As expected, the mutant DJAs had little effect on the Tom20-dependent import of ISP (Figure 4F). For ANT and PiC, it seems that the effect of each dominant DJA mutant cannot be overcome by the other DJA proteins or J-domain cochaperones present in the lysate. These data suggest that each of the DJAs function in the Tom70 import pathway.
Hsc70–Preprotein Interactions
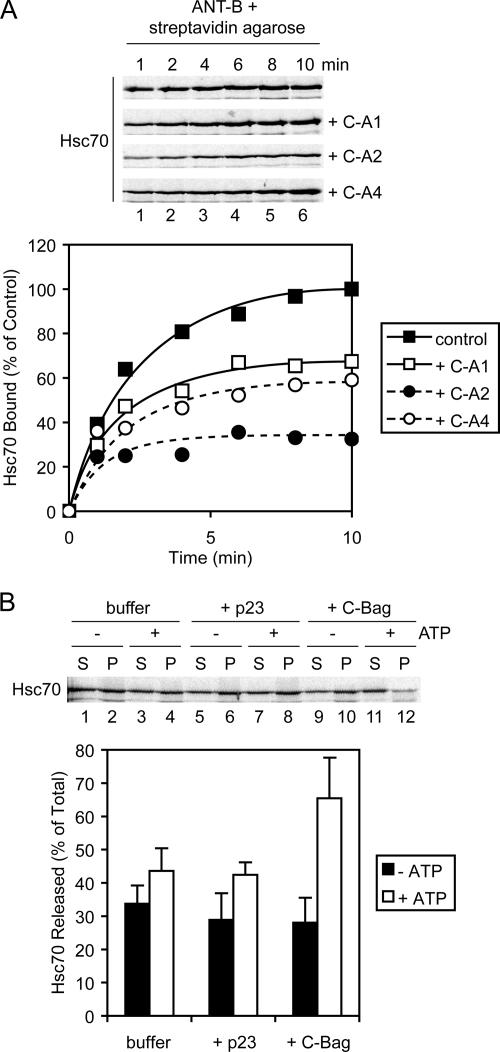
To further test the importance of the DJAs for Hsc70 binding to preprotein, the binding of Hsc70 to purified ANT was monitored over time. As established in previous work on Hsc70 and Hsp90 interactions with the glucocorticoid receptor (Young and Hartl, 2000; Sondermann et al., 2001; Brychzy et al., 2003), radiolabeled Hsc70 generated by cell-free translation was used for ease and accuracy of quantitation. Although unlabeled Hsc70 is in excess in the binding reactions, the radiolabeled protein acts as a tracer and matches the behavior of unlabeled protein. At different times after addition to ANT-B, the preprotein was recovered with streptavidin-agarose, and bound Hsc70 was quantified. Hsc70 bound with a fast initial increase and reached a maximum at ∼10 min (Figure 5A). When C-A1, C-A2, and C-A4 were added to the reactions at 20 μM, binding of Hsc70 was clearly diminished (Figure 5A). Moreover, C-A2 was the most inhibitory, reducing Hsc70 binding as early as 1 min into the reaction, with a final level of ∼30% of the control reaction. C-A1 and C-A4 were slightly more moderate in their effect, with inhibition visible by 2–4 min and a final Hsc70 level of ∼60% of the control. Thus, the three truncation mutants directly inhibit Hsc70 interactions with preprotein, albeit to different degrees.
Figure 5.
Hsc70-preprotein interactions. (A) Top, Hsc70 was radiolabeled by cell-free translation, incubated with ANT-B immobilized on streptavidin-agarose, and coprecipitated after the indicated times. Reactions contained either no addition, or 20 μM purified C-A1, C-A2, or C-A4. Recovered material was analyzed by SDS-PAGE and autoradiography. Bottom, quantitation of time courses of Hsc70 binding is shown. (B) Top, cell-free translated Hsc70 was coprecipitated with ANT-B as described in A but after 15 min of binding. Samples were resuspended in dissociation reactions with or without ATP, containing buffer alone, or 5 μM purified p23 or C-Bag. After 10 min, released and remaining bound fractions were separated and analyzed. Bottom, quantitation of dissociation reactions.
The dominant mutants seem to compete with wild-type DJAs for polypeptide, preventing the wild-type cochaperones from initiating Hsc70 binding (Cintron and Toft, 2006). The partial effects of the mutants suggested that competition with the wild-type DJAs was not complete. This in turn implied that the DJAs have distinct sets of binding sites that only partly overlap with each other. Interestingly, C-A2 was the strongest inhibitor of Hsc70 binding (Figure 5A), but it seemed to bind less overall preprotein than C-A1 or C-A4 (Figure 4D). We suggest that C-A2 might bind fewer sites within the polypeptides, but these sites are relatively more important for Hsc70 binding. In contrast, C-A1 and C-A4 might bind more, but less distinctly important, sites. The overall result is that the three mutants inhibit preprotein import to a similar degree (Figure 4F).
To confirm that the Hsc70–preprotein complexes observed in these experiments were formed normally, their nucleotide dependence was assayed. In previous experiments with the glucocorticoid receptor, the C-Bag nucleotide exchange factor promoted Hsc70 dissociation from isolated complexes in an ATP-dependent manner (Sondermann et al., 2001). Following a similar approach, Hsc70–ANT complexes were isolated after 15 min of binding, and then they were incubated under varying conditions in the absence or presence of ATP. Dissociated and bead-bound fractions were analyzed separately. As observed previously, ATP alone did not promote complex dissociation greatly over buffer without ATP (Figure 5B). As another negative control, the purified Hsp90-specific cochaperone p23 also did not strongly dissociate Hsc70. However, C-Bag markedly promoted Hsc70 dissociation but only in the presence of ATP (Figure 5B). Thus, the Hsc70–preprotein complexes inhibited by the DJA mutants seem to be normally functional complexes.
