Abstract
Global amphibian decline by chytridiomycosis is a major environmental disaster that has been attributed to either recent fungal spread or environmental change that promotes disease. Here, we present a population genetic comparison of Batrachochytrium dendrobatidis isolates from an intensively studied region of frog decline, the Sierra Nevada of California. In support of a novel pathogen, we find low diversity, no amphibian-host specificity, little correlation between fungal genotype and geography, local frog extirpation by a single fungal genotype, and evidence of human-assisted fungus migration. In support of endemism, at a local scale, we find some diverse, recombining populations. Therefore neither epidemic spread nor endemism alone explains this particular amphibian decline. Recombination raises the possibility of resistant sporangia and a mechanism for rapid spread as well as persistence that could greatly complicate global control of the pathogen.
Keywords: chytridiomycosis, global spread, recombination, enigmatic amphibian decline, emerging infectious disease
Amphibian biodiversity is declining globally, and chytridiomycosis is one of the factors attributed for this loss (1, 2). The disease perplexes scientists because the mechanism of death, the mode of spread, and the origin of the disease are unknown. What is known is that susceptible amphibians of many species die soon after their skin is infected by the fungus, Batrachochytrium dendrobatidis (3, 4). First identified in 1998 (3) and described in 1999 (5), the fungus has now been found on museum specimens dating back to African Xenopus laevis collections made in the 1930s (6). Two competing hypotheses exist regarding the origin of chytridiomycosis. The first is that the fungus is endemic to many regions and disease is due to recent environmental change; the second, the novel pathogen theory, suggests that disease is the result of recent spread of the fungus (perhaps with increased virulence) into new geographic areas where it encountered naive hosts (reviewed in ref. 7).
Arguing for endemism is the fact that B. dendrobatidis does not seem adapted for broad-scale dispersal yet frogs carrying it have been recorded on every continent except Antarctica (7). B. dendrobatidis is a waterborne pathogen, transmitted among frogs via a free-swimming zoospore stage that lacks a resistant cell wall and is easily desiccated (5). For many fungi, including other chytrids (e.g., Chytriomyces hyalinus), a sexual phase precedes the production of resistant sporangia, which aid in persistence and dispersal (8). However, no sexual phase or resistant stage has yet been found for B. dendrobatidis; thus, dispersal is assumed to be via infected frogs, zoospore-contaminated water, or an as yet unidentified reservoir host. Although X. laevis from southern Africa has been suggested as the original source of B. dendrobatidis (6), X. laevis was exported around the world from the 1930s to the 1950s for human pregnancy testing (9), well before the first known amphibian declines from B. dendrobatidis commenced in the 1970s (7). Furthermore, other African anurans remain susceptible to the disease. Another troubling observation is that declines are occurring in remote and protected preserves and national parks. If humans are mediating the spread of the disease, one would expect frogs living in close proximity to humans to be more affected than those in protected wilderness, which does not seem to be the case. Theories consistent with endemism that have been put forward to explain the sudden increase in global amphibian declines because of chytridiomycosis include host immunosuppression (10) and global warming (11).
In support of B. dendrobatidis as a novel pathogen (12) are mass mortalities in susceptible amphibian populations that suggest that the hosts are naive or that a new, virulent fungal genotype has spread through existing fungal populations. Intensive field surveys of frog populations in Australia and North, Central, and South America indicate that apparently uninfected populations are becoming infected and succumbing to the disease as it spreads through the region (2, 4, 13). Two widespread and well known species, the African clawed frog, X. laevis, and the American bullfrog, Rana catesbeiana, can carry the disease asymptomatically, resulting in speculation that these exotic species may have aided in the intercontinental spread of chytridiomycosis (6, 14). To date, screening of global B. dendrobatidis isolates for genetic differences has found very few variable markers, indicating the pathogen may have recently moved between continents or that a newly derived genotype has displaced older fungal populations (15). Collections made of global isolates have tried to span geographic distance and variable host species; as such they are limited to one or at most a few individuals per population.
Two factors set apart our study. First, we focus on a single, well sampled geographic area, the Sierra Nevada of California, where frog decline associated with chytridiomycosis has been carefully documented and where the population biology and genetics of the resident frogs has been studied in detail. Second, we can address population structure, migration and reproductive mode because we have developed sufficient polymorphic genetic markers in B. dendrobatidis populations from this region and have scored them in nearly 100 local isolates. At the extremes, a single panmictic population of B. dendrobatidis would suggest recent global spread, whereas geographically defined populations would indicate a longer evolutionary association with an area. The inclusion of alternative hosts from California and four global isolates provides a baseline of information on the continuum of variability one might expect over greater geographic and biological scales. Resolving the origin of chytridiomycosis will have direct implications for the development of global amphibian trade practices and local resource management for the prevention of further extinctions.
Sierra Nevada and the Mountain Yellow-Legged Frog Model.
