Abstract
Cyclin E/Cdk2 is a key regulator in G1-S transition. We have identified a novel cyclin E/Cdk2 substrate called Ankrd17 (ankyrin repeat protein 17) using the TAP tag purification technique. Ankrd17 protein contains two clusters of a total 25 ankyrin repeats at its N terminus, one NES (nuclear exporting signal) and one NLS (nuclear localization signal) in the middle, and one RXL motif at its C terminus. Ankrd17 is expressed in various tissues and associates with cyclin E/Cdk2 in an RXL-dependent manner. It can be phosphorylated by cyclin E/Cdk2 at 3 phosphorylation sites (Ser1791, Ser1794, and Ser2150). Overexpression of Ankrd17 promotes S phase entry, whereas depletion of Ankrd17 expression by small interfering RNA inhibits DNA replication and blocks cell cycle progression as well as up-regulates the expression of p53 and p21. Ankrd17 is localized to the nucleus and interacts with DNA replication factors including MCM family members, Cdc6 and PCNA. Depletion of Ankrd17 results in decreased loading of Cdc6 and PCNA onto DNA suggesting that Ankrd17 may be directly involved in the DNA replication process. Taken together, these data indicate that Ankrd17 is an important downstream effector of cyclin E/Cdk2 and positively regulates G1/S transition.
Progression through the cell cycle is driven by the sequential and periodic activation of cyclin/Cdk complexes. Cyclin D/Cdk4/64 complexes are active throughout the G1 phase, cyclin E/Cdk2 at the G1/S boundary, cyclin A/Cdk2 during S phase, and cyclin A/Cdk1 and cyclin B/Cdk1 during the G2/M transition. Cyclin E/Cdk2 plays a central role in coordinating both the onset of S phase and centrosome duplication in cell cycle (1–4). It presumably exerts most of its biologic activities by phosphorylating its substrates, most frequently through the RXL motif in the substrate that interacts with the cyclin box (5–8). A number of RXL-containing proteins are themselves cell cycle regulatory proteins. In the case of pRb, cyclin E/Cdk2-mediated Rb phosphorylation inactivates Rb by derepressing E2F transcription factors (9, 10), whereas in the case of p27, phosphorylation of p27 by cyclin E/Cdk2 stimulates its degradation by the SCF-Skp2 ubiquitin ligase (11–15).
There are a number of proteins identified as cyclin E/Cdk2 substrates that regulate cell division. NPAT is a transcription factor that controls cell cycle-dependent histone gene transcription. The phosphorylation of NPAT by cyclin E/Cdk2 promotes histone transcription (16–19). Cells without NPAT fail to enter S phase from quiescence (20). CBP/p300 is another protein phosphorylated by cyclin E/Cdk2 at G1/S transition to activate its histone acetyltransferase activity (21) and may function as a cofactor for many transcription factors including E2F (22). Interestingly, cyclin E/Cdk2 phosphorylates E2F-5 and increases its transcriptional activity through CBP/p300 recruitment (23). Cyclin E/Cdk2 also phosphorylates centrosomal proteins that regulate centrosome duplication. Phosphorylation of nucleophosmin (NPM) by cyclin E/Cdk2 promotes NPM release from the centrosome, thereby stimulating centrosomal duplication (24, 25). Phosphorylation on Thr (199) by cyclin E (and A)/Cdk2 targets NPM to nuclear speckles, and enhances the RNA binding activity of NPM (26). CP110 is another centrosomal cyclin E/Cdk2 substrate that plays an essential, but poorly understood role in the centrosome cycle (27). Cyclin E/Cdk2 cooperates with Cdc6 to allow the formation of prereplication complexes during the G1-S transition (28). In addition, cyclin E/Cdk2 was found to be involved in the phosphorylation of MCM family members MCM2 and MCM3 that play essential roles in eukaryotic DNA replication (29, 30). The FOXM1 forkhead protein, originally identified as a proliferation-associated transcriptional regulator, was found as a novel cyclin E/Cdk2 substrate that functions in cell cycle progression, genetic stability, and tumorigenesis (31).
Although cyclin/Cdks are orderly activated during cell cycle progression, it is their phosphorylation substrates that carry out the ordered cellular changes that lead to cell division. However, many of these substrates remain unknown. To better understand the mechanism of cell cycle regulation, we took the approach of TAP tag purification to identify the new substrates of cyclin E/Cdk2. Here we report that human ankyrin repeat protein 17 (Ankrd17) is a novel substrate for cyclin E/Cdk2. Ankrd17 positively regulates cell cycle progression.
EXPERIMENTAL PROCEDURES
TAP Tag Purification of Cdk2-associated Proteins—pMSCV-c-FLAG-HA-Cdk2 retroviral vector was transfected into the virus packaging cell line phoenix, the viruses were harvested 48 h after transfection and subjected to infect 293T cells. The infected cells were selected with puromycin (1 μg/ml). About 2 × 109 cells were lysed with cross-linking lysis buffer (40 mm Hepes, pH 7.5, 120 mm NaCl, 1 mm EDTA, 10 mm pyrophosphate, 10 mm β-glycerophosphate, 50 mm sodium fluoride, 1.5 mm Na3VO4, 1% Triton X-100, protease inhibitor (Roche), dithiobissulfosuccinimidyl propionate (DTSSP) (0.25 μg/ml) (Pierce)). Cell lysates were incubated with anti-FLAG beads (Sigma), washed with TAP buffer (20 mm Tris-HCl, pH 8.0, 10% glycerol, 5 mm MgCl2, 100 mm KCl, 0.1% Tween 20), and eluted with FLAG peptide (Sigma) (0.25 mg/ml). The elution was incubated with anti-HA beads (Sigma), washed with TAP buffer, and eluted with HA peptide (0.25 mg/ml). For the control purpose, we used cell lysates transfected with empty vector and similarly immunoprecipitated with anti-FLAG beads followed by anti-HA beads (data not shown). The elution was run on a 4–12% Nupage BisTris gel (Invitrogen), stained with Sigma EZ blue staining solution, and subjected to mass spectrometry analysis.
Plasmids and Antibodies—The mammalian expression vectors for cyclin E and Cdk2 have been described previously (32). The Ankrd17 expression vectors (pCMV-FLAG-Ankrd17) were made by cloning the full-length cDNA into pCMV-FLAG vector at KpnI/NotI. The Ankrd17AQA mutant was generated by site-directed mutagenesis at the RQL (amino acids 1745–1747) site. The pET41b-Ankrd17RQL was made by cloning the Ankrd17 cDNA (nucleotides 5041–5463) into pET41b (Novagen) at the BamHI/XhoI site. The pET41b-Ankrd17AQA was made from pET41b-Ankrd17RQL by site-directed mutagenesis (R → A, L → A). Rabbit anti-Ankrd17 polyclonal antibody was prepared using GST fusion protein containing amino acids 2074–2352 of Ankrd17 as the immunogen. Rabbit anti-Ankrd17 polyclonal antibody was generated against GST-Ankrd17 (amino acids 1681–1821). Rabbit anti-MCM3 and MCM5 polyclonal antibodies were generated against GST-MCM3 (amino acids 490–807) and GST-MCM5 (amino acids 644–734), respectively.
The following primary antibodies were used: polyclonal rabbit antibody to cyclin E, cyclin D, cyclin A, p53, Cdc45, and β-actin (Santa Cruz Biotechnology); monoclonal antibodies to cyclin E, p21, p27, Cdc6, PCNA, MCM7, and Myc (9E10) (Santa Cruz Biotechnology), and FLAG (Sigma).
