Abstract
Although landscapes of several histone marks are now available for Arabidopsis thaliana and Oryza sativa, such profiles remain static and do not provide information about dynamic changes of plant epigenomes in response to developmental or environmental cues. Here, we analyzed the effects of light on four histone modifications (acetylation and trimethylation of lysines 9 and 27 on histone H3: H3K9ac, H3K9me3, H3K27ac, and H3K27me3, respectively). Our genome-wide profiling of H3K9ac and H3K27ac revealed that these modifications are nontransposable element gene-specific. By contrast, we found that H3K9me3 and H3K27me3 target nontransposable element genes, but also intergenic regions and transposable elements. Specific light conditions affected the number of modified regions as well as the overall correlation strength between the presence of specific modifications and transcription. Furthermore, we observed that acetylation marks not only ELONGATED HYPOCOTYL5 and HY5-HOMOLOG upon deetiolation, but also their downstream targets. We found that the activation of photosynthetic genes correlates with dynamic acetylation changes in response to light, while H3K27ac and H3K27me3 potentially contribute to light regulation of the gibberellin metabolism. Thus, this work provides a dynamic portrait of the variations in histone modifications in response to the plant's changing light environment and strengthens the concept that histone modifications represent an additional layer of control for light-regulated genes involved in photomorphogenesis.
INTRODUCTION
Plants display a high degree of developmental plasticity in response to the dynamics of changing environmental conditions. This plasticity enables plant cells to integrate intrinsic and extrinsic signals to optimize their developmental patterns in a way that maximizes the chances of survival and reproduction for the organism as a whole (Kendrick and Kronenberg, 1994). Of all the environmental signals to which plants have to respond, light is probably the single most important cue. To integrate light signals, higher plants have evolved a sophisticated photosensory system that detects the quality, quantity, direction, and duration of light (Jiao et al., 2007). This photosensory system in turn triggers morphological and developmental changes. For example, dark-grown (skotomorphogenic) seedlings are characterized by elongated hypocotyls, closed cotyledons on an apical hook, and nonphotosynthetic etioplasts. By contrast, light-grown (photomorphogenic) seedlings have short hypocotyls, expanded cotyledons, and photosynthetically active chloroplasts (von Arnim and Deng, 1994).
To initiate proper morphological and developmental changes in response to ambient light, higher plants rely heavily on light-responsive nuclear genes, which direct appropriate growth and developmental responses. Therefore, developmental patterns are mediated primarily by changes in light-regulated gene expression (Terzaghi and Cashmore, 1995; Puente et al., 1996). The majority of light-regulated genome expression is attributable to CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) activity (Ma et al., 2002). COP1, a central switch in light signal transduction, acts as an E3 ubiquitin ligase to repress light signaling by targeting photoreceptors and downstream transcription factors such as LONG HYPOCOTYL5 (HY5) and HY5-HOMOLOG (HYH) for ubiquitylation and degradation in darkness (Osterlund et al., 2000; Holm et al., 2002). On the other hand, HY5 and HYH are positive regulators of photomorphogenesis that bind to specific motifs in light-inducible promoters (Holm et al., 2002). The full range of HY5 target genes was recently unveiled using a specific antibody and DNA chip hybridization with the chromatin DNA bound as a probe (ChIP-chip; Lee et al., 2007). This study alone identified ∼3800 binding targets of HY5 that include many transcription factors. Thus, it is not surprising that the expression of several thousand genes is influenced by light (Ma et al., 2001; Jiao et al., 2005). This massive reprogramming of genome activity during plant photomorphogenesis is likely to involve chromatin-level regulation.
Extensive high-resolution studies have established that chromatin remodeling plays an important role in regulating chromatin states that affect transcription (Zhang et al., 2006, 2007; Zilberman et al., 2007; Li et al., 2008; Wang et al., 2008, 2009; Schones et al., 2008). These genome-wide studies provided a glimpse of the overall eukaryotic chromatin architecture. The knowledge accumulated so far points to functional chromatin domains differentiated by posttranslational histone modifications, histone variants, and DNA methylation that define levels of chromatin organization and gene activity (Henikoff, 2008).