DJA Heterocomplex Formation
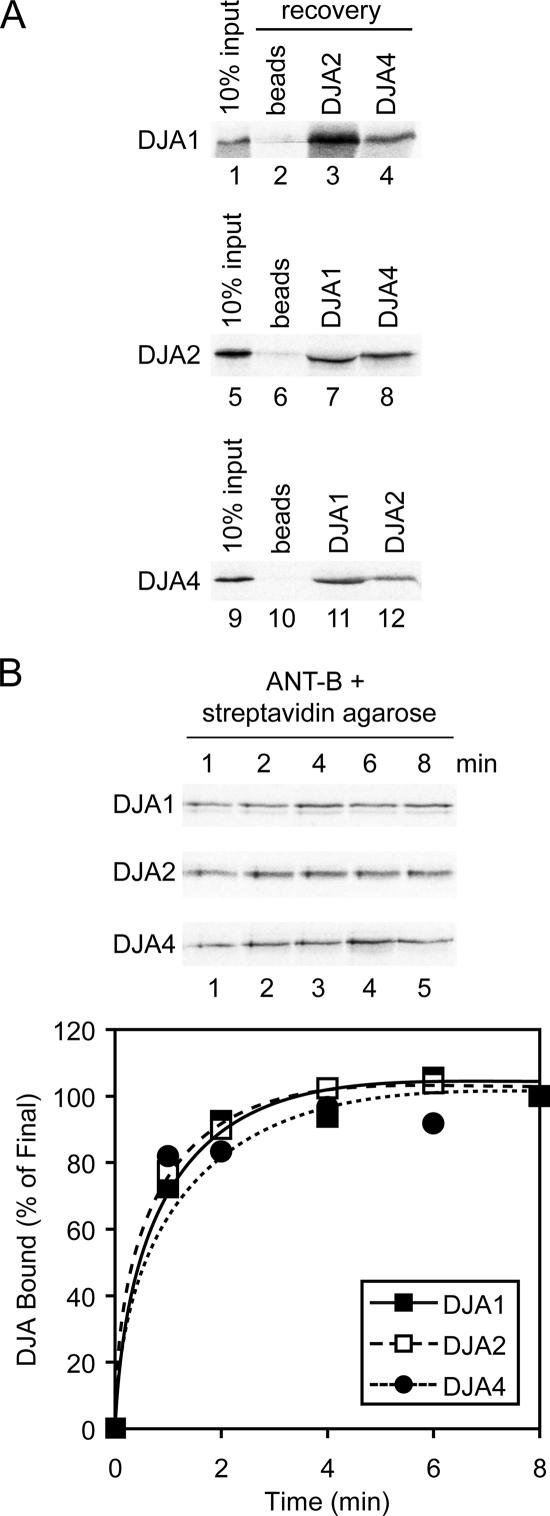
To further examine the formation of DJA–preprotein complexes, we asked whether different DJAs could bind to a polypeptide simultaneously. Experimental approaches described above were modified for this purpose. Chaperone complexes with ANT-B were formed by its dilution into RL with ATP, as performed for the gel filtration analysis of Figure 2B, but with the addition of different cell-free translated, radiolabeled DJA proteins into the binding reaction. Each reaction was then allowed to bind to a different purified His-tagged DJA protein identically to Figure 4A. Complexes were reisolated with nickel-Sepharose and analyzed for the presence of the radiolabeled DJA. In this manner, DJA1 was detected in chaperone–ANT complexes recovered with DJA2 and DJA4, at levels above the low background binding (Figure 6A). Similar, DJA2 was recovered with DJA1 and DJA4, and DJA4 with DJA1 and DJA2. Therefore, binding of each DJA cochaperone does not seem to be exclusive of the others, and they can form heterocomplexes with substrate.
Figure 6.
DJA heterocomplex formation. (A) DJA1, DJA2, and DJA4 were radiolabeled by cell-free translation (input, lanes 1, 5, and 9). ANT-B was diluted into reactions containing RL, ATP, and each radiolabeled DJA, as described in Figure 2B. Complexes were coprecipitated (recovery) with nickel-Sepharose alone (beads, lanes 2, 6, and 10) or nickel-Sepharose and the indicated His-tagged DJA protein, as described in Figure 4A. Recovered material was analyzed by autoradiography. (B) Top, DJA1, DJA2, and DJA4 were radiolabeled by cell-free translation and coprecipitated with ANT-B as described in Figure 5A. Bottom, averaged quantitation of time courses of DJA binding are shown.
It was also possible that the different DJA cochaperones bound polypeptide substrate in a defined order, or with a characteristic kinetic. To investigate this, ANT-B complexes with chaperones were formed in RL, and they were reisolated at different times as in Figure 5A; radiolabeled DJA proteins generated by cell-free translation were used to monitor their binding to complexes. All three of the DJA proteins showed the same fast initial rate of binding, with complex formation essentially complete in <5 min (Figure 6B). Binding of the DJAs to the preprotein was as fast, if not faster than, the rate of Hsc70 binding (compare Figure 6B with 5A), consistent with the expected mechanism in which the Hsp40-related cochaperones activate substrate binding by Hsc70. Together with the formation of DJA heterocomplexes (Figure 6A), these results suggest that the DJA cochaperones do not have a sequential binding mechanism where one DJA displaces another previously bound DJA. So, it is likely that the DJAs act independently of each other to bind substrate and activate Hsc70.