RNA Analyses—Total RNAs were prepared using TRIzol (Invitrogen). Reverse transcriptase-PCR analyses were done by following the manufacturer's instructions (Invitrogen). Primers for Ankrd17 were: upstream, 5′-TCA TCA TCA CCA GTG GTT TC TTC-3′; downstream, 5′-GGT AGG ACA CAT AAT CTT CT TG-3′. Primers for hGAPDH as internal control were: upstream, 5′-GGT CAT CCC TGA GCT GAA CG-3′, downstream, 5′-TCC GTT GTC ATA CCA GGA AAT-3′.
GST Pull-down Assays and Co-immunoprecipitation—GST fusion proteins were purified from bacteria with glutathione-Sepharose beads (Amersham Biosciences). GST fusion proteins immobilized on glutathione beads were incubated with the lysates from 293T cells transfected with Myc-cyclin E and Myc-Cdk2 in lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.5% Triton X-100, and protease inhibitor) (Roche Applied Science). The beads then washed with lysis buffer, boiled in SDS loading buffer, and the proteins were resolved on 10% SDS-PAGE, followed by immunoblotting with Myc (9E10) antibody. For coimmunoprecipitation experiments, total cell lysates were incubated with antibodies at 4 °C for 2 h followed by the addition of protein A-agarose beads. After another 2 h of incubation at 4 °C, the beads were washed with lysis buffer. The immune complexes were separated by SDS-PAGE followed by Western blot analyses with chemiluminescence (Pierce).
Cell Synchronization—U2OS cells were synchronized at the G1/S transition by a double thymidine/mimosine block as previously described (33). In brief, U2OS cells were incubated for 16 h in Complete medium with 2 mm thymidine (Sigma) released in fresh medium for 9 h and incubated a second time with 2 mm thymidine (Sigma) for 16 h. At this point, >90% of cells were in G1/S as assessed by flow cytometry. For S phase synchronization, cells were released from this second block for 4 h. After 8 h, most of the cells were in G2 phase. To obtain cells synchronized in mitosis, cells were incubated for 18 h in the presence of 2 mm thymidine, and then released for 6 h in fresh medium; then 40 ng/ml nocodazole (Sigma) was added for 6 h. Mitotic cells were collected from this population by the shake-off procedure, to avoid contamination with G2 cells. Cells in G1 phase were obtained from this pool of cells by reseeding them in fresh medium and allowing them to end mitosis for 3–5 h, and discarding the non-attached cells by shake-off.
To synchronize HeLa cell in prometaphase, 50 ng/ml nocodazole was added in culture medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum). After 24 h, mitotic cells were shaken off, washed with phosphate-buffered saline, and then resuspended in prewarmed fresh medium without nocodazole. Cells were collected at the indicated time to prepare total cell extracts and chromatin-bound protein. A total of 106 cells from each fraction were used for cytofluorometric analysis of DNA content.
Cell Fractionation—Cells were incubated in hypotonic lysis buffer (10 mm Tris, pH 8.0, 10 mm NaCl, 3 mm MgCl2, and 1 mm EGTA with protease inhibitors) for 10 min, followed by 20 strokes of a Dounce B homogenizer. The lysates were spun down for 5 min at 500 × g, and the supernatant was designated the cytosolic fraction. The nuclear pellet was washed three times in wash buffer (hypotonic lysis buffer with 0.1% Nonidet P-40) and then treated with nuclear lysis buffer (20 mm Hepes, pH 8.0, 25% glycerol, 0.42 m NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, and 0.5 mm dithiothreitol with a mixture of protease and phosphatase inhibitors) for 30 min. The sample was spun at 14,000 × g for 30 min, and the supernatant designated the nuclear fraction. Western blots were probed with nuclear (MCM3), cytosolic (β-tubulin) markers. To obtain chromatin-enriched fractions, cells were separated into Triton X-100-soluble and -insoluble fractions. Briefly, cells were lysed in cytoskeleton (CSK) buffer (10 mm PIPES, pH 6.8, 100 mm NaCl, 1.5 mm MgCl2, 300 mm sucrose), containing 0.5% Triton X-100, 1 mm ATP, 1 mm dithiothreitol, and protease inhibitors for 20 min on ice. Protein extracts were then centrifuged for 5 min at 1000 × g. The supernatant was collected and labeled as the soluble fraction. The pellet was washed once with CSK buffer for 5 min on ice, centrifuged for 4 min at 1000 × g, and resuspended in 2× SDS sample buffer. For MNase or NaCl extraction, the pellet was resuspended in 100 μl of CSK buffer supplemented with the indicated amounts of NaCl or MNase (Fermentas) with 1 mm CaCl2 and incubated at the indicated temperature. After incubation, insoluble and soluble materials were separated by centrifugation. Insoluble fraction was resuspended in 2× SDS sample buffer.
Immunofluorescence Assays—For bromodeoxyuridine (BrdUrd) incorporation, cells were pulse-labeled with BrdUrd for 10 min, fixed, and incubated in 3 n HCl for 10 min, and stained with Alexa Fluor 594-conjugated mouse anti-BrdUrd antibody (Molecular Probes). Nuclear DNA was revealed with 4′,6-diamidino-2-phenylindole staining. The cells were observed under a Leica confocal microscope.
In Vitro Kinase Assay—1 μg of GST-Ankrd17 protein was incubated with or without GST-cyclin E or GST-cyclin A and GST-Cdk2 in kinase buffer (50 mm Tris-Cl, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol) with 1 mm cold ATP, 1 μCi of [γ-32P]ATP at 30 °C for 30 min. The reaction was terminated with SDS-PAGE loading buffer. The samples were subjected to 10% SDS-PAGE gel and analyzed by autoradiography.
Mass Spectrometry—pCMV-FLAG-Ankrd17 was transfected into 293T cells using Lipofectamine 2000 (Invitrogen). Subsequently, FLAG-Ankrd17 was enriched using immobilized FLAG antibodies and further isolated using SDS-PAGE under reducing conditions by 8% (w/v) gels. Protein bands were stained by Colloidal Blue (Invitrogen). Gel bands correlated to Ankrd17 protein were excised, and proteins were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin (Promega, Madison, WI). Ankrd17 phosphopeptides were further enriched using a commercial immobilized metal affinity chromatography disc (Pierce). After immobilized metal affinity chromatography, the peptides were desalted with ZiptipC18 (Millipore), and eluted by 7 μl of 50% acetonitrile, 0.1% formic acid and loaded into a PicoTip EMITTER (tip: 1 μm) (new objective). The mass spectrometry analysis was performed on a QTRAP3200 instrument equipped with a nano-ESI source (Applied Biosystems). Peptides were sequenced in an off-line tuning mode. In short, potential phosphopeptides were detected by precursor ion scan (for m/z 79 in negative mode), then the enhanced resolution scan was performed to determine the charge state of the peptide ion, and enhanced product ion scan was performed to obtain collision-induced dissociation MS/MS spectrum of the peptide. The mass spectral data were searched against a data base (SwissProt) using the Mascot searching algorithm (Matrix Science). Phosphopeptides of Ankrd17 were identified based on the following search criteria: trypsin used with up to two missed cleavages, phosphorylation of Ser, Thr, or Tyr, carbamidomethylation of cysteine, oxidized methionine. All amino acid modifications were set as variable modifications.