Mountain yellow-legged frogs, recently redescribed as two species Rana muscosa and Rana sierrae, are endemic to the mountains of California (16). During the past century, these frogs have disappeared from >90% of their historic range in large part because of the historical introductions of predatory nonnative fish (17) and more recently an epidemic of chytridiomycosis (4, 18). Frog population surveys dating back to 1995 (19, 20) and frog population genetic studies (refs. 16 and 21 and V.T.V., unpublished data) make the Californian R. muscosa and R. sierrae populations the best current system for studying B. dendrobatidis populations. Geographical subdivision within the Sierra Nevada, with R. sierrae in the north and R. muscosa in the south, suggests an historical barrier to gene flow ≈2.2 million years ago (16, 21). Representatives of other amphibian and reptilian taxa (reviewed in ref. 21) also separate into distinct north and south clades within the Sierra Nevada. Genetic structure within each frog species reflects isolation within river drainages separated by impassable mountain ranges. If B. dendrobatidis has an ancient association with mountain yellow-legged frog populations then geographic structuring of B. dendrobatidis populations should also be evident. We sampled and cultured B. dendrobatidis isolates from frogs and tadpoles collected from each of six populations over the summers of 2003 and 2004 (for a total of 97 B. dendrobatidis isolates); populations were from three R. sierrae lakes in the north and three R. muscosa lakes in the south.
Finding Genetic Footprints.
In B. dendrobatidis, nuclear ribosomal DNA internal transcribed spacer sequences display as much within-individual variability as they do between-individuals, making them unsuitable for population comparisons (J.A.T.M., unpublished data). Mitochondrial DNA is too conserved among B. dendrobatidis isolates to be useful [J. N. Busby and R. T. M. Poulter (2005); direct submission to GenBank, accession no. AY859488]. For this reason, we targeted microsatellite sequences and single nucleotide polymorphisms (SNPs). Sequences containing microsatellites were found by mining a genomic DNA library of 7,600 sequences and an EST (expressed sequence tag) library of 1,500 sequences, together representing ≈20% of the B. dendrobatidis genome. Microsatellite motifs were rare and short, none were found longer than eight repeats. Of 76 microsatellite loci developed, only 12 varied among a subset of global isolates. The variation was either in the number of microsatellite repeat elements or in SNPs in the DNA sequence flanking microsatellites. These 12 variable loci plus 3 variable published loci (15) were screened for 104 B. dendrobatidis isolates (Table 1).
Table 1.
Isolates of Batrachochytrium dendrobatidis included in this study, with abbreviations used
| Host species | Collection site | Abbrev. | n | Culture reference |
|---|---|---|---|---|
| Rana sierrae North | Mono Pass, CA | MP | 20 | JAM011-030 |
| Summit Meadow, CA | SM | 17 | JAM033-049 | |
| Little Indian Valley, CA | LIV | 20 | JAM081-100 | |
| Rana muscosa South | Hitchcock Lakes, CA | HL | 17 | JAM050-054, 057–068 |
| Woods Lake, CA | WL | 19 | JAM102, 106, 108, 110, 112, 115, 117, 119, 203, 205–212 | |
| Laurel Creek, CA | LC | 4 | LJR089, LJR091, LJR134, LJR137 | |
| Rana draytonii | Point Reyes, CA | PR-Red | 1 | LJR299 |
| Rana catesbeiana | Trinity River, CA | TR-Bul | 1 | JAM234 |
| Rana catesbeiana | Point Reyes, CA | PR-Bul | 1 | JEL271 |
| Dendrobates azures | National Zoological Park, Washington, DC | Wash Zoo | 1 | JEL197 |
| Limnodynastes dumerilii | Melbourne, Victoria, Australia | Aus | 1 | JEL253 |
| Xenopus tropicalis | Imported to USA from Ghana | Ghana | 1 | JEL245 |
| Xenopus laevis | Imported to UC Berkeley from South Africa in the 1980s | SAf | 1 | JP005 |
Results
Genetic divergence among isolates was extremely low; no more than two alleles were found in 14 of the 15 loci, for all individuals. At one locus, a rare third allele was identified from an isolate cultured from a long-term U.S. laboratory colony of Xenopus laevis maintained at the University of California, Berkeley (Table 2). Although chytrids have traditionally been thought of as haploid, genetic work (15) suggests that this species is diploid. Within the Sierra Nevada, heterozygosity ranged from 20% at Laurel Creek to 87% at Mono Pass. Global isolates and long-term cultures fell within this range (Table 3). Fourteen of the 15 loci displayed heterozygote excess within at least one population; however, with clone-corrected data, tests for heterozygote excess were not significant [supporting information (SI) Text]. Similarly, by using clone-corrected data, tests to determine whether alleles were in Hardy–Weinberg equilibrium were not significant (Fisher's Exact test; SI Text). The high level of heterozygosity is likely to be a consequence of clonal reproduction because selfing would promote homozygosity. Clonal reproduction in B. dendrobatidis is implicated in the decline of two of the study populations of R. muscosa in the Sierra Nevada, Woods Lake and Laurel Creek. These sites have been driven close to extinction by single, but different, genotypes of B. dendrobatidis. Although alleles were not divergent at a global scale, genotypes were; 24 distinct genotypes and 42 haplotypes were identified for the 104 B. dendrobatidis isolates screened. Similarly, although a single genotype could dominate a site, no genotype was found at more than one site.