DJA Activation of Hsc70
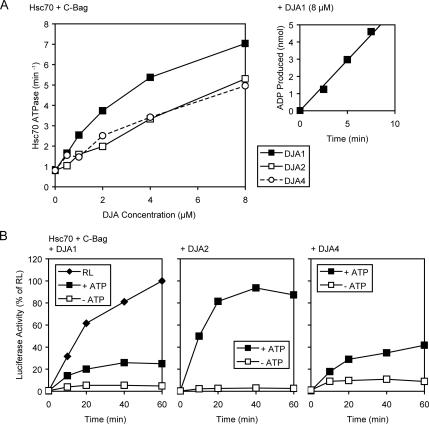
In addition to binding polypeptides, the DJAs should activate the Hsc70 ATPase through their J domains. To further explore differences between them, we measured the steady-state Hsc70 ATPase rates supported by the three full-length DJAs. J-domains specifically stimulate ATP hydrolysis to ADP by Hsc70, but not the exchange of ADP for ATP, so an excess of the C-Bag exchange factor was used to ensure that ATP hydrolysis was rate limiting (Sondermann et al., 2001; Mayer and Bukau, 2005). Purified Hsc70 at 4 μM was supplemented with 20 μM C-Bag and increasing concentrations of the DJAs. All DJAs could stimulate Hsc70 ATPase activity above its basal rate of <1 min−1 (Figure 7A). Interestingly, at all concentrations above 1 μM, DJA1 was clearly more efficient at ATPase stimulation than DJA2 and DJA4, which were similar to each other (Figure 7A). At 8 μM DJA1, Hsc70 activity reached around 7 min−1, in line with previously reported rates, whereas DJA2 and DJA4 at the same concentrations supported rates ∼5 min−1. Our results are in the same range as reported previously (Terada and Mori, 2000; Hafizur et al., 2004), with the difference that in our hands, DJA4 is somewhat more active than was thought. Although the difference between DJA1 and the other two may not seem great, in a finely balanced biological system, the functional consequences of such variation may be amplified. Also, differences between DJA activation of Hsc70 seem to be greatest (at least 2-fold) in the concentration range of 2–4 μM DJAs, which lies closest to the estimated cellular concentration of the DJAs, and the estimated ratio to Hsc70 in cells.
Figure 7.
DJA activation of Hsc70. (A) Steady-state ATPase rates of purified Hsc70 were measured in reactions containing 1 mM ATP, 4 μM Hsc70, 20 μM C-Bag, and the indicated concentrations of DJA1, DJA2, or DJA4. Right inset, representative example of the data from which linear rates were calculated. Reactions contained α-[32P]ATP and the amount of ADP produced at each time point was determined by separation on TLC and phosphorimager quantitation. (B) Refolding of guanidine-denatured luciferase was monitored in reactions containing 50% RL and 2 mM ATP, or 4 μM Hsc70, 0.5 μM C-Bag, and 4 μM DJA1, DJA2, or DJA4 as indicated, with or without 2 mM ATP. The activity of luciferase refolded in RL reactions at 60 min was set to 100%.
Another property of the DJAs was their expected ability to support polypeptide refolding by Hsc70. A polypeptide substrate commonly used to study Hsc70 function is firefly luciferase, which has a sensitive and reproducible enzymatic assay. Purified Hsc70 has been shown to assist luciferase refolding in numerous studies (Hohfeld et al., 1995; Freeman and Morimoto, 1996; Minami et al., 1996; Hohfeld and Jentsch, 1997; Terada et al., 1997; Luders et al., 2000; Terada and Mori, 2000). However, in most cases the stress-inducible cochaperone Hsp40 was used, and a direct comparison between all three DJAs with Hsc70 has not yet been reported. Thus, we tested the refolding of luciferase after dilution out of 6 M guanidine into reactions containing 4 μM Hsc70 and 4 μM of each DJA in turn, as well as 0.5 μM C-Bag, which was found to be optimal for refolding at this concentration (data not shown). Refolding reactions contained 1 mM ATP, whereas negative control reactions contained no ATP, and they were treated with apyrase to destroy any endogenous ATP. 50% RL with ATP served as a positive control. Surprisingly, DJA1 seemed to be the least effective in assisting refolding (Figure 7B), despite providing the strongest stimulation of the Hsc70 ATPase activity (Figure 7A). Refolding with DJA1 reached slightly above 20% of the RL control after 60 min (Figure 7B). In marked contrast, DJA2 was very efficient at promoting luciferase refolding, supporting a refolding level essentially the same as the RL control. DJA4, in contrast, was also relatively weak in promoting refolding, although the final activity reached was ∼40% of the RL control and somewhat higher than with DJA1. In all cases, the negative control reactions lacking ATP produced no significant refolding (Figure 7B). Overall, the divergent refolding function of the DJAs may result from a more complex combination of differences in polypeptide binding, Hsc70 ATPase stimulation, and perhaps other factors. In an earlier report, DJA1 and DJA2 promoted luciferase refolding to similar degrees (Terada and Mori, 2000), and the divergence from our results seems due to apparently small differences in experimental conditions. It might be that DJA2 is more robust and able to support refolding under a broader range of conditions. The variability of the results may also be taken as further evidence that minor differences between the DJAs might be amplified depending on the biological context.