Transient Transfection and Flow Cytometry Analysis—Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transfections were done on 60-mm dishes using the indicated plasmid DNA, together with pCMV-CD20 plasmid for the selection of transfected cells. Cells were harvested at the indicated time after transfection, fixed in ethanol, and stained with propidium iodide. Flow cytometry analysis was performed with a FACScan instrument.
RNA Interference—RNA interference was carried out using double strand RNA purchased from Invitrogen. The synthetic siRNA duplexes corresponding to the Ankrd17 mRNA sequences 5′-UUA GAA UGC GUA UUA AUG CCA GCC C-3′ (Ankrd17 siRNA1), 5′-UUA ACU GAG UGA CAU CUG GUG GAG G-3′ (Ankrd17 siRNA2) and 5′-UGU ACA AGU AGA UAG AGA AUU ACU G-3′ (Ankrd17 siRNA3) were used to inhibit Ankrd17 protein expression. A synthetic siRNA duplex (Cat#12935-300, Invitrogen) was used as a negative control. Transfection of siRNA into cells was performed according to the manufacturer's instructions. U2OS cells were plated in a six-well plate and transfected with 40 nm siRNA using the Lipofectamine 2000 transfection reagent (Invitrogen).
RESULTS
Association of Ankrd17 with Cdk2 in Vitro and in Vivo—To identify potential Cdk2-associated proteins, a TAP tag purification procedure was performed. 293T cells were transduced with Cdk2 expression retrovirus pMSCV-c-FLAG-HA-Cdk2. Empty retrovirus pMSCV-c-FLAG-HA, which does not express Cdk2, was also included in the experiment as a control. The transduced cells were lysed and subjected to double immunoprecipitation, first with anti-FLAG beads and then with anti-HA beads. The elution was separated on a 4–12% Nupage SDS-PAGE and resolved proteins were visualized by staining with Sigma EZ blue (Fig. 1A). The proteins were excised from the gel and subjected to trypsin digestion followed by liquid chromatographytandem mass spectrometry analysis. The mass spectrum results showed that some proteins were known Cdk2 interacting proteins such as cyclin E, cyclin A, p21, and pRb, but also some new proteins were identified as potential Cdk2-associated partners (Fig. 1B). One of them is a novel ankyrin repeat protein 17 (Ankrd17), which was also identified as a relevant cyclin E/Cdk2 substrate with the use of expression cloning (data not shown).
FIGURE 1.
Identification of Cdk2-associated proteins. A, 293T cells were transduced with Cdk2 expression retrovirus pMSCV-c-FLAG-HA-Cdk2. Empty retrovirus pMSCV-c-FLAG-HA was used as a control. The transduced cells were lysed and subjected to double immunoprecipitation, first with anti-FLAG beads and then with anti-HA beads. The elution was separated on a 4–12% Nupage SDS-PAGE and resolved proteins were visualized by staining with Sigma EZ blue and excised and subjected to mass spectrometry analysis. B, the list of Cdk2-associated proteins were identified by mass spectrometry. C, GST pull-down assay. 293T cells were transfected with Myc-tagged cyclin E and Myc-tagged Cdk2 plasmids. The cell lysates were incubated with GST, GST-Ankrd17, and GST-Ankrd17AQA, respectively. The associated proteins were pulled down by glutathione beads and analyzed by immunoblotting (IB) with Myc antibody. D, 293T cells were transfected with FLAG-tagged Ankrd17 or Ankrd17AQA, in which the RQL motif was mutated to AQA. The cell lysates were immunoprecipitated (IP) with FLAG antibody and immunoblotted with Cdk2 and cyclin E antibodies, respectively. E, the lysates from 293T cells were immunoprecipitated with normal rabbit serum and anti-Ankrd17 antibody, respectively, and immunoblotted with Cdk2 and Ankrd17 antibodies. F, the cell lysates from 293T cells were immunoprecipitated with normal rabbit serum or cyclin E antibody and immunoblotted with Ankrd17 antibody.
To confirm the direct interaction of Ankrd17 with Cdk2, GST-Ankrd17 (amino acids 1681–1819) was used for the GST pull-down experiments (Fig. 1C). In addition, because cyclin E was known to complex with Cdk2, we also examined the interaction of Ankrd17 with cyclin E. The result showed that Ankrd17 interacts with cyclin E/Cdk2 (Fig. 1C, lane 3). The presence of RXL motifs in Ankrd17, together with interaction with cyclin E/Cdk2, suggested that Ankrd17 is a substrate for cyclin E/Cdk2, as RXL motifs are often found in substrates of cyclin kinase and are involved in the interaction between cyclins and their substrates (5–8). To see if the RXL motif in Ankrd17 was responsible for the interaction with cyclin E/Cdk2, RXL mutants were generated, such as GST-Ankrd17AQA and pCMV-FLAG-Ankrd17AQA and tested in GST pull-down and coimmunoprecipitation assays, respectively. Results showed that in both assays, whereas wild type Ankrd17 was found to be associated with cyclin E/Cdk2, the Ankrd17AQA mutants failed to interact with the kinase complex (Fig. 1, lane 4 of panel C; lane 6 of panel D). These data suggested that the RXL motif of Ankrd17 is critical for cyclin E/Cdk2 binding. To further demonstrate the interaction between Ankrd17 and Cdk2 as well as cyclin E in vivo, expression vectors for FLAG-tagged Ankrd17 (pCMV-FLAG-Ankrd17) were introduced into 293T cells by transfection. The cell lysates were immunoprecipitated with FLAG antibody and immunoblotted with cyclin E and Cdk2 antibodies. The tagged Ankrd17 protein was found to be associated with cyclin E/Cdk2 (Fig. 1D, lane 5), indicating that Ankrd17 interacts with the cyclin E/Cdk2 complex in cells. To further confirm the interaction between endogenous Ankrd17 and cyclin E/Cdk2 in vivo, the cell lysates from 293T cells were immunoprecipitated with Ankrd17 antibody and immunoblotted with Cdk2 antibody. The result showed that endogenous Ankrd17 binds to Cdk2 (Fig. 1E). The cell lysates were also immunoprecipitated with cyclin E antibody and immunoblotted with Ankrd17 antibody. The data showed that endogenous Ankrd17 was associated with cyclin E (Fig. 1F).
Characterization of Ankrd17—Ankrd17 is encoded by a non-annotated gene located at human chromosome 4q13.3. Ankrd17 has two isoforms (a and b). Full-length cDNA of Ankrd17 of isoform b has an open reading frame of 7059 bp with a calculated molecular mass of 260 kDa (2352 amino acids) (Fig. 2A). It was ubiquitously expressed in all human tissues examined by reverse transcriptase-PCR (Fig. 2B). Predicted Ankrd17 protein consists of 2 clusters with a total of 25 ankyrin repeats at its N-terminal, one NES site, one NLS site, one KH domain, and one RXL motif at its C-terminal.
FIGURE 2.
Characteristics of Ankrd17 protein. A, structure of ankyrin repeat protein Ankrd17. B, Ankrd17 expressed in different tissues. RNA from different human tissues were isolated and 1 μg of RNA was used for reverse transcriptase-PCR with Ankrd17 primers and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers as internal control. C, Ankrd17 was localized in the nucleus in U2OS and HeLa cells. U2OS and HeLa cells were fractionated as cytosolic supernatant (C) and nuclear fraction (N). The lysates were subjected to immunoblot with Ankrd17, β-tubulin, and MCM antibodies. TCE refers to total cell lysate. D, the phosphorylation sites of Ankrd17 identified by mass spectral analysis. 293T cells were transfected with pCMV-FLAG-Ankrd17, pCS2mt-cyclin E, and pCMV-Myc-Cdk2. Cell lysates were harvested and Ankrd17 protein was immunoprecipitated with FLAG beads and subjected for mass spectrometry analysis.