Table 2.
Variable loci identified for B. dendrobatidis isolates and their primer sequences
| Locus | Length | Type - position | Alleles | Primers | GenBank accession nos. |
|---|---|---|---|---|---|
| 1 | 179 | deln - 57 | 2 | F-CATAAGAGTCATACTGCGGTAAATCGA | EF069391 |
| 6164Y2 | A4 / A5 | R-GATCTACCGGTGTGCAAAAGTTCC | EF069392 | ||
| 2 | 199 | msat - 144 | 2 | F-GTTGGTATTACTCAACGTCCATACAC | EF069393 |
| 9893X2 | TA4 / TA5 | R-ACTCAGTCGTACGTAGCTAGTTTG | EF069394 | ||
| 3 | 219 | msat - 139 | 2 | F-GAAAACATGGCATGCAGTGG | EF069395 |
| mb-b13-8b | AT5 / AT6 | R-CGGCGAAGCTCTCGCTAC | EF069396 | ||
| 4 | 314 | complex - 92..102..103–109 | 2 | F-TGTCTGAATGATTTTCCCTCGG | EF069397 |
| b7-10c | C..A——/G..TACTACC | R-GGTAGCTCAGTAGTTCCATGC | EF069398 | ||
| 5 | 395 | complex - throughout | 3 | F-TGCTGACAATGGTGCCAGCTAT | EF069399 |
| 9908X2 | 13 SNPs..3 x deln.. ACT3/4 | R-TAGCCGTTTCGACAGTGGTGGC | EF069401 | ||
| 6 | 190 | SNPs - 32..44 | 2 | F-CACCAACGGAGGATGATCGCACA | EF069402 |
| 6677X2 | C..T / T..C | R-CTTGAAAAACCAAGCCACAGTCCTAG | EF069403 | ||
| 7 | 337 | SNPs - 110..244..279 | 2 | F-TCGTCCTGATGAGATGCAAACCAG | EF069404 |
| 6873X2 | A..C..C / G..A..T | R-GAGTTTCCAGGCAAGTGTTTTGCT | EF069405 | ||
| 8 | 342 | SNPs - 58..194..212..303..315 | 2 | F-TCGTGAAGAGCTTGGAAAGTCG | EF069406 |
| 8009X2 | G..G..A..G..T / A..T..G..A..C | R-AGTTCTGTCGTCAATGCTGTAGGG | EF069407 | ||
| 9 | 336 | SNP - 114 | 2 | F-CTGAATCTTGCCTCGTCTAGTAGC | EF069408 |
| 8329X2 | A / G | R-TATCAAGGTCTTTTGGCAAGACCG | EF069409 | ||
| 10 | 261 | SNP - 94 | 2 | F-CATCGGGTTTGTCATTGCCTGC | EF069410 |
| 8392X2 | A / T | R-TATGGCATGTGGTCTACTCTGTCC | EF069411 | ||
| 11 | 289 | SNPs - 205 | 2 | F-AAGTGTAGACATGGCACCCGAGTT | EF069412 |
| 8667Y2 | AA / GG | R-ATACATGTACAAGACCAAGAAGGTCG | EF069413 | ||
| 12 | 245 | SNP - 61 | 2 | F-GGATCTGCCAGTTTCGATCTACTCG | EF069414 |
| 8702X2 | C / T | R-GAATATGGCATGGGAGAAGTAGCC | EF069415 | ||
| 13 | 553 | SNP - 399 | 2 | F-ACCAACTATAACATCATCAAG | EF069416 |
| ctsynl* | A / G | R-CGAATATCAGTCAACGCAAGC | EF069417 | ||
| 14 | 688 | SNP - 181 | 2 | F-ATCCCTGTGGTAACTTTTCTG | EF069418 |
| lsu35 (cy)* | T / C | R-ACGGACATGGGGAATCTGACT | EF069419 | ||
| 15 | 496 | deln - 447 | 2 | F-CTATCTGCGCTCCCGTGTCAA | EF069389 |
| r6046 | T / C | R-ACGGACATGGGGAATCTGACT | EF069419 | ||
| 15 | 496 | deln - 447 | 2 | F-CTATCTGCGCTCCCGTGTCAA | EF069389 |
| r6046** | CA / - | R-AGGGCTGCAACAACTGGATTT | EF069390 |
Table 3.