DJA Function in Cells
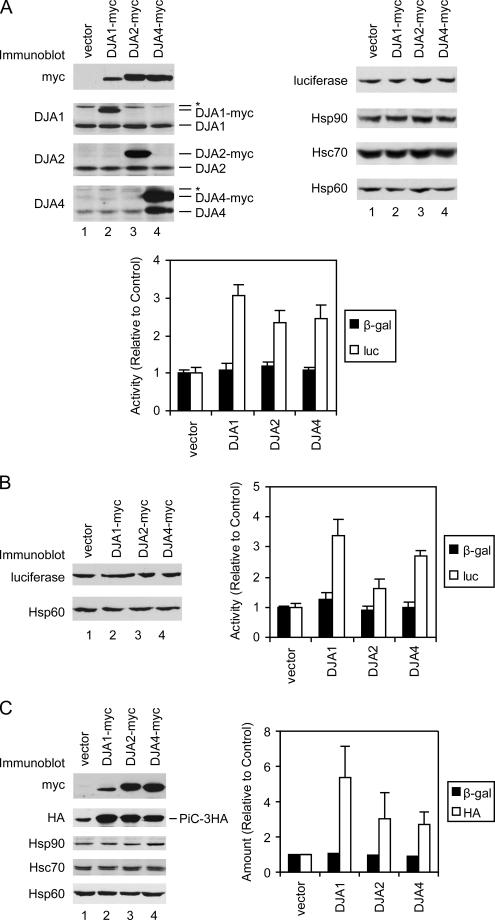
The refolding of a guanidine-denatured protein can be distinct from the cellular folding of the same protein during its synthesis on ribosomes. Experiments in mammalian and S. cerevisiae cells suggested that Hsc70 is important for the folding of firefly luciferase during or shortly after its synthesis. Thus, the DJA proteins were compared in their effects on the cellular folding of luciferase. A vector expressing luciferase from a constitutive cytomegalovirus (CMV) promoter was transfected into HeLa cells simultaneously with transient overexpression of each of the DJA proteins. Immunoblots showed that the levels of transfected myc-tagged DJA1 and DJA2 were in the same range as those of the endogenous proteins, although normal levels of DJA4 were low compared with DJA1 and DJA2 (Figure 8A). Levels of other chaperones such as Hsp90, Hsc70, and mitochondrial matrix Hsp60 were unaffected by DJA expression. Cells expressing DJA1 produced markedly higher levels of luciferase enzymatic activity relative to control cells transfected with noncoding vector, around a threefold increase (Figure 8A). Expression of DJA2 and DJA4 also enhanced activity around 2.5-fold above the control. The activity of cotranfected β-galactosidase controlled by a simian virus 40 (SV40) promoter showed comparatively little variation whether the DJAs were expressed (Figure 8A), and it was used to correct for transfection efficiency. It was notable that the amount of luciferase protein detected by immunoblot seemed to be the same between the control and DJA-expressing cells (Figure 8A). This suggested that the DJAs increased the enzymatic activity of the luciferase relative to its total amount, that is, the specific activity of luciferase. Such an effect would be expected if the folding of the model protein is enhanced. The folding of luciferase newly synthesized in the cells is also distinct from its refolding from the guanidine-denatured state observed in Figure 7B. The change in apparent activity but not absolute expression also argues against a nonspecific effect on transcription or translation.
Figure 8.
DJA function in cells. (A) HeLa cells were transfected with plasmids expressing luciferase from a CMV promoter, β-galactosidase from an SV40 promoter, and either empty vector or the indicated myc-tagged DJA proteins under a CMV promoter. Two days after transfection, cells were harvested and equivalent amounts of lysate analyzed by immunoblots for the indicated proteins (top) and by enzymatic activity assays (bottom). In blots for DJA1, DJA2, and DJA4, positions of the endogenous and overexpressed myc-tagged proteins are marked. Bands cross-reacting with the antibodies are marked with an asterisk; in cells transfected with myc-tagged DJA4 and detected with antibody against DJA4, a band of similar size to endogenous DJA4 seems to be a proteolytic fragment of the myc-tagged protein. To compare enzymatic activities, the average activities of cells transfected with luciferase (luc), β-galactosidase (β-gal), and empty vector was set to 1. The β-galactosidase activities were then used to correct luciferase activities for variations in transfection. As in all figures, standard deviations were determined from at least three independent experiments. (B) HeLa cells were transfected with plasmids expressing luciferase from a GRE-controlled promoter, constitutive GR-ΔLBD and β-galactosidase, and either empty vector or the indicated myc-tagged DJA proteins as described above. Two days after transfection, cells were analyzed as described above. (C) HeLa cells were transfected with plasmids expressing 3HA-tagged PiC from a chicken β-actin promoter, constitutive β-galactosidase, and either empty vector or the indicated myc-tagged DJA proteins as described above. Two days after transfection, cells were analyzed as described above. Quantitation of the HA immunoblot signal is shown, with that of cells transfected with β-galactosidase and empty vector set to 1.
To confirm these observations, cells were transfected with a vector expressing luciferase controlled by a GRE, together with a constitutively expressed glucocorticoid receptor lacking the chaperone-dependent ligand-binding domain (GR-ΔLBD). This combination is known to provide constitutive, hormone-, and Hsp90-independent expression of the luciferase gene (Hollenberg et al., 1987; Hollenberg and Evans, 1988). When the DJA cochaperones were overexpressed, a pattern of luciferase activity was observed similar to that of the above-mentioned experiments. All three produced significant increases in luciferase activity, with DJA1 being most effective at more than threefold of the control (Figure 8B). DJA2 and DJA4 were somewhat less effective at ∼1.8- and 2.7-fold of the control, respectively. As mentioned above, immunoblots seemed to detect similar absolute amounts of luciferase as well as the other control proteins. These results argue against effects derived from the luciferase expression vectors or their promoters. Overall, it seems that each of the DJA cochaperones can promote polypeptide folding in the native cellular environment, albeit with different degrees of efficiency.
We next addressed the effect of the DJAs on a Tom70-dependent mitochondrial preprotein in cells. The mitochondrial import of a transiently transfected 3HA-tagged PiC was previously shown to be sensitive to inhibition of Hsp90 by geldanamycin, which reduced the levels of mature, proteolytically processed PiC (Young et al., 2003). So, the same protein was constitutively expressed along with transient overexpression of each DJA cochaperone in HeLa cells. When levels of mature PiC were tested by immunoblot, noticeably more protein was detected upon expression of DJA1 relative to control cells (Figure 8C). More PiC was also observed with expression of DJA2 and DJA4. Again, the β-galactosidase transfection control showed relatively little variation (Figure 8C). The increased accumulation of PiC was most likely due to increased efficiency of import. The PiC detected by immunoblots corresponded in size to the mature form of the protein that is proteolytically processed after import, suggesting that in the vector control, excess PiC that could not be imported was removed by degradation, possibly by proteosomes. Furthermore, levels of mitochondrial Hsp60 remained unchanged upon DJA expression (Figure 8C), arguing against an effect on the Tom20-dependent import pathway used by Hsp60, or a general expansion of the mitochondria. These data are consistent with a function of the DJAs in promoting the import of the transfected PiC.