In a nucleosome assembly state, DNA is very compact and the histone proteins block gene expression by preventing the association of transcription factors with their binding sites and obstructing the transcription machinery from moving along the DNA strands. A diverse array of posttranslational covalent modifications (e.g., methylation, acetylation, and phosphorylation) of the histone tails can influence nucleosome compaction and access to the DNA (Rice and Allis, 2001). Specifically, active chromatin is typically enriched in acetylated lysines on histones H3 and H4. Lys has a positively charged amino group in its side chain. This amino group can be acetylated, which neutralizes its charge and therefore reduces its potential for electrostatic interactions with negatively charged DNA and negatively charged regions of the histone complex. Thus, acetylation of histone tails fosters nucleosome unwrapping and mobility such that transcription complexes can bind (Rice and Allis, 2001). On the other hand, methylation renders these specific lysines immune to acetylation and can signal either activation or repression. Thus, different modifications form a complex regulatory network fundamental to normal development (Strahl and Allis, 2000; Rice and Allis, 2001; Margueron et al., 2005; Berger, 2007), which, we hypothesize, would allow for the existence of a specific regulatory program for photomorphogenesis.
Analyzing the effects of light on histone modifications has only just begun. Previous studies have demonstrated that light-regulated expression of the pea (Pisum sativum) plastocyanin gene (PetE) is specifically associated with the acetylation of histones H3 and H4 (Chua et al., 2001, 2003). Furthermore, results from genetic analyses of histone acetyltransferase and histone deacetylase mutants in Arabidopsis thaliana suggest a role of histone acetylation in light-activated gene expression (Bertrand et al., 2005; Benhamed et al., 2006; 2008). Moreover, it has recently been shown that changes in acetylation of Lys-9 on histone H3 (H3K9ac) in four representative genes in plants grown under different light conditions were an important component of light-regulated gene transcription during seedling development in Arabidopsis (Guo et al., 2008).
However, a systematic genome-wide analysis of the regulation of histone modifications during photomorphogenesis is not available to date. Here, we describe high-resolution mapping of the acetylation and trimethylation patterns of lysines 9 and 27 on histone H3 (H3K9ac, H3K9me3, H3K27ac, and H3K27me3) in response to changing light conditions in Arabidopsis. We compare global histone modifications patterns of dark-grown seedlings with dark-grown seedlings moved to light. This in-depth, genome-scale analysis provides new and comprehensive insights into the dynamics of histone modification in response to the plant's changing light environment.
RESULTS
Light Regulates Genes in a Function-Dependent Manner
A recent study presented evidence for a relationship between mRNA steady state levels and H3K9ac levels at four representative light-regulated loci in Arabidopsis (Guo et al., 2008). This relationship was particularly pronounced when dark-grown seedlings were moved to white light for 6 h. To extend these pioneering observations to a dynamic picture at the whole-genome level, we used dark-grown wild-type Arabidopsis seedlings that were exposed to either 0 or 6 h of white light (Figure 1A). These seedlings will be called D and D to L, respectively, hereafter. Etiolated plants presented the typical phenotypic traits associated with skotomorphogenic development (i.e., long hypocotyls, closed cotyledons, and inhibition of chlorophyll biosynthesis). Upon exposure to light, signs of photomorphogenic development were clearly visible as green cotyledons started to unfold. Using Affymetrix ATH1 arrays, which include 24,000 Arabidopsis genes, we performed a genome wide transcriptome analysis to verify the extent to which changes in gene expression are associated with these obvious phenotypic differences.
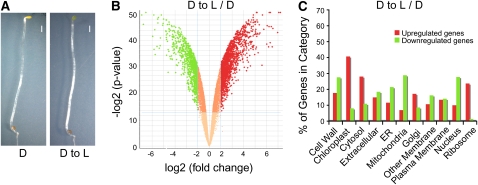
Figure 1.
Light-Regulated Morphological and Genome Expression Changes in Arabidopsis.
(A) Phenotypes of wild-type seedlings under dark and light conditions (D, wild-type plants dark grown for 5 d; D to L, wild-type plants dark grown for 4.75 d and transferred to white light for 6 h). Photographs were taken at the same magnification. All seedlings were harvested at the same time of day for analysis. Bars = 1 mm.
(B) Volcano plots illustrating the log2 of the fold changes (i.e., the ratio of means for each gene) and inverse significance (i.e., log2-transformed P value) in gene expression differences between dark-grown wild-type seedlings transferred to white light (D to L) and dark-grown wild-type seedlings (D). Genes with statistically different expression (P value < 0.0001) and fold changes above 2 were considered to be induced genes and are shown in red. Genes with statistically different expression (P value < 0.0001) and fold changes below −2 were considered to be repressed genes and are shown in green.
(C) Gene Ontology annotation of differentially expressed genes. Upregulated (red bars) and downregulated (green bars) genes were analyzed using WEGO software (Ye et al., 2006). Genes were classified on the basis of cellular components.