Genetic diversity within Californian B. dendrobatidis populations and samples compared with global isolates
| Host | Site | n | Haplotypes | Genotypes | Heterozygous loci |
|---|---|---|---|---|---|
| R. sierrae | LIV | 20 | 6 | 5 | 5 (33%) |
| SM | 17 | 4 | 2 | 12 (80%) | |
| MP | 20 | 7 | 4 | 13 (87%) | |
| R. muscosa | WL | 19 | 2 | 1 | 7 (47%) |
| HL | 17 | 7 | 4 | 12 (80%) | |
| LC | 4 | 2 | 1 | 3 (20%) | |
| Cumulative Total | 97 | 28 | 17 | 14 (93%) | |
| R. catesbeiana | TR-Bul | 1 | 2 | 1 | 8 (53%) |
| R. catesbeiana | PR-Bul | 1 | 2 | 1 | 7 (47%) |
| R. draytonii | PR-Red | 1 | 2 | 1 | 11 (73%) |
| X. laevis | SAf Xen | 1 | 2 | 1 | 6 (40%) |
| D. azures | Wash Zoo | 1 | 2 | 1 | 4 (27%) |
| X. tropicalis | Ghana Xen | 1 | 2 | 1 | 10 (67%) |
| L. dumerilii | Aus | 1 | 2 | 1 | 5 (33%) |
| Cumulative Total | 104 | 42 | 24 | 15 (100%) |
Structured Populations: Migration or Multiple Introductions?
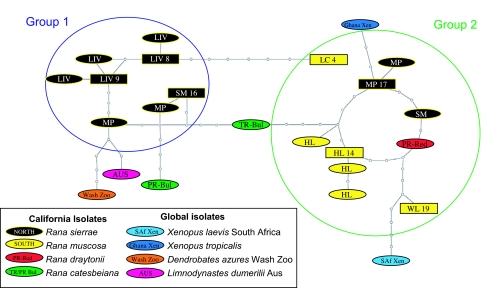
The hypothesis that B. dendrobatidis is a novel pathogen is supported by the global multilocus genotype network (Fig. 1) in which genotypes are separated by relatively few steps and international and North American isolates do not dissociate. In some cases, more genetic variation was observed among isolates from a single Californian lake than between isolates from different continents. Coevolution of amphibian host and fungus in North America is not supported because no clades of B. dendrobatidis isolates are exclusive to R. muscosa, R. sierrae, R. catesbeiana, or Xenopus spp.
Fig. 1.
Multilocus genotype network of B. dendrobatidis isolates. Boxes signify more than one isolate (number given) from the same location with the same genotype. Circled in blue and green are groups 1 and 2 as predicted by Structure for the Sierra Nevada B. dendrobatidis population samples. The key indicates frog host and number of samples with site abbreviations following those in Table 1. Each step in the network represents a change in genotype at a single locus.
In contrast, geographic structure occurs at a local population scale in the Sierra Nevada. Isolates from Little Indian Valley form an exclusive clade, as do isolates from Hitchcock Lakes. This local geographic structure does not extend to broader regions, because populations from neighboring sites do not group together and isolates from R. sierrae in the northern Sierra Nevada do not form a clade distinct from southern R. muscosa isolates.
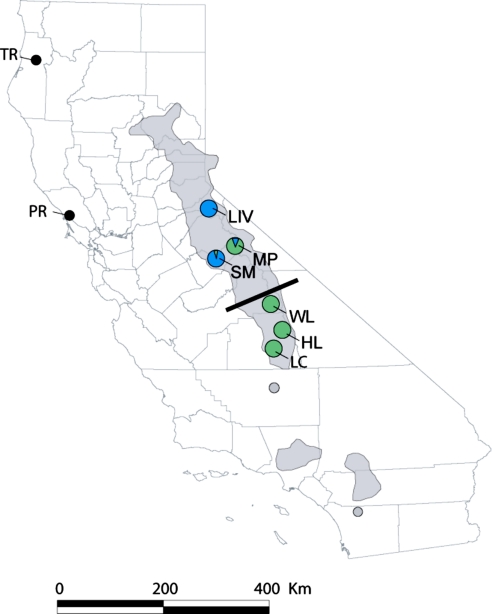
Inferred population composition [using Structure software, version 2.1 (22)] for the B. dendrobatidis isolates from mountain yellow-legged frogs supports two introductions into the Sierra Nevada followed by subsequent divergence and spread. Isolates from the northern Sierra Nevada (Little Indian Valley and Summit Meadow) generally fall into one group (group 1) and those from the southern Sierra Nevada (Laurel Creek, Hitchcock Lakes, Woods Lake, and Mono Pass) fall into a second group (group 2; Fig. 2). Genotypes from both groups are found at Summit Meadow and Mono Pass. These two sites are on opposite sides of Yosemite National Park separated by a distance of 40 km, although they are close to, and connected by, the main Yosemite traffic route, which carries >4 million tourists per year (www.yosemite.org). The observed distribution of genotypes between these two sites (Fig. 2) is evidence for recent movement of B. dendrobatidis by people. Further evidence of recent spread that was probably human mediated is seen at Point Reyes National Seashore, a coastal National Park just north of San Francisco (Fig. 2). Two genetically distant isolates were collected from this site, one from a native endangered red-legged frog, Rana draytonii, which falls among group 1 isolates, and the other, which is closer to the group 2 isolates (Fig. 1), from an introduced bullfrog, R. catesbeiana.