In the live cell experiments, transient overexpression of DJA1 was typically less than that of DJA2 and DJA4, as judged by detection of the myc-tags (Figure 8, A and C). DJA1 still showed the greatest effect on luciferase activity and the accumulation of PiC, within the limits of immunoblot quantitation. Overall, our in vitro and live cell results suggest that the DJA proteins differ in separate aspects of their function—polypeptide binding, Hsc70 ATPase stimulation, assistance of protein folding or refolding, and mitochondrial import—and that no single DJA is superior in all aspects.
Hsp90–Preprotein Interactions
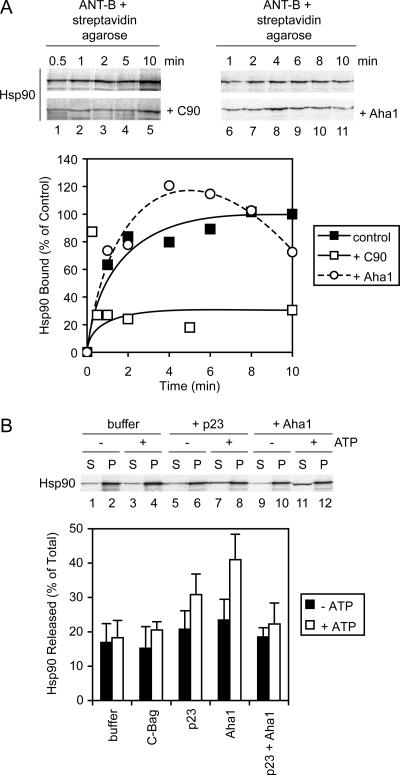
In the analysis of chaperone–preprotein complexes, Hsp90 and its cochaperone p23 were positively identified. Hsp90 seems to have a lesser, although still significant role in the import of ANT, and we next addressed the characteristics of its complex formation. Performed as described above for Hsc70 (Figure 5A), the binding of Hsp90α to ANT-B was monitored over time by using cell-free translated radiolabeled Hsp90α as a tracer. Like Hsc70, Hsp90 bound with a fast initial phase and reached its maximum level by 10 min (Figure 9A). In models of Hsp90 function based on steroid receptor maturation, a complex mediated by Hop acts to load substrate polypeptide from Hsc70 onto Hsp90 (Chen and Smith, 1998; Pratt and Toft, 2003). However, in other cases such as with some kinases, Hop mediation seems dispensable for Hsp90 entry into complexes (Lee et al., 2002; Lee et al., 2004). Because Hop was positively identified as a component of chaperone–ANT complexes (Figure 3 and Table 1), the importance of Hsp90 interactions with Hop for complex formation was assayed. The C-90 fragment competes with full-length Hsp90 for the binding site on Hop (Scheufler et al., 2000), and when added at 20 μM to Hsp90 binding reactions with ANT-B, it markedly reduced the amount of Hsp90 bound to <30% of the control (Figure 9A). This suggests that a large fraction of Hsp90 requires the interaction with Hop for recruitment to preprotein complexes. The three DJA truncation mutants lacking J domains also partially reduced Hsp90 binding (data not shown), in further agreement with Hop-mediated recruitment of Hsp90 to Hsc70-bound preprotein. Nevertheless, there is some level of Hop-independent binding of preprotein by Hsp90 (Figure 9A), most likely by direct recognition of polypeptide.
Figure 9.
Hsp90–preprotein interactions. (A) Top, Hsp90 was radiolabeled by cell-free translation and coprecipitated with ANT-B as described in Figure 5A. Reactions contained either no addition, or 20 μM purified C-90 or 4 μM Aha1. Bottom, quantitation of time courses of Hsp90 binding is shown. (B) Top, cell-free translated Hsp90 was coprecipitated with ANT-B, and the dissociation of complexes monitored as described in Figure 5B. Dissociation reactions, with or without ATP, contained buffer alone, or 5 μM purified p23 or C-Bag, or 4 μM Aha1, or 5 μM p23 and 4 μM Aha1. Bottom, quantitation of dissociation reactions.
The Hsp90 cochaperone Aha1 was recently found to stimulate the Hsp90 ATPase activity by binding to the central region of the chaperone (Panaretou et al., 2002; Lotz et al., 2003). Aha1 has been hypothesized to promote dissociation of Hsp90 from polypeptide substrate, but no direct experimental demonstration of this has yet been reported. Although Aha1 was not found to be a component of our chaperone–ANT complexes (Table 1; data not shown), it was possible that a dissociation factor would not be stably bound due to its function. We therefore asked what effect, if any, Aha1 would have on Hsp90 binding reactions with ANT. Intriguingly, 4 μM Aha1 caused a transient increase in the amount of Hsp90 bound, followed by a progressive decline (Figure 9A). The peak of Hsp90 binding in the presence of added Aha1 varied between 20 and 60% above the final level of control reactions, and it was reached by 4–6 min; because the reason for the increase is unclear, a conservative representative time course experiment is shown. Final levels of Hsp90 binding with Aha1 continued to drop below 70% of the control after 10 min. Addition of p23 had no effect on Hsp90 binding (data not shown). These data agree in part with a function of Aha1 in dissociating Hsp90 complexes; however, the two-phase Hsp90 binding curve suggests that the dynamic equilibrium of the complexes may be changed over time by Aha1.