Previous studies estimated that light affects the expression of 19 to 32% of all detectable genes in Arabidopsis (Ma et al., 2001; Jiao et al., 2005). Examination of the ratio of gene expression in D to L seedlings versus that in D seedlings indicated that 15% (2421 out of 16,344) of all genes whose expression could be detected were regulated by light at this developmental stage (see Supplemental Data Set 1 online). Specifically, we found that 1468 genes were induced and 953 genes were repressed by at least twofold, with a P value below 0.0001 (Figure 1B; see Supplemental Data Set 1 online).
To determine the nature of these light-regulated genes, Gene Ontology (Ashburner et al., 2000) terms were assigned and comparisons for both downregulated and upregulated genes were performed. The annotations were then grouped according to ontologies reflecting cellular components (Figure 1C). When wild-type D to L seedlings were compared with D seedlings, upregulated genes were enriched in chloroplast, cytosol, and ribosome categories, while genes downregulated by light were mostly enriched in cell wall, endoplasmic reticulum, mitochondria, and nucleus categories (Figure 1C). This means that light affects gene expression not only genome wide, but also in a function-dependent manner.
Light-Regulated Histone Modification Patterns Correlate with Gene Expression in Euchromatic Regions of Arabidopsis Chromosomes
After establishing that the phenotypic variations observed after the light treatment were associated with changes in gene expression, we sought to determine if the global patterns of four histone modifications were also altered in response to light. To accurately map histone modifications on Arabidopsis chromosomes, we used Affymetrix Arabidopsis tiling array 1.0R. This array contains >3.2 million of 25-mer probe pairs tiled over the complete nonrepetitive Arabidopsis genome at a median resolution of 35 bp.
Genomic regions associated with modified histones were recovered by chromatin immunoprecipitation (ChIP) with specific antibodies that recognize H3K9ac, H3K9me3, H3K27ac, and H3K27me3, respectively (see Supplemental Figure 1 online). The recovered DNA fragments were labeled and hybridized to tiling arrays using nucleosomal DNA as a control, which allowed specific histone modifications to be quantified for each probe. To identify genomic regions significantly enriched in H3K9ac, H3K9me3, H3K27ac, and H3K27me3, we used a two-state hidden Markov model (TileMap) based on probe-level t statistic included in the CisGenome package (Ji et al., 2008). Although TileMap was originally developed for transcription factor binding site analysis, this model has proven to be effective and reliable to map DNA methylation and H3K27me3 regions in Arabidopsis (Zhang et al., 2006, 2007).
The overall distribution patterns of H3K9ac, H3K9me3, H3K27ac, and H3K27me3 modifications, gene expression, and gene distribution over the five Arabidopsis chromosomes are shown in Figure 2. The four histone modifications were highly enriched in the euchromatic arms pointing away from pericentromeric regions. This pattern resembled the distribution of genes and is consistent with results from transcription (this study; Yamada et al., 2003), mapping (J. Zhou, X. Wang, X. He, J.-B.F. Charron, A.A. Elling, and X.W. Deng, unpublished data; Zhang et al., 2007), and immunofluorescence (Naumann et al., 2005) studies.
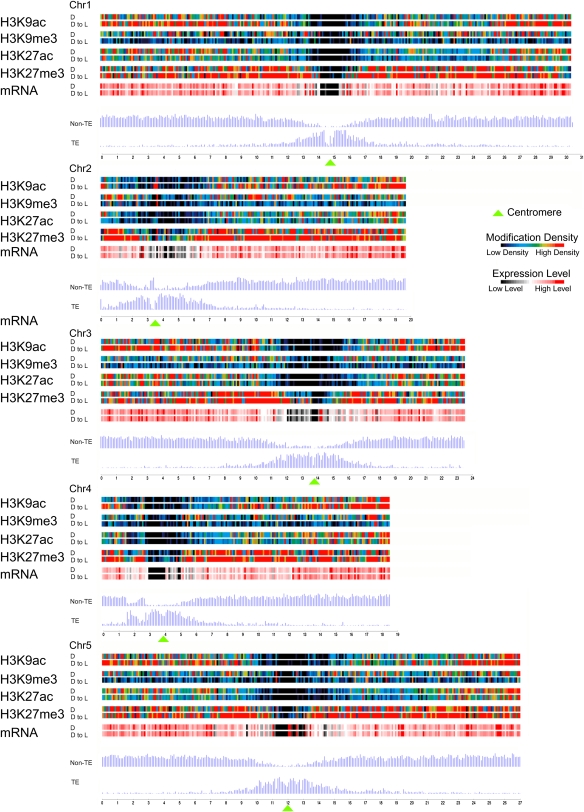
Figure 2.