Fig. 2.
Map of California showing B. dendrobatidis collection sites and the inferred genetic groupings in the Sierra Nevada (blue, group 1; green, group 2). Gray shading corresponds to the distribution of the mountain yellow-legged frog species complex. The black bar marks the approximate boundary between R. sierrae in the north (LIV, SM, MP) and R. muscosa in the south (WL, LC, HL). Other Californian B. dendrobatidis isolates were from Trinity River (TR-Bul, R. catesbeiana) and Point Reyes (PR-Bul, R. catesbeiana and PR-Red, R. draytonii).
Genetic subdivision within and among populations as well as between the two inferred groups was tested by using AMOVA (analysis of molecular variance) in Arlequin [version 2.0 (23)]. Irrespective of how the populations are grouped, >50% of the observed genetic variance occurs within sites (Table 4). The inferred two-group split accounts for a greater proportion of the variance (18.01%) than a species-based north to south split (10.2%) although significant genetic structure was detected for both divisions. With ≈30% of variance attributable to the collection site, a random sample can be assigned to an individual lake with some confidence; the software GMA [Genetic Mixture Analysis, version 1.0 (24)] assigned 23 of the 25 genotypes to the correct site with 95–100% probability. Genetic subdivision within a site was not significant.
Table 4.
AMOVA results for Sierra Nevada populations of B. dendrobatidis grouped by host (R. sierrae LIV, SM, & MP and R. muscosa WL, HL & LC) or the two predicted Structure groups (group 1 SM & LIV; group 2 MP, HL, WL, & LC)
| Source of variation | df | Sum of squares | Variance components | % of variation | Fixation indices | P value |
|---|---|---|---|---|---|---|
| R. sierrae vs R. muscosa | 1 | 82.811 | 0.38095 Va | 10.20 | FST: 0.44257 | 0.0000 |
| Within host spp. | 4 | 164.059 | 1.27141 Vb | 34.05 | FSC: 0.37923 | 0.0000 |
| Within populations | 188 | 391.259 | 2.08116 Vc | 55.74 | FCT: 0.10203 | 0.20039 |
| Total | 193 | 638.129 | 3.73352 | |||
| Group 1 vs group 2 | 1 | 105.914 | 0.69728 Va | 18.01 | FST: 0.48255 | 0.0000 |
| Within groups | 4 | 140.929 | 1.09388 Vb | 28.25 | FSC: 0.34453 | 0.0000 |
| Within populations | 188 | 391.259 | 2.08116 Vc | 53.74 | FCT: 0.18007 | 0.06061 |
| Total | 193 | 638.129 | 3.87233 |
Association, Recombination, Sex, and a Possible Resistant Stage.
The importance of clonal versus sexual reproduction was assessed by calculating index of association measures (IA and r̄d) for each population separately with clone-corrected data [Table 5; Multilocus, version 1.3 (25)]. These association measures test for multilocus linkage disequilibrium and are zero if alleles are recombining in the population (no linkage disequilibrium). The dominance of single genotypes within a population leaves no doubt that wild isolates of B. dendrobatidis undergo clonal reproduction via mitotically produced zoospores, however, recombination could not be ruled out in two populations, Little Indian Valley and Hitchcock Lakes (Table 5). At these sites, sexual reproduction may be occurring. Sexual reproduction in Chytridiomycota initiates the production of resistant meiosporangia (8), which would provide both a propagule for long-distance dispersal and a resting stage to maintain the fungus in the absence of hosts.
Table 5.
Comparison of Sierra Nevada B. dendrobatidis populations and groupings plus index of association measures
| Frog population | Genotypes | Theta | IA | rd | Repro |
|---|---|---|---|---|---|
| LIV | 5 | 1.76 | 0.24 | 0.12 | Sex |
| SM | 2 | 1.0 | —* | —* | Clonal |
| MP | 4 | 1.28 | 1.5† | 0.21† | Clonal |
| WL | 1 | 0.79 | —* | —* | Clonal |
| HL | 4 | 1.41 | 0.68 | 0.14 | Sex |
| LC | 1 | 1.0 | —* | —* | Clonal |
| Total | 17 | 11.76 | 1.68† | 0.13† | Clonal |
| R. sierrae (LIV, SM, MP) | 11 | 6.23 | 1.24† | 0.11† | Clonal |
| R. muscosa (WL, HL, LC) | 6 | 3.82 | 2.78† | 0.22† | Clonal |
| Group 1 (SM, LIV) | 7 | 3.76 | 2.28† | 0.22† | Clonal |
| Group 2 (MP, HL, WL, LC) | 10 | 6.32 | 1.86† | 0.15† | Clonal |
Site abbreviations match Table 1.
*Too few genotypes to calculate index of association.
†P value <0.05 indicating significant linkage disequilibrium.