Next, the dissociation of Hsp90–preprotein complexes was assayed. Hsp90 complexes with ANT-B were formed for 15 min as described above and isolated, and then they were incubated in the presence of various purified proteins as described for Hsc70. The absence or presence of ATP had no effect on Hsp90 dissociation when no protein was added (Figure 9B). As a negative control, the Hsc70-specific cochaperone C-Bag also had no effect, independent of nucleotide presence. The cochaperone p23 at 5 μM favored some dissociation of Hsp90 in the presence of ATP (Figure 9B). This effect was less pronounced than previously observed with glucocorticoid receptor complexes (Young and Hartl, 2000; Brychzy et al., 2003), under somewhat different experimental conditions. Aha1 at 4 μM had a slightly stronger ability to promote Hsp90 dissociation, again requiring ATP (Figure 9B). Combined treatment with p23 and Aha1 seemed to reduce dissociation to the level of the buffer control (Figure 9B), suggesting there was no synergy of function between the cochaperones and that they might counteract each other. This would be consistent with their overlapping binding sites on Hsp90 (Ali et al., 2006). The C-90 fragment had no dissociation effect on Hsp90–ANT complexes (data not shown), indicating that once Hsp90 bound preprotein its binding became independent of Hsc70 and Hop. Our results suggest that Aha1 may indeed act as a substrate dissociation factor for Hsp90. p23 may also aid dissociation in addition to stabilizing Hsp90 in chaperone–preprotein complexes.
DISCUSSION
Our results identify a basic set of proteins complexed with Tom70-dependent mitochondrial preproteins in the cytosol before import: Hsp90 and Hsc70, Hop, Tpr2, DJA1, DJA2, DJA4, and p23. These components seem to be specifically bound, because a number of other chaperones and cochaperones are excluded from such complexes. The cochaperones bound outline what is thought to be a core Hsc70–Hsp90 system or pathway (Young et al., 2004), starting with the Hsp40-related DJAs that initiate Hsc70 binding, progressing via Hop to Hsp90 binding, which is stabilized by p23. The core system components were also found in a recent proteomic analysis of chaperone-associated misfolded CFTR chloride channels, with several additional cochaperones including Cdc37 and FKBP8 (Wang et al., 2006). In our chaperone–preprotein complexes, the presence of multiple DJA proteins may be seen as elaborations to the core system (only DJA1 was associated with the misfolded CFTR). The DJA proteins bind polypeptides themselves, and probably they contribute to the solubility of preproteins as well as initiate Hsc70 binding. We propose that the advantage of having three DJAs probably comes from the partial specialization of each, as discussed in more detail below. Tpr2 is a further addition to the core system, and it is usually not observed as a major component of Hsp90 complexes. It is likely that Tpr2 helps maintain the amount of Hsc70 bound to preproteins, in addition to Hsp90.
Interestingly, many cochaperones that are excluded from the chaperone–preprotein complexes are thought to associate specifically with certain classes of Hsp90-bound substrates at later stages of their chaperone pathway. For example, FKBP52 is commonly associated with Hsp90-bound steroid hormone receptors, Cdc37 with folding intermediates of kinases, and UNC-45 (which is found in mammals as well as the original Caenorhabditis elegans source) with myosins (Pratt and Toft, 2003; Young et al., 2004). The exclusion of such cochaperones from preprotein complexes may reflect biological parsimony. Preproteins do not need to be assisted to a specific native state, so the specialized chaperoning functions of the substrate-specific cochaperones would not be required. Rather, preproteins must be maintained in a soluble form capable of being imported into mitochondria. This requirement may be fulfilled largely by the Hsc70 and Hsp90 chaperones themselves. Targeting commences with the docking of Hsc70 and Hsp90 to the Tom70 import receptor, and ATPase cycling by Hsp90 and most likely Hsc70 assist preprotein translocation across the outer membrane. Thus, the cytosolic cochaperones complexed with preproteins would act mainly to support the binding of Hsc70 and Hsp90 in a balance optimal for import.
A conceptual function not found in the chaperone–preprotein complexes is a mitochondrial-specific sorting factor. This is perhaps not a great surprise: a mitochondrial equivalent of the secretory Signal Recognition Particle has proven elusive over many years. Most earlier approaches to finding new cytosolic factors focused on Tom20-dependent preproteins with classical leader peptides. However, the major import receptor Tom20 seems designed to recognize the structural features of amphipathic α-helical leader peptides themselves (Schleiff et al., 1997; Abe et al., 2000; Yano et al., 2004). The situation is less clear for Tom70-dependent preproteins, as the targeting information read out is still largely undefined. There is evidence that Tom70 contacts preproteins directly, and a recent crystal structure of S. cerevisiae Tom70 revealed a potential preprotein-binding groove (Brix et al., 2000; Wu and Sha, 2006). Our results support the idea that sorting of Tom70-dependent preproteins from other polypeptides is accomplished solely by Tom70 within the outer membrane translocase.
The discovery of the three cytosolic DJA cochaperones in preprotein complexes suggested either that one DJA was particularly suited to aiding preprotein import and the others were less important or that each DJA might have characteristics favorable to import of the preproteins. Our results are most consistent with the latter idea. The truncated DJAs lacking J domains seem to act as dominant inhibitors and to impair mitochondrial import in vitro to the same level (Figure 4, E and F). When different biochemical properties of the DJAs are compared, as summarized in Table 2, variations between them become apparent, but without one standing out consistently above the others. DJA1 is the strongest activator of the Hsc70 ATPase, and it binds the greatest amounts of the preproteins. DJA2 is most efficient at promoting luciferase refolding, and its truncation mutant blocks Hsc70–preprotein binding most severely. DJA4 discriminates between ANT and PiC in the levels of preprotein bound, and moderately promotes luciferase refolding. In live cells, DJA1 is more effective in promoting new luciferase activity and mitochondrial import of PiC. The distribution of properties forms a unique profile for each of the DJA proteins. DJA1 might be more important, based on new luciferase folding in live cells; in contrast, DJA2 might be considered the most biochemically active, if the luciferase refolding function reflects general effectiveness. However, it seems more likely that different polypeptides will be optimally handled by different DJAs, or combinations of DJAs.