To examine the localization of Ankrd17, the U2OS and HeLa cell lysates were fractionated into nuclear and cytosolic fractions and immunoblotted with Ankrd17, β-tubulin, and MCM3 antibodies. The result indicated that Ankrd17 was localized in the nucleus in both cell lines (Fig. 2C).
To examine the phosphorylation sites of Ankrd17, Ankrd17 purified from 293T cells after transfection with a pCMV-FLAG-Ankrd17 expression plasmid was subjected to a mass spectrometry analysis (Fig. 2D). Three phosphorylation sites in Ankrd17 at Ser1791, Ser1794, and Ser2150 were identified in this assay.
Phosphorylation of Ankrd17 by Cyclin E/Cdk2 in Vitro—The interaction of Ankrd17 with the cyclin E/Cdk2 complex and involvement of the RXL motif in the interaction suggested that Ankrd17 is a substrate of cyclin E/Cdk2. To examine whether Ankrd17 can be phosphorylated by cyclin E/Cdk2, GST-Ankrd17 (amino acids 1681–1819) and GST-Ankrd17AQA protein were incubated with [γ-32P]ATP in the absence or presence of purified GST-cyclin E and Cdk2. As shown in the Fig. 3A, GST-Ankrd17 can be phosphorylated by cyclin E/Cdk2, indicating that the Ankrd17 is a substrate for cyclin E/Cdk2. In contrast, GST-Ankrd17AQA was not efficiently phosphorylated by cyclin E/Cdk2. The GST-cyclin A/Cdk2 was also included in the kinase assay, the result showed that Ankrd17 was phosphorylated more specifically by cyclin E/Cdk2 than cyclin A/Cdk2 (Fig. 3B). According to the mass spectrometry data as shown in Fig. 2D, Ser1791 and Ser1794 in Ankrd17 were phosphorylated in vivo. To test whether Ankrd17 Ser1791 and Ser1794 were phosphorylated by cyclin E/Cdk2, GST Ankrd17 S1737A/S1740A/S1791A/S1794A and triple mutants were generated and subjected to in vitro kinase assay, the result showed that the phosphorylation of Ankrd17 were decreased when Ser1791 and Ser1794 were mutated to Ala, whereas its phosphorylation was recovered to 86 and 93% when they were changed back to Ser, respectively (Fig. 3C, lanes 5 and 6). Interestingly, Ser1740 in Ankrd17 was also important for its phosphorylation (Fig. 3C, lane 4). Because GST-Ankrd17 S1737A/S1740A/S1791A/S1794A still could be phosphorylated, it implied that there were other sites of serine or threonine in this region that can be phosphorylated by cyclin E/Cdk2 at certain levels.
FIGURE 3.
In vitro kinase assay. A, 1 μg of GST, GST-Ankrd17, or GST-Ankrd17AQA were incubated with [γ-32P]ATP in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of purified GST-cyclin E and GST-Cdk2. The reactions were subjected for SDS-PAGE and kinase activity was visualized by autoradiography of the gel. B, GST, GST-Ankrd17, or GST-Ankrd17AQA were incubated with [γ-32P]ATP in the presence of GST-cyclin E and GST-Cdk2 (Lanes 1–3) or GST-cyclin A and GST-Cdk2 (lanes 4–6). The reactions were subjected to SDS-PAGE and the phosphorylation of Ankrd17 was visualized by autoradiography of the gel. C, the GST-Ankrd17 (amino acids 1729–1795) and its Ser/Ala mutants were subjected to in vitro kinase assay as described in A. Lanes 1–7 were GST, GST-Ankrd17, GST-Ankrd17 S1740A/S1791A/S1794A, GST-Ankrd17 S1737A/S1791A/S1794A, GST-Ankrd17 S1737A/S1740A/S1794A, GST-Ankrd17 S1737A/S1740A/S1791A, and GST-Ankrd17 S1737A/S1740A/S1791A/S1794A, respectively.
Effects of Ankrd17 on U2OS Cell Cycle Distribution—To gain further insight into the function of Ankrd17, we examined the effect of Ankrd17 overexpression on cell cycle distribution. When transfected into U2OS cells, overexpressed Ankrd17 caused a significant increase in the fraction of cells in S phase and a concomitant decrease in the G1 population in a dose-dependent manner (Fig. 4A). To examine whether the increase of S phase population was due to the S phase blockage caused by overexpression of Ankrd17, we used nocodazole to block the cell cycle at the M phase and analyzed the cell cycle distribution of Ankrd17-overexpressed cells and control cells. As shown in Fig. 4B, Ankrd17-overexpressed cells could proceed from S phase to the M phase as well as the control cells, indicating that the increase of S phase population in Ankrd17-overexpressed cells was not the result of the S phase blockage. Furthermore, U2OS cells were cotransfected with cyclin E, Cdk2, and Ankrd17 and subjected to flow cytometry analysis, the result showed that Ankrd17 coordinated with cyclin E/Cdk2 to promote cell cycle progression (Fig. 4C). These results suggested that Ankrd17 is a positive regulator for the cell cycle progression.
FIGURE 4.
Overexpression of Ankrd17 promotes cell cycle progression. A, the U2OS cells were transfected with pcDNA3.1 as control or the indicated amount of pCMV-FLAG-Ankrd17 together with pCMV-CD20 plasmid for the selection of transfected cells. Cells were harvested at 48 h after transfection and subjected for flow cytometry analysis. The results represent the means from triplicate samples. B, U2OS cells were transfected with 7.5 μg of pCMV-FLAG-Ankrd17 or pcDNA3.1 as control. 24 h after transfection, cells were then treated with DMSO (mock) or 100 ng/ml nocodazole for 22 h and harvested for flow cytometry. C, U2OS cells were transfected with pCMV-FLAG-Ankrd17 alone or with pCMV-Myc-Cdk2 and pCMV-Myc-cyclin E as indicated and harvested 48 h after transfection and subjected to flow cytometry analysis.
Effects of Depletion of Ankrd17 on DNA Replication and Cell Cycle Progression—To address whether Ankrd17 is required for cell cycle progression, we used RNA interference assays with synthetic siRNA duplexes to inhibit the expression of Ankrd17 protein in U2OS cells and examined the effect of the depletion of Ankrd17 expression on cell cycle distribution. As shown in Fig. 5A, 3 sets of siRNA of Ankrd17 (1, 2, and 3) were tested for eliminating Ankrd17 protein in U2OS cells. Although all 3 siRNAs were capable to knockdown Ankrd17 protein significantly at 48 h after transfection, siRNA1 and siRNA3 are more efficient at an earlier time point (12 h). Flow cytometry data showed that all 3 sets of siRNA were able to significantly block the cell entering S phase when compared with the control (Fig. 5B). Interestingly, siRNA1 and siRNA3 were found more effective in blocking S phase entry than siRNA2, which is consistent with the different knockdown efficiencies found for these siRNAs. These observations suggested that Ankrd17 has an essential role in regulating S phase entry.
FIGURE 5.