The clustering of similar genotypes at Little Indian Valley (five genotypes, six distinct haplotypes) and Hitchcock Lakes (four genotypes, seven different haplotypes) suggests that they have evolved from a common ancestor rather than being the result of independent introductions. These sites may have maintained stable B. dendrobatidis populations for a longer period, a scenario consistent with local endemism. The absence of new alleles intimates that the different genotypes evolved through recombination of existing alleles rather than through mutation. We propose that B. dendrobatidis has had a limited number of introductions into the Sierra Nevada, two based on our analysis. Initial observations of enigmatic frog declines in the Sierra Nevada were made 25 years ago. Could sexual reproduction and recombination have produced the observed genotypes in that time frame? Yes, provided that at least some frogs survived the initial onslaught of infection or provided reservoir hosts were present. Heterozygosity now ranges from 80% (12 heterozygous loci) in isolates from Summit Meadow and Mono Pass down to 20% (3 heterozygous loci) at Laurel Creek. Through time, selfing could be increasing the proportion of isolates with homozygous loci. The absence of shared genotypes between sites indicates that recombination may be an important mechanism for spreading the disease through the generation of long-lived resistant stages.
Conclusions
One of our two important findings is that the agent of chytridiomycosis seems to have entered the Sierra Nevada as a novel pathogen but is now rapidly showing signs of local endemism. In support of the novel pathogen hypothesis, frog populations have collapsed after infection by a single B. dendrobatidis genotype. In addition sites having easy public access share similar fungus genotypes. Globally, endemism is not consistent with some Sierran genotypes being more closely related to international genotypes than to other Sierran genotypes. In contrast, sites within the Sierra Nevada now show signs of local endemism; no two sites share the same genotype and some sites contain several related genotypes with evidence of recombination. This result is consistent with studies of human fungal diseases (e.g., coccidioidomycosis or histoplasmosis) where the present distribution of the fungus is best explained by a combination of endemism associated with natural animal hosts and human assisted migration (26, 27). Unlike these fungi that can cause human disease, there is no suggestion that B. dendrobatidis has coevolved with particular amphibian hosts, or that it can associate with nonamphibian hosts (J.A.T.M., unpublished data). Over the past decade, gorgonian sea fan corals in the Caribbean have suffered mass mortality events from aspergillosis, resulting from infection by the terrestrial fungal pathogen Aspergillus sydowii. The prevalence of A. sydowii is affected by sea fan size, species and density (28). We could be seeing a similar trend with B. dendrobatidis; as a generalist, it infects many amphibian species and spreads rapidly. However, the effect of chytridiomycosis on a particular species may be site-dependent due to factors such as temperature, host density, and water movement. Sites where the disease persists have time for multiple introductions, and the fungus has the opportunity to recombine genes.
Our other important finding is that the diversity of B. dendrobatidis cannot be explained by clonal reproduction alone. Although evidence is abundant for clonal, mitotic reproduction, in the form of sites with a single genotype of B. dendrobatidis and fixed heterozygosity at a majority of loci at sites with multiple genotypes, the null hypothesis of recombination could not be rejected at two genotypically diverse sites. Although we have no measure of the relative importance of clonal and recombining reproduction, even low rates of recombination can have profound consequences for pathogen spread and persistence (29). The most likely mechanism for recombination is sexual reproduction, and, in chytrids, sexual reproduction typically results in thick-walled, resistant sporangia (8, 30). Such sporangia would be a boon both to long distance dispersal of the fungus and to its persistence at sites where amphibians have become extinct; persistence of the fungus would complicate efforts to reintroduce amphibians to sites after local extinction.
The evidence for recombination, which raises the possibility of sexual reproduction, underscores how little we know about B. dendrobatidis ecology. Discovery of direct evidence for sexual reproduction, finding of resistant sporangia, and surveying for other hosts would all improve efforts to model chytridiomycosis and bring about its control. Attempts to find the fungus in environmental samples have also been fruitless, although B. dendrobatidis has been shown in laboratory studies to survive up to 3 months when grown with soil or feathers (15).
Our study provides evidence of a mechanism for spreading the disease, that one genotype can cause an epidemic and send a population extinct, and that local endemism can occur at persisting sites with the potential for recombination. These conclusions could be made only by studying well sampled local populations from one geographic region, yet they can be used to understand the global spread of emerging infectious disease. Our use of variable microsatellite and SNP markers and the current efforts to completely sequence the genomes of two B. dendrobatidis isolates indicate that access to sufficiently polymorphic markers will not limit further study of fungus population genetics. A critical next step is to challenge hypotheses arising from our work by using similar studies in other geographic regions, especially from areas where infected frog populations show no overt disease, from Africa where B. dendrobatidis may have originated, and from Asia where the disease has only recently been reported by the Japanese Ministry of the Environment.