Table 2.
| DJA1 | DJA2 | DJA4 | |
|---|---|---|---|
| Mutant inhibition of import | + | + | + |
| ANT binding | ++ | + | ++ |
| PiC binding | ++ | + | + |
| Mutant blocking of Hsc70 binding | + | ++ | + |
| Hsc70 ATPase activation | ++ | + | + |
| Hsc70-mediated luciferase refolding | + | +++ | ++ |
| New luciferase activity in cells | ++ | + | + |
| PiC accumulation in cells | + | + | + |
Plus signs are assigned according to the rank order of each DJA for quantitative activity measured.
We propose that this partial specialization between the DJA cochaperones provides an advantage above a single Hsp40-related factor. For each polypeptide substrate, whether it needs to be folded or transported, an optimal combination of DJA properties might be postulated: on- and off-rates of polypeptide binding, sites of binding, rates of transfer to Hsc70. Unlike with a single DJA, the three DJAs will permit a larger, more complex range of polypeptides, as found in the cytosol of higher animals, to be productively chaperoned. Binding of each DJA to a polypeptide seems to be independent of other DJAs, suggesting that they can act in any combination. Some polypeptides may be relatively unrestricted as to which DJA can assist their folding. Other polypeptides may have a stringent requirement for one apart from the others. Still other polypeptides may require two or all three of the DJAs. Further insight might eventually be derived from more comprehensive analyses of the range of polypeptides bound by each DJA.
The fourth J domain protein in the chaperone–preprotein complexes was Tpr2. Unlike the DJAs, Tpr2 does not have a substrate-binding domain or intrinsic chaperone activity. Tpr2 is thought to trigger Hsc70 binding to substrate while promoting dissociation of Hsp90–substrate interactions, resulting in the retrograde transfer of substrate between the chaperones (Brychzy et al., 2003). The cochaperone Hop seems to be important for recruiting Hsp90 to Hsc70–bound preprotein but not for maintaining Hsp90 binding (Figure 9), similar to the expected chaperone pathway. Hop and Tpr2 thus form an opposing pair. They bind both Hsp90 and Hsc70 through their TPR clamp domains, and they likely function to maintain the optimal balance between the chaperones in preprotein complexes. Docking of the chaperones onto the TPR clamp domain of Tom70 would displace both Hop and Tpr2 from preprotein complexes. Replacement of the cytosolic TPR cochaperones with the membrane-integrated Tom70 may help switch the function of the chaperones from preprotein targeting to preprotein translocation across the mitochondrial outer membrane.
The Hsp90 cochaperones p23 and Aha1 may form another opposing pair. Both seem to favor the closed ATP-bound form of Hsp90, but p23 slows the ATPase cycle, whereas Aha1 accelerates it (Pearl and Prodromou, 2006). The functional consequences of these regulatory cochaperones are less clear. p23 enhances Hsp90-dependent folding, and it is thought to stabilize substrate-binding by Hsp90, yet after ATP hydrolysis, p23 promotes Hsp90 dissociation from substrate (Young and Hartl, 2000; Sullivan et al., 2002; Morishima et al., 2003). Aha1 also enhances Hsp90 function in vivo, but it has been proposed to also dissociate Hsp90 from substrate (Panaretou et al., 2002; Lotz et al., 2003). Our results now suggest that Aha1 is more efficient than p23 at triggering the ATP-dependent dissociation of Hsp90–preprotein complexes. However, the unexplained effect of Aha1 on the kinetics of Hsp90 binding to preprotein suggests it has an additional effect on the nature of Hsp90 complexes over time. Regardless, both p23 and Aha1 seem to regulate Hsp90 interactions with polypeptides, in our case with preproteins. In parallel to the DJAs, they may provide another example of how minor differences in an apparently similar function are combined for optimal biological efficiency.
ACKNOWLEDGMENTS
We thank Christoph Kloeckner for preliminary experiments, Megan Kooistra for assistance with protein purifications, and François Bouthillier for technical support. This work was supported by a Canadian Institutes of Health Research operating grant, the Canadian Foundation for Innovation, and the Ministry of Education of Quebec. A.C.Y.F. is supported by the Canadian Institutes of Health Research Training Program in Chemical Biology. J.C.Y. is a Canada Research Chair in Molecular Chaperones.
Abbreviations used:
- ANT
adenine nucleotide transporter
- GA
geldanamycin
- Hsc70
70-kDa heat-shock cognate protein
- Hsp90
90-kDA heat shock protein
- ISP
Rieske iron-sulfur protein
- PiC
phosphate carrier
- PK
proteinase K
- RL
reticulocyte lysate
- TOM
translocase of the mitochondrial outer membrane
- TPR
tetratricopeptide repeat.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0088) on June 27, 2007.