Effects of depletion of Ankrd17 on U2OS cell cycle distribution. A, analysis of the knockdown efficiency of three Ankrd17 siRNAs. U2OS cells were transfected with control siRNA and Ankrd17 siRNA1, -2, and -3. Cells were harvested at 12 or 48 h after transfection and the protein level of Ankrd17 were assessed by immunoblotting. B, U2OS cells were transfected with control siRNA and Ankrd17 siRNA1, -2, and -3 as in A, 48 h after transfection cells were harvested and analyzed for flow cytometry. C, U2OS cells were transfected with control siRNA and Ankrd17 siRNA1 for 24 h and arrested by serum starvation for 48 h and released for 16 h, and flow cytometry was performed before and after serum stimulation. The results represent the means from triplicate samples. D, depletion of Ankrd17 regulates the expression of p53 and p21. U2OS cells were transfected with control siRNA and Ankrd17 siRNA. The cell lysates were harvested and subjected to immunoblot with the antibodies as indicated.
To further confirm that depletion of Ankrd17 blocked S phase entry, the U2OS cells transfected with Ankrd17 siRNA were starved without serum and release for 16 h, the cell cycle profiles were analyzed by flow cytometry. As seen in Fig. 5C, after serum release, the fraction of Ankrd17-depleted cells entering the S/G2/M phase (46.6%) was much lower than that of control cells (69.8%) indicating depletion of Ankrd17 inhibited cell cycle progression.
To determine whether depletion of Ankrd17 regulates proteins involved in cell cycle, U2OS cells were transfected with Ankrd17 siRNA. Immunoblot analysis showed that silencing Ankrd17 was associated with increases in p53 and p21 and decreases in cyclin A as compared with that in cells transfected with control siRNA, whereas the level of cyclin D, cyclin E, and p27 were not changed (Fig. 5D). It is known that activation of p53 induces the expression of various genes including p21, which mediates cell cycle arrest (34). This result suggested that depletion of Ankrd17 may activate the p53-p21 pathway and block cell cycle progression.
To examine the effect of the inhibition of Ankrd17 expression on DNA replication, we monitored the incorporation of BrdUrd in U2OS cells transfected with siRNA duplexes of Ankrd17. The U2OS cells were labeled with BrdUrd at 48 and 72 h after transfection. The percentage of BrdUrd-labeled cells was scored by immunofluorescence (Fig. 6, A–C). The percentage of BrdUrd positive cells in Ankrd17-depleted cells was lower than that in control cells, which was consistent with the results from flow cytometric analysis.
FIGURE 6.
Depletion of Ankrd17 inhibits DNA replication. U2OS cells were transfected with control siRNA and Ankrd17 siRNA1. The cells were pulse labeled with BrdUrd for 10 min at 48 or 72 h after transfection. The cells were fixed and stained with anti-BrdUrd antibody (red) and DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) (A for 48 h, B for 72 h). The percentage of BrdUrd positive cells were counted from 600 cells (C). The mean results from two independent experiments are depicted. D, the U2OS cells were transfected with siRNA and starved for 48 h, then released for 2–20 h as indicated and labeled with BrdUrd. The BrdUrd-positive cells were counted.
Following transfection with siRNA duplexes of Ankrd17, U2OS cells were starved for 48 h and released for 2–20 h at 2-h intervals and labeled with BrdUrd. As shown in Fig. 6D, BrdUrd incorporation in the cells transfected with siRNA of Ankrd17 decreased dramatically compared with that in the cells transfected with control siRNA. These results further indicated that depletion of Ankrd17 blocked the DNA replication and the S phase entry.
Ankrd17 Localized on Chromatin in a Cell Cycle-dependent Manner—As shown in Fig. 2C, Ankrd17 was predominantly localized in the nucleus. To further investigate its localization in the nucleus, U2OS cells were separated into CSK-soluble and -insoluble fractions. As shown in Fig. 7A, Ankrd17 was detected both in CSK-soluble and -insoluble fractions. The CSK-insoluble fraction was further digested with micrococcal nuclease. Unlike MCM3, Ankrd17 did not dissociate from the CSK-insoluble fraction treated with micrococcal nuclease (MNase) indicating Ankrd17 may be either a nuclear matrix protein or chromatin-bound protein that is resistant to MNase. To determine this, the CSK-insoluble fraction was treated with different concentrations of NaCl. As shown in Fig. 7A (right panel), most of the Ankrd17 dissociated at a salt concentration of 0.3 m NaCl, which shared the similar pattern with the origin recognition complex protein Orc1, Cdc6, and MCM10 (35, 36). Lamin B, as a nuclear matrix protein, was not extracted by 0.3 m NaCl. These results strongly indicate that the CSK-insoluble fraction of Ankrd17 binds to chromatin, not a nuclear matrix protein.
FIGURE 7.
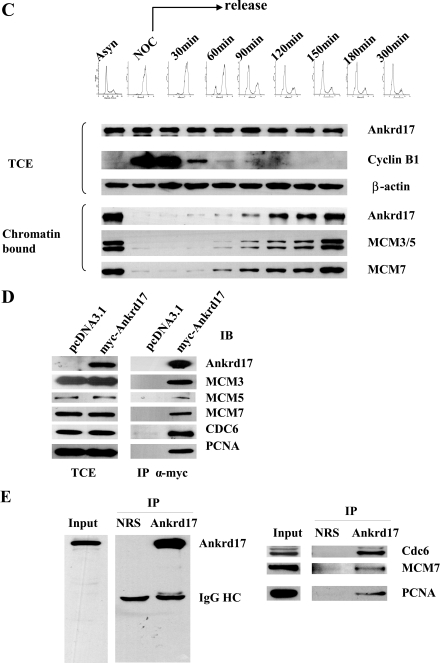
Ankrd17 bound to chromatin and interacted with proteins in DNA pre-replication complex. A, U2OS cells were lysed in CSK buffer; the CSK-insoluble (CSKP) fraction was then treated with micrococcal nuclease (MNase) or the indicated concentration of NaCl. The soluble (S) and insoluble (P) fractions were separated by centrifugation. The total cell extract (TCE) and all fractions were subjected to immunoblot with Ankrd17 and MCM3 antibodies. B, U2OS cells were synchronized at G1, G1/S, S, S/G2, and M phases assessed by flow cytometry (upper panel) as described under “Experimental Procedures.” Total cell extracts and chromatin-bound proteins were prepared for immunoblotting with the indicated antibodies. C, HeLa cells were synchronized at M phase with nocodazole and harvested by mitotic shake-off. Cells were collected at different time points after release from the block and the total cell extract and CSK-insoluble fraction (chromatin-bound) were prepared and subjected to immunoblot with the indicated antibodies and cyclin B as the G2/M phase marker. The DNA contents of cells were determined by flow cytometry (upper panel). D, 293T cells were transfected with pCMV-Myc-Ankrd17 or pcDNA3.1 as control. After 48 h the cells were harvested and cell lysates were immunoprecipitated with Myc antibody and immunoblotted with MCM3, MCM5, MCM7, Cdc6, and PCNA antibodies, respectively. E, 293T cell lysates were immunoprecipitated (IP) with Ankrd17 antibody and normal rabbit serum as control and immunoblotted with indicated antibodies.
To examine the protein level of Ankrd17 during the cell cycle, U2OS cells were synchronized in G1, G1/S, S, S/G2, and M phases, respectively. The cells were collected and their progression through cell cycle was monitored by flow cytometry. The total cell extract and chromatin-bound fraction from different phases were prepared and analyzed by immunoblotting with Ankrd17 as well as MCM3, -5, -7, Cdc6, and PCNA antibodies. As shown in Fig. 7B, Ankrd17 in the chromatin-bound fraction was much lower in M phase compared with that in other phases, whereas Ankrd17 in the total cell extract was expressed at similar levels in all phases of cell cycle. A binding profile similar to that of chromatin was also found for MCM3, MCM5, and MCM7. They were absent during mitosis, and they reassociated with chromatin in early G1 phase, similar to results described previously. Very similar results have been obtained in HeLa cells (data not shown).