Methods
B. dendrobatidis Collection and Culture.
At each site, 20 infected R. muscosa or R. sierrae tadpoles (31) were collected into individual containers. The tadpoles were housed in individual 5-liter tanks containing 4 liters of charcoal-filtered water at the animal housing facility at the University of California, Berkeley until processed; ethics were approved by the Office of Laboratory Animal Care. B. dendrobatidis was isolated and cultured as described (5) and cryopreserved as described (32). Cultures and their ethanol preserved hosts have been submitted to the University of California, Berkeley Museum of Vertebrate Zoology (MVZ accession no. 14139).
Screening of Loci.
DNA was extracted from B. dendrobatidis cultures by using Prepman Ultra (Applied Biosystems, Foster City, CA) as described (33). Preliminary screening of eight isolates (JAM059, JAM081, JAM234, JEL197, JEL245, JEL253, JEL271, and JP005; see Table 1) involved PCR amplification with high fidelity Pfu DNA polymerase (Stratagene, La Jolla, CA), Topo TA cloning (Invitrogen, Carlsbad, CA) after A-extension with TaqDNA polymerase, wizard minipreps (Qiagen, Valencia, CA) and sequencing (Big Dye, version 3.1; Applied Biosystems, Foster City, CA) of at least five clones per isolate. Based on this screen, 12 of the 76 loci were found to vary. The 12 variable loci plus 3 variable published loci (15) were then screened for the 104 B. dendrobatidis isolates listed in Table 1. Loci with length variants, either microsatellites or deletions, were PCR amplified with a fluorescently labeled primer. Then, the products were separated on an ABI3100 automated sequencer, and their sizes were scored by comparison with internal size markers by using Genotyper software (Applied Biosystems). Loci with SNP mutations were PCR amplified, and products were direct sequenced and scored for single or double peaks at the SNP site.
Genetic Structure and Distribution.
Hardy–Weinberg probability tests per population and per locus were conducted in Genepop [version 3.4 (34)]. A multilocus genotype network was constructed by using statistical parsimony in TCS [version 1.13 (35)]; a pairwise genotype distance matrix was constructed by treating each locus as a single character and the three possible genotypes at each locus as unique character states as described (15). Thus, the two homozygous genotypes were considered to be two steps from each other but only one step from the heterozygous state.
Inferring Genotypic Populations.
Population composition was inferred for the B. dendrobatidis isolates from R. muscosa and R. sierrae by using the program Structure [version 2.1 (22)]. Structure estimates the log probability of the data for each value of K (number of clusters or populations). A series of independent runs were performed by using K = 1–12 populations, a burn-in of 40,000 Markov chain Monte Carlo (MCMC) iterations, and a data collection period of 1 million MCMC iterations. Each simulation of K was replicated 20 times. To predict the true population size, the rate of change in the log probability of the data between successive K values (ΔK) was calculated and plotted against K following ref. 36 (see SI Figs. 3 and 4).
Testing for Genetic Subdivision and Index of Association.
Analysis of molecular variance (AMOVA) was used to test for genetic subdivision within and between the six geographic populations from the Sierra Nevada [Arlequin, version 2.000 (23)]. Index of association measures IA and r̄d were calculated for clone-corrected data by using the program Multilocus [version 1.3 (25)].
Supplementary Material
Acknowledgments
We thank Jim Bristow, Chris Detter, and Eileen Dalin (Joint Genome Institute, Walnut Creek, CA) for constructing and sequencing the genomic DNA library; Mary Berbee (University of British Columbia, Vancouver, BC, Canada) and Tim James (Duke University, Durham, NC) for providing early access to the B. dendrobatidis EST library; James Bettaso (U.S. Fish and Wildlife, Arcata, CA) for sick frogs; Andy Li Ren Chiang (University of California, Berkeley, CA) for his assistance culturing isolates; and Austin Burt and Matt Fisher (Imperial College, London, U.K.) for their comments on the recombination analysis. This work was supported by National Institutes of Health (NIH) Grant R01 ES12067 (to C.J.B.) from the National Institute of Environmental Health Sciences as part of the National Science Foundation/NIH Ecology of Infectious Disease program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF069389–EF069419) and the University of California, Berkeley Museum of Vertebrate Zoology (accession no. 14139).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701838104/DC1.