REFERENCES
- Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges J. C., Fischer H., Craievich A. F., Ramos C. H. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 2005;280:13671–13681. doi: 10.1074/jbc.M408349200. [DOI] [PubMed] [Google Scholar]
- Bose S., Weikl T., Bugl H., Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- Brix J., Ziegler G. A., Dietmeier K., Schneider-Mergener J., Schulz G. E., Pfanner N. The mitochondrial import receptor Tom 70, identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 2000;303:479–488. doi: 10.1006/jmbi.2000.4120. [DOI] [PubMed] [Google Scholar]
- Brychzy A., Rein T., Winklhofer K. F., Hartl F. U., Young J. C., Obermann W. M. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Smith D. F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- Cintron N. S., Toft D. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J. Biol. Chem. 2006;281:26235–26244. doi: 10.1074/jbc.M605417200. [DOI] [PubMed] [Google Scholar]
- de Marcos-Lousa C., Sideris D. P., Tokatlidis K. Translocation of mitochondrial inner-membrane proteins: conformation matters. Trends Biochem. Sci. 2006;31:259–267. doi: 10.1016/j.tibs.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Fan A. C., Bhangoo M. K., Young J. C. Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J. Biol. Chem. 2006;281:33313–33324. doi: 10.1074/jbc.M605250200. [DOI] [PubMed] [Google Scholar]
- Fan C. Y., Lee S., Ren H. Y., Cyr D. M. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B. C., Morimoto R. I. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Freeman B. C., Toft D. O., Morimoto R. I. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- Hafizur R. M., Yano M., Gotoh T., Mori M., Terada K. Modulation of chaperone activities of Hsp70 and Hsp70–2 by a mammalian DnaJ/Hsp40 homolog, DjA4. J. Biochem. 2004;135:193–200. doi: 10.1093/jb/mvh023. [DOI] [PubMed] [Google Scholar]
- Hohfeld J., Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohfeld J., Minami Y., Hartl F. U. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Evans R. M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Giguere V., Segui P., Evans R. M. Colocalization of DNA-binding and transcriptional activation functions in the human glucocorticoid receptor. Cell. 1987;49:39–46. doi: 10.1016/0092-8674(87)90753-7. [DOI] [PubMed] [Google Scholar]
- Iwahashi J., Yamazaki S., Komiya T., Nomura N., Nishikawa S., Endo T., Mihara K. Analysis of the functional domain of the rat liver mitochondrial import receptor Tom20. J. Biol. Chem. 1997;272:18467–18472. doi: 10.1074/jbc.272.29.18467. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Craig E. A. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Rutherford S. L., Miyata Y., Yahara I., Freeman B. C., Yue L., Morimoto R. I., Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- Klingenberg M., Aquila H., Riccio P. Isolation of functional membrane proteins related to or identical with the ADP, ATP carrier of mitochondria. Methods Enzymol. 1979;56:407–414. doi: 10.1016/0076-6879(79)56039-x. [DOI] [PubMed] [Google Scholar]
- Koehler C. M. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- Lee P., Rao J., Fliss A., Yang E., Garrett S., Caplan A. J. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J. Cell Biol. 2002;159:1051–1059. doi: 10.1083/jcb.200210121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P., Shabbir A., Cardozo C., Caplan A. J. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell. 2004;15:1785–1792. doi: 10.1091/mbc.E03-07-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach K. R., Graf L. J., Dunn A. R., Hoogenraad N. J. Effect of deletions within the leader peptide of pre-ornithine transcarbamylase on mitochondrial import. Eur. J. Biochem. 1986;161:19–23. doi: 10.1111/j.1432-1033.1986.tb10119.x. [DOI] [PubMed] [Google Scholar]
- Lotz G. P., Lin H., Harst A., Obermann W. M. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- Luders J., Demand J., Papp O., Hohfeld J. Distinct isoforms of the cofactor BAG-1 differentially affect Hsc70 chaperone function. J. Biol. Chem. 2000;275:14817–14823. doi: 10.1074/jbc.275.20.14817. [DOI] [PubMed] [Google Scholar]
- Mayer M. P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y., Hohfeld J., Ohtsuka K., Hartl F. U. Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J. Biol. Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- Morishima Y., Kanelakis K. C., Murphy P. J., Lowe E. R., Jenkins G. J., Osawa Y., Sunahara R. K., Pratt W. B. The hsp90 cochaperone p23 is the limiting component of the multiprotein hsp90/hsp70-based chaperone system in vivo where it acts to stabilize the client protein: hsp90 complex. J. Biol. Chem. 2003;278:48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- Panaretou B., et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trezeguet V., Lauquin G. J., Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- Pratt W. B., Toft D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Qiu X. B., Shao Y. M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Reichert A. S., Neupert W. Mitochondriomics or what makes us breathe. Trends Genet. 2004;20:555–562. doi: 10.1016/j.tig.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Shore G. C., Goping I. S. Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J. Biol. Chem. 1997;272:17784–17789. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sollner T., Griffiths G., Pfaller R., Pfanner N., Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- Sollner T., Pfaller R., Griffiths G., Pfanner N., Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Sullivan W. P., Owen B. A., Toft D. O. The influence of ATP and p23 on the conformation of hsp90. J. Biol. Chem. 2002;277:45942–45948. doi: 10.1074/jbc.M207754200. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Maeda M., Mihara K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 2002;115:1895–1905. doi: 10.1242/jcs.115.9.1895. [DOI] [PubMed] [Google Scholar]
- Terada K., Kanazawa M., Bukau B., Mori M. The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J. Cell Biol. 1997;139:1089–1095. doi: 10.1083/jcb.139.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K., Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J. Biol. Chem. 2000;275:24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- Vijayasarathy C., Bhat N. R., Avadhani N. G. Intramitochondrial fatty acylation of a cytoplasmic imported protein in animal cells. J. Biol. Chem. 1989;264:7772–7775. [PubMed] [Google Scholar]
- Walsh P., Bursac D., Law Y. C., Cyr D., Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Wu Y., Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- Yano M., Kanazawa M., Terada K., Namchai C., Yamaizumi M., Hanson B., Hoogenraad N., Mori M. Visualization of mitochondrial protein import in cultured mammalian cells with green fluorescent protein and effects of overexpression of the human import receptor Tom20. J. Biol. Chem. 1997;272:8459–8465. doi: 10.1074/jbc.272.13.8459. [DOI] [PubMed] [Google Scholar]
- Yano M., Terada K., Mori M. Mitochondrial import receptors Tom20 and Tom22 have chaperone-like activity. J. Biol. Chem. 2004;279:10808–10813. doi: 10.1074/jbc.M311710200. [DOI] [PubMed] [Google Scholar]
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Young J. C., Hartl F. U. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C., Hoogenraad N. J., Hartl F. U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Zara V., Palmieri F., Mahlke K., Pfanner N. The cleavable presequence is not essential for import and assembly of the phosphate carrier of mammalian mitochondria but enhances the specificity and efficiency of import. J. Biol. Chem. 1992;267:12077–12081. [PubMed] [Google Scholar]