We further analyzed the chromatin association of Ankrd17 in cells that were released from nocodazole block to the G1 phase. Because the U2OS cell line enters the G1 phase very rapidly (within 20 min) after release from nocodazole-mediated mitotic arrest, it was very difficult to obtain consistent data to carry out the experiment to determine the Ankrd17 loading onto chromatin using U2OS cells. To overcome this technical problem, we utilized HeLa cells, which showed similar expression patterns of Ankrd17 in different cell phases (data not shown) and had ∼3 h to exit mitosis after release from nocodazole-induced mitotic arrest. As shown in Fig. 7C, Ankrd17 loading onto chromatin was gradually increased, whereas cyclin B as the M phase marker was decreased as cells exit M phase. Interestingly, the chromatin-bound Ankrd17 accumulated at the M to G1 phase transition, which showed a similar pattern as MCM3, -5, and -7.
Ankrd17 Interacted with Proteins Involved in DNA Replication—The cell cycle-dependent chromatin loading of Ankrd17 leads us to ask whether it directly participates in DNA replication. The interaction between Ankrd17 and MCM family members, Cdc6, and PCNA were examined. 293T cells were transfected with pCMV-FLAG-Ankrd17. The cell lysates were immunoprecipitated with FLAG antibody and immunoblotted with MCM3, MCM5, MCM7, Cdc6, and PCNA antibodies, respectively. As shown in Fig. 7D, Ankrd17 can interact with MCM3, MCM5, MCM7, Cdc6, and PCNA. To further confirm the interaction between Ankrd17 and the MCM family member, Cdc6, and PCNA, the cell lysates of 293T cells were immunoprecipitated with Ankrd17 antibody or normal rabbit serum as control and immunoblotted with MCM7, Cdc6, and PCNA antibodies. The result showed that endogenous Ankrd17 can interact with MCM7, Cdc6, and PCNA (Fig. 7E).
Depletion of Ankrd17 Decreases the Loading of CDC6 and PCNA onto the Chromatin—To analyze the role of Ankrd17 on the replication machinery, we used a well characterized fractionation method to determine the loading of Ankrd17 to chromatin; wherein actively engaged replication proteins are tethered to chromatin, replication proteins that are not involved in replication are in a soluble nucleoplasmic pool (33). The siRNA-transfected U2OS cell lysates were separated as CSK-soluble and -insoluble parts. The total cell extracts and CSK-insoluble fraction were immunoblotted with MCM3, -5, -7, Cdc6, Cdc45, and PCNA antibodies. As seen in Fig. 8A, Cdc6 levels were decreased both in total cell extract and the CSK-insoluble fraction in Ankrd17-depleted cells. PCNA was found to be significantly less only in the CSK-insoluble fraction (chromatin-bound fraction) in Ankrd17 knockdown cells, but not the total amount of PCNA. In contrast, the levels of MCM3, -5, -7, and Cdc45 remained unchanged in both the total cell extracts and CSK-insoluble fraction in Ankrd17-depleted cells. Because depletion of Ankrd17 caused a dramatic decrease of chromatin-bound CDC6 and PCNA, this suggested that assembly of the replication complex was deregulated with the loss of Ankrd17. These results were further validated by immunofluorescence data, wherein U2OS cells were transfected with control siRNA (control) and Ankrd17 siRNA (Ankrd17) or untransfected (untreated). These cells were extracted with or without CSK buffer, fixed, and followed by immunostaining for PCNA (red) and MCM3 (green) antibodies, respectively (Fig. 8B). The PCNA and MCM3 positive cells were scored and the results showed that the ratio of PCNA positive cells in CSK-extracted cells was significantly lower in the Ankrd17-depleted cells when compared with controls (Fig. 8C, left panel), whereas the percentage of PCNA positive cells did not change in unextracted cells. The percentage of MCM3 positive cells was not altered in Ankrd17-depleted cells in both CSK-treated and untreated cells (Fig. 8C, right panel). The results were consistent with the immunoblotting data (Fig. 8A). Collectively, our data suggested that Ankrd17 interacts with proteins involved in DNA replication. Loss of Ankrd17 affects Cdc6 and PCNA loading onto DNA.
FIGURE 8.
Depletion of Ankrd17 decreases Cdc6 and PCNA loading on chromatin. A, U2OS cells were transfected with control siRNA and Ankrd17 siRNA1 and extracted with CSK buffer. Total cell extract and CSK-insoluble proteins were subjected for immunoblotting with the indicated antibodies. B, the untransfected or transfected cells were extracted with or without CSK buffer (unextracted), fixed, and immunostained for PCNA (red) and MCM3 (green) antibodies, DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). C, untransfected and transfected cells were subjected to in situ extraction with CSK buffer or mock extraction (unextracted), fixed, and immunostained for PCNA (left) or MCM3 (right) antibodies as B. The percentages of nuclei staining positive cells for PCNA or MCM3 were counted.
DISCUSSION
Our results suggest that Ankrd17 is a novel substrate of cyclin E/Cdk2 with potent cell cycle regulatory properties. Its overexpression promotes cell cycle progression, whereas depletion blocks DNA replication and regulates the level of p53 and p21. The predicted amino acid sequence analysis reveals five characteristic features of the Ankrd17 protein (see Fig. 2A), including 25 ankyrin repeats, NES, NLS, KH, and RXL motif. The RXL motif was demonstrated to be essential for Ankrd17 binding to the cyclin E/Cdk2 complex (see Fig. 1, C and D). Ankyrin repeats are a conserved sequence of 33 amino acids found in many proteins with a wide variety of functions, ranging from cell cycle control, transcription regulation, and cell differentiation, to cell anchorage (37–39). One ankyrin repeat consists of a β-hairpin loop and two α-helices. These repeats form a higher order complex where protein-protein interacts via the exposed β-hairpin loops. It has been found that some ankyrin repeat proteins may bind to multiple targets to form functional complexes. For example, p16 not only binds to Cdk4 and Cdk6, but also to transcription factor NF-κB and c-Jun kinase under physiological conditions (40, 41). Another example of the multipartner ankyrin protein is gankyrin, which is overexpressed in esophageal carcinoma and hepatoma (39). Its physiological partners include Cdk4, Rb, and S6 ATPase of the 26 S proteasome and HDM2 (42, 43). Here we demonstrate that Ankrd17 can bind cyclin E/Cdk2, MCM family members (MCM3, MCM5, and MCM7), Cdc6, and PCNA. However, more studies are required to address whether Ankrd17 protein may bind to different target proteins and function in different pathways.
A proportion of Ankrd17 is located in chromatin regions that are resistant to nuclease digestion. It can be released from the nuclease-resistant fraction by high salt concentrations, which shares the property of other nuclease-resistant chromatin-bound proteins, such as Orc1, MCM10, Cdc6, transcribing RNA polymerase, and SWI/SNF chromatin remodeling complex (35, 36, 44, 45). This observation suggests that Ankrd17 is involved in DNA pre-replication complex formation, chromatin remodeling, and transcribing chromatin. Like the MCM2–7 subunits, Ankrd17 is found in both the soluble and chromatin-bound fractions in the nucleus. In mammalian cells, the MCM subunits assemble on DNA beginning in late mitosis and remain associated with the chromatin until DNA synthesis is completed. The chromatin-bound Ankrd17 showed a similar pattern as MCMs, suggesting that Ankrd17 may be involved in DNA replication. The observation that Ankrd17 interacts with MCM family members, Cdc6, and PCNA supports this hypothesis, as does the decreased association of Cdc6 and PCNA with chromatin when Ankrd17 is depleted. Unlike the MCMs, dissociation of Ankrd17 from chromatin appears to occur slightly later in the cell cycle (early M as opposed to G2 phase) than for the MCM subunits, which could imply that Ankrd17 might play a role in other aspects besides DNA replication.