References
- 1.Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW. Science. 2004;306:1783–1786. doi: 10.1126/science.1103538. [DOI] [PubMed] [Google Scholar]
- 2.Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, Carey C, Livo L, Pessier AP, Collins JP. Proc Natl Acad Sci USA. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, Slocombe R, Ragan MA, Hyatt AD, McDonald KR, et al. Proc Natl Acad Sci USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rachowicz LJ, Knapp RA, Morgan JAT, Stice MJ, Vredenburg VT, Parker JM, Briggs CJ. Ecology. 2006;87:1671–1683. doi: 10.1890/0012-9658(2006)87[1671:eidaap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Longcore JE, Pessier AP, Nichols DK. Mycologia. 1999;91:219–227. [Google Scholar]
- 6.Weldon C, du Preez LH, Hyatt AD, Muller R, Speare R. Emerg Infect Dis. 2004;10:2100–2105. doi: 10.3201/eid1012.030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rachowicz LJ, Hero J-M, Alford RA, Taylor J, Morgan JAT, Vredenburg VT, Collins JP, Briggs CJ. Conserv Biol. 2005;19:1441–1448. [Google Scholar]
- 8.Miller CE, Dylewski DP. Am J Bot. 1981;68:342–349. [Google Scholar]
- 9.Hansen KL. Herpetologica. 1960;16:33–38. [Google Scholar]
- 10.Carey C. Conserv Biol. 1993;7:355–362. [Google Scholar]
- 11.Pounds AJ, Bustamante MR, Coloma LA, Consuegra JA, Fogden MPL, Foster PN, La Marca E, Masters KL, Merino-Viteri A, Puschendorf R, et al. Nature. 2006;439:161–167. doi: 10.1038/nature04246. [DOI] [PubMed] [Google Scholar]
- 12.Alford RA. In: Developing Management Strategies to Control Amphibian Diseases: Decreasing the Risks Due to Communicable Diseases. Speare R, editor. Townsville, Australia: School of Public Health and Tropical Medicine, James Cook University; 2001. p. 20. [Google Scholar]
- 13.Laurance WF, McDonald KR, Speare R. Conserv Biol. 1996;10:406–413. [Google Scholar]
- 14.Daszak P, Strieby A, Cunningham AA, Longcore JE, Brown CC, Porter D. Herpetol J. 2004;14:201–207. [Google Scholar]
- 15.Morehouse EA, James TY, Ganley ARD, Vilgalys R, Berger L, Murphy PJ, Longcore JE. Mol Ecol. 2003;12:395–403. doi: 10.1046/j.1365-294x.2003.01732.x. [DOI] [PubMed] [Google Scholar]
- 16.Vredenburg VT, Bingham R, Knapp R, Morgan JAT, Moritz C, Wake D. J Zool. 2007;271:361–374. [Google Scholar]
- 17.Vredenburg VT. Proc Natl Acad Sci USA. 2004;101:7646–7650. doi: 10.1073/pnas.0402321101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellers GM, Green DE, Longcore JE. Copeia. 2001;2001:945–953. [Google Scholar]
- 19.Knapp RA, Matthews KR. Conserv Biol. 2000;14:428–438. [Google Scholar]
- 20.Knapp RA. Biol Conserv. 2005;121:265–279. [Google Scholar]
- 21.Macey JR, Stasburg J, Brisson J, Vredenburg VT, Jennings M, Larson A. Mol Phylogenet Evol. 2001;19:131–143. doi: 10.1006/mpev.2000.0908. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard JK, Stephens P, Donnelly P. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider S, Roessli D, Excoffier L. Arlequin, A Software for Population Genetics Data Analysis. Geneva: Genetics and Biometry Laboratory, Univ of Geneva; 2000. Version 2.000. [Google Scholar]
- 24.Kalinowski ST. Genetic Mixture Analysis. Bozeman, MT: Department of Ecology, Montana State Univ; 2003. Version 1.0; www.montana.edu/kalinowski. [Google Scholar]
- 25.Agapow PM, Burt A. Mol Ecol Notes. 2001;1:101–102. [Google Scholar]
- 26.Fisher MC, Koenig GL, White TJ, San-Blas G, Negroni R, Gutierrez Alvarez I, Wanke B, Taylor JW. Proc Natl Acad Sci USA. 2001;98:4558–4562. doi: 10.1073/pnas.071406098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasuga T, White TJ, Koenig GL, McEwen J, Restrepo A, Castaneda E, de Silva Lacaz C, Heins-Vaccari EM, de Freitas RS, Zancope-Oliveira RM, et al. Mol Ecol. 2003;12:3383–3401. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- 28.Mullen KM, Harvell CD, Alker AP, Dube D, Jordán-Dahlgren E, Ward JR, Petes LE. Marine Biol. 2006;149:1355–1364. [Google Scholar]
- 29.Halkett F, Simon J-C, Balloux F. Trends Ecol Evol. 2005;20:194–201. doi: 10.1016/j.tree.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Sparrow FK. Aquatic Phycomycetes. 2nd Rev Ed. Ann Arbor, MI: Univ of Michigan Press; 1960. [Google Scholar]
- 31.Knapp RA, Morgan JAT. Copeia. 2006;2006:188–197. [Google Scholar]
- 32.Boyle DG, Hyatt AD, Daszak P, Berger L, Longcore JE, Porter D, Hengstberger SG, Olsen V. Dis Aquat Org. 2003;56:59–64. doi: 10.3354/dao056059. [DOI] [PubMed] [Google Scholar]
- 33.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. Dis Aquat Org. 2004;60:141–148. doi: 10.3354/dao060141. [DOI] [PubMed] [Google Scholar]
- 34.Raymond M, Rousset F. J Hered. 1995;86:248–249. [Google Scholar]
- 35.Clement M, Posada D, Crandall KA. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 36.Evanno G, Regnaut S, Goudet J. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.