DNA replication initiation proceeds in the presence of a pre-replicative complex (pre-RC) at origins of replication and the licensing cofactor Cdt1 and Cdc6 that recruit the MCM2–7 complex. The formation of a replication fork is promoted by subsequent recruitment of additional factors such as MCM10, Cdc45, and the GINS complex, and activation of S phase cyclin-dependent kinases (Cdks) and the Cdc7-Dbf4 kinase (DDK), MCM2–7 complex together with associated factors recruit DNA polymerases and other factors required for DNA synthesis, such as PCNA (46, 47). In this study, we have shown that depletion of Ankrd17 affects the loading of PCNA, but not MCMs, onto the chromatin. MCMs are components of the pre-RC, whereas PCNA is a component of the replication complex that synthesizes DNA at the replication fork. This observation might suggest that Ankrd17 function is dispensable for the loading of MCMs onto chromatin to form the pre-RC, but is needed for the formation of replicative complexes. However, additional analyses are required to accurately determine whether Ankrd17 directly or indirectly affects the loading of PCNA. Previous researches suggest that Cdc6 is essential for MCM loading onto the chromatin. Intriguingly, depletion of Ankrd17 causes the decrease of Cdc6 loading onto chromatin but has no effects on the MCM family. This may be due to the fact that a lower level of Cdc6 can be sufficient enough to mediate MCM loading in Ankrd17-depleted cells.
It has been found that phosphorylation of MCM family members by cyclin/Cdks regulates their assembly into the prereplication complex and their helicase activity, which is essential for DNA replication (30, 48, 49). Ankrd17 can be phosphorylated by cyclin E/Cdk2, but whether phosphorylation of Ankrd17 regulates its association with the components in the pre-replication complex and loading onto chromatin and controls the activity of pre-RC needs to be further elucidated. In addition, Ankrd17 affects the loading of Cdc6 and PCNA onto chromatin, whereas the connection between the phosphorylation of Ankrd17 and the recruitment of Cdc6 and PCNA still need to be addressed in the future.
To date, studies in mice suggest that Ankrd17 is involved in liver development, but the detailed mechanism is unclear (50). Ankrd17 was serologically defined as breast cancer antigen NY-BR-16 and its peptide APRVPVQAL was identified as a major histocompatibility complex-associated tumor peptide (51, 52) but its relevance to cancer is currently unknown. Recent study using kinase engineering strategy combined with chemical enrichment and mass spectrometry showed that Ankrd17 was a potential substrate of cyclin A/Cdk2 (53). Given the interesting and complicated structural features of Ankrd17, it is tempting to speculate that Ankrd17 could play pivotal roles in cell cycle regulation and DNA replication. Further studies are warranted to unmask the link between functional domains of Ankrd17 and their potential role in cell cycle progression and tumorigenesis.
Acknowledgments
We thank Dr. W. J. Harper (Harvard Medical School) for helpful suggestions on this work and Dr. S. Gygi (Harvard Medical School) for kind help with mass spectrometry. We are grateful to Dr. D. Li (Shanghai Jiaotong University), Y. Yuan (University of Pennsylvania), and G. Nalepa (Indiana University) for critical reading of the manuscript.
This work was supported by National Natural Science Foundation of China Grants 30771098 and 30871238 and the “100 Talents Program” of the Chinese Academy of Sciences. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Cdk, cyclin-dependent kinase; BrdUrd, bromodeoxyuridine; pre-RC, prereplicative complex; MNase, micrococcal nuclease; GST, glutathione S-transferase; NPM, nucleophosmin; HA, hemagglutinin; PCNA, proliferating cell nuclear antigen; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; PIPES, 1,4-piperazinediethanesulfonic acid; siRNA, small interfering RNA.
References
- 1.Hwang, H. C., and Clurman, B. E. (2005) Oncogene 24 2776-2786 [DOI] [PubMed] [Google Scholar]
- 2.Prior, M., Lehmann, S., Sy, M. S., Molloy, B., and McMahon, H. E. (2007) J. Virol. 81 11195-11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reed, S. I. (1997) Cancer Surv. 29 7-23 [PubMed] [Google Scholar]
- 4.Sherr, C. J. (1994) Cell 79 551-555 [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. R., Nobel, M., Endicott, J. A., and Johnson, L. N. (1999) Nat. Cell Biol. 1 438-443 [DOI] [PubMed] [Google Scholar]
- 6.Schulman, B. A., Lindstrom, D. L., and Harlow, E. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 10453-10458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall, C., Nelson, D. M., Ye, X., Baker, K., DeCaprio, J. A., Seeholzer, S., Lipinski, M., and Adams, P. D. (2001) Mol. Cell. Biol. 21 1854-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams, P. D., Sellers, W. R., Sharma, S. K., Wu, A. D., Nalin, C. M., and Kaelin, W. G., Jr. (1996) Mol. Cell. Biol. 16 6623-6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson, N. (1998) Genes Dev. 12 2245-2262 [DOI] [PubMed] [Google Scholar]
- 10.Trimarchi, J. M., and Lees, J. (2002) Nat. Rev. Mol. Cell. Biol. 3 11-20 [DOI] [PubMed] [Google Scholar]
- 11.Carrano, A. C., Eytan, E., Hershko, A., and Pagano, M. (1999) Nat. Cell Biol. 1 193-199 [DOI] [PubMed] [Google Scholar]
- 12.Ganoth, D., Bornstein, G., Ko, T. K., Larsen, B., Tyers, M., Pagano, M., and Hershko, A. (2001) Nat. Cell Biol. 3 321-324 [DOI] [PubMed] [Google Scholar]
- 13.Hara, T., Kamura, T., Nakayama, K., Oshikawa, K., Hatakeyama, S., and Nakayama, K. (2001) J. Biol. Chem. 276 48937-48943 [DOI] [PubMed] [Google Scholar]
- 14.Nakayama, K., Nagahama, H., Minamishima, Y. A., Matsumoto, M., Nakamichi, I., Kitagawa, K., Shirane, M., Tsunematsu, R., Tsukiyama, T., Ishida, N., Kitagawa, M., Nakayama, K., and Hatakeyama, S. (2000) EMBO J. 19 2069-2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagano, M. (2004) Mol. Cell 14 414-416 [DOI] [PubMed] [Google Scholar]
- 16.Ma, T., Van Tine, B. A., Wei, Y., Garrett, M. D., Nelson, D., Adams, P. D., Wang, J., Qin, J., Chow, L. T., and Harper, J. W. (2000) Genes Dev. 14 2298-2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao, J., Kennedy, B. K., Lawrence, B. D., Barbie, D. A., Matera, A. G., Fletcher, J. A., and Harlow, E. (2000) Genes Dev. 14 2283-2297 [PMC free article] [PubMed] [Google Scholar]
- 18.Wei, Y., Jin, J., and Harper, J. W. (2003) Mol. Cell. Biol. 23 3669-3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deran, M., Pulvino, M., Greene, E., Su, C., and Zhao, J. (2008) Mol. Cell. Biol. 28 435-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye, X., Wei, Y., Nalepa, G., and Harper, J. W. (2003) Mol. Cell. Biol. 23 8586-8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ait-Si-Ali, S., Polesskaya, A., Filleur, S., Ferreira, R., Duquet, A., Robin, P., Vervish, A., Trouche, D., Cabon, F., and Harel-Bellan, A. (2000) Oncogene 19 2430-2437 [DOI] [PubMed] [Google Scholar]
- 22.Wang, H., Larris, B., Peiris, T. H., Zhang, L., Le Lay, J., Gao, Y., and Greenbaum, L. E. (2007) J. Biol. Chem. 282 24679-24688 [DOI] [PubMed] [Google Scholar]
- 23.Morris, L., Allen, K. E., and La Thangue, N. B. (2000) Nat. Cell Biol. 2 232-239 [DOI] [PubMed] [Google Scholar]
- 24.Okuda, M., Horn, H. F., Tarapore, P., Tokuyama, Y., Smulian, A. G., Chan, P. K., Knudsen, E. S., Hofmann, I. A., Snyder, J. D., Bove, K. E., and Fukasawa, K. (2000) Cell 103 127-140 [DOI] [PubMed] [Google Scholar]
- 25.Tokuyama, Y., Horn, H. F., Kawamura, K., Tarapore, P., and Fukasawa, K. (2001) J. Biol. Chem. 276 21529-21537 [DOI] [PubMed] [Google Scholar]
- 26.Tarapore, P., Shinmura, K., Suzuki, H., Tokuyama, Y., Kim, S. H., Mayeda, A., and Fukasawa, K. (2006) FEBS Lett. 580 399-409 [DOI] [PubMed] [Google Scholar]
- 27.Chen, Z., Indjeian, V. B., McManus, M., Wang, L., and Dynlacht, B. D. (2002) Dev. Cell 3 339-350 [DOI] [PubMed] [Google Scholar]
- 28.Coverley, D., Laman, H., and Laskey, R. A. (2002) Nat. Cell Biol. 4 523-528 [DOI] [PubMed] [Google Scholar]
- 29.Montagnoli, A., Valsasina, B., Brotherton, D., Troiani, S., Rainoldi, S., Tenca, P., Molinari, A., and Santocanale, C. (2006) J. Biol. Chem. 281 10281-10290 [DOI] [PubMed] [Google Scholar]
- 30.Lin, D. I., Aggarwal, P., and Diehl, J. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8079-8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luscher-Firzlaff, J. M., Lilischkis, R., and Luscher, B. (2006) FEBS Lett. 580 1716-1722 [DOI] [PubMed] [Google Scholar]
- 32.Ye, X., Nalepa, G., Welcker, M., Kessler, B. M., Spooner, E., Qin, J., Elledge, S. J., Clurman, B. E., and Harper, J. W. (2004) J. Biol. Chem. 279 50110-50119 [DOI] [PubMed] [Google Scholar]
- 33.Braden, W. A., Lenihan, J. M., Lan, Z., Luce, K. S., Zagorski, W., Bosco, E., Reed, M. F., Cook, J. G., and Knudsen, E. S. (2006) Mol. Cell. Biol. 26 7667-7681 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Bartek, J., and Lukas, J. (2001) FEBS Lett. 490 117-122 [DOI] [PubMed] [Google Scholar]
- 35.Kreitz, S., Ritzi, M., Baack, M., and Knippers, R. (2001) J. Biol. Chem. 276 6337-6342 [DOI] [PubMed] [Google Scholar]
- 36.Fujita, M., Yamada, C., Goto, H., Yokoyama, N., Kuzushima, K., Inagaki, M., and Tsurumi, T. (1999) J. Biol. Chem. 274 25927-25932 [DOI] [PubMed] [Google Scholar]
- 37.Mosavi, L. K., Cammett, T. J., Desrosiers, D. C., and Peng, Z. Y. (2004) Protein Sci. 13 1435-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, J., Mahajan, A., and Tsai, M. D. (2006) Biochemistry 45 15168-15178 [DOI] [PubMed] [Google Scholar]
- 39.Ortiz, C. M., Ito, T., Tanaka, E., Tsunoda, S., Nagayama, S., Sakai, Y., Higashitsuji, H., Fujita, J., and Shimada, Y. (2008) Int. J. Cancer 122 325-332 [DOI] [PubMed] [Google Scholar]
- 40.Wolff, B., and Naumann, M. (1999) Oncogene 18 2663-2666 [DOI] [PubMed] [Google Scholar]
- 41.Choi, B. Y., Choi, H. S., Ko, K., Cho, Y. Y., Zhu, F., Kang, B. S., Ermakova, S. P., Ma, W. Y., Bode, A. M., and Dong, Z. (2005) Nat. Struct. Mol. Biol. 12 699-707 [DOI] [PubMed] [Google Scholar]
- 42.Higashitsuji, H., Itoh, K., Nagao, T., Dawson, S., Nonoguchi, K., Kido, T., Mayer, R. J., Arii, S., and Fujita, J. (2000) Nat. Med. 6 96-99 [DOI] [PubMed] [Google Scholar]
- 43.Dawson, S., Apcher, S., Mee, M., Higashitsuji, H., Baker, R., Uhle, S., Dubiel, W., Fujita, J., and Mayer, R. J. (2002) J. Biol. Chem. 277 10893-10902 [DOI] [PubMed] [Google Scholar]
- 44.Reyes, J. C., Muchardt, C., and Yaniv, M. (1997) J. Cell Biol. 137 263-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izumi, M., Yatagai, F., and Hanaoka, F. (2004) J. Biol. Chem. 279 32569-32577 [DOI] [PubMed] [Google Scholar]
- 46.Blow, J. J., and Dutta, A. (2005) Nat. Rev. Mol. Cell. Biol. 6 476-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moldovan, G. L., Pfander, B., and Jentsch, S. (2007) Cell 129 665-679 [DOI] [PubMed] [Google Scholar]
- 48.Komamura-Kohno, Y., Karasawa-Shimizu, K., Saitoh, T., Sato, M., Hanaoka, F., Tanaka, S., and Ishimi, Y. (2006) FEBS J. 273 1224-1239 [DOI] [PubMed] [Google Scholar]
- 49.Wheeler, L. W., Lents, N. H., and Baldassare, J. J. (2008) Cell Cycle 7 2179-2188 [DOI] [PubMed] [Google Scholar]
- 50.Watt, A. J., Jones, E. A., Ure, J. M., Peddie, D., Wilson, D. I., and Forrester, L. M. (2001) Mech. Dev. 100 205-215 [DOI] [PubMed] [Google Scholar]
- 51.Hofmann, S., Gluckmann, M., Kausche, S., Schmidt, A., Corvey, C., Lichtenfels, R., Huber, C., Albrecht, C., Karas, M., and Herr, W. (2005) Mol. Cell Proteomics 4 1888-1897 [DOI] [PubMed] [Google Scholar]
- 52.Scanlan, M. J., Gout, I., Gordon, C. M., Williamson, B., Stockert, E., Gure, A. O., Jager, D., Chen, Y. T., Mackay, A., O'Hare, M. J., and Old, L. J. (2001) Cancer Immun. 1 4. [PubMed] [Google Scholar]
- 53.Chi, Y., Welcker, M., Hizli, A. A., Posakony, J. J., Aebersold, R., and Clurman, B. E. (2008) Genome Biol. 9 R149. [DOI] [PMC free article] [PubMed] [Google Scholar]