Genome-Wide Identification of H3K9ac, H3K9me3, H3K27ac, and H3K27me3 Landscapes.
Map showing the distribution of histone modifications, expressed genes, and gene (TE and non-TE) density along chromosomes of wild-type Arabidopsis grown in darkness (D) and after transfer to light conditions (D to L). Color-coded bars represent the percentage of probes within each 100-kb bin that were detected for the indicated modification. Average expression level (mRNA) within each bin represents the percentage of signal probes detected in the genome-wide transcription analysis presented in Figure 1. Green triangles mark the position of centromeres within each chromosome.
Interestingly, we found differences between D and D to L seedlings in the densities of modified regions observed. In general, more H3K9ac and H3K27me3 regions were detected in D to L seedlings, while more H3K9me3 and H3K27ac regions were detected in D seedlings. The number of H3K9ac-modified regions considerably increased after deetiolation (4551 versus 7039; Table 1). Conversely, H3K9me3 regions were reduced by half in D to L compared with D seedlings. The number of H3K27ac regions was comparable between D and D to L seedlings, while 2000 more H3K27me3 regions were detected in D to L seedlings than in D seedlings (Table 1). Taken together, the variation in the number of modified regions detected in D and D to L seedlings (Table 1) implies an adjustment of histone modification patterns in response to the light signal.
Table 1.
Genome-Wide Identification of Modified Regions
| Histone Modification | Growth Condition | Number of Regionsa | Average Length (bp)b | Genome Coverage (Mb)c | Genome Coverage (%)d |
|---|---|---|---|---|---|
| H3K9ac | D | 4551 | 788 | 3.6 | 3.0 |
| D to L | 7039 | 793 | 5.6 | 4.6 | |
| H3K9me3 | D | 3892 | 935 | 3.6 | 3.0 |
| D to L | 2046 | 730 | 1.5 | 1.2 | |
| H3K27ac | D | 7669 | 657 | 5.0 | 4.1 |
| D to L | 6296 | 621 | 3.9 | 3.2 | |
| H3K27me3e | D | 6238 | 1106 | 6.9 | 5.7 |
| D to L | 8395 | 1169 | 9.8 | 8.1 |
Number of detected regions with the indicated modification.
Average length of detected regions with the indicated modification.
Portion of the genome covered by the indicated modification in megabases.
Proportion of the genome covered by the indicated modification.
Modification previously mapped by Zhang et al. (2007) using 10- to 14-d-old plants grown under long-day conditions with the following results: 8979 regions, 770 bp, 6.9 Mb, and 5.7%.
Light Modulates the Correlation Strength between Specific Histone Modifications and Gene Expression
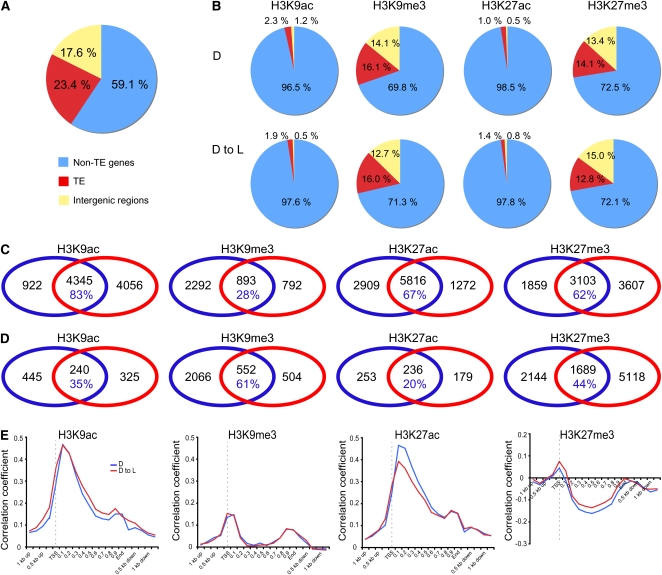
To characterize the histone modifications of interest at the gene level, we defined the territory of a gene as its body (annotated transcribed region) plus its putative promoter region reaching 1 kb upstream of the annotated transcription start site (TSS), while DNA between gene territories was designated as intergenic regions. Based on our definition of a gene territory and according to The Arabidopsis Information Resource (TAIR) Arabidopsis genome annotation 8.0 release, the Arabidopsis genome is composed of 59.1% nontransposable element (TE) protein-coding genes, 23.4% TEs, and 17.6% intergenic regions (Figure 3A). As shown in Figure 3B, acetylated regions were found almost exclusively in non-TE gene territories, while H3K9me3 and H3K27me3 regions were found in non-TE genes but also over intergenic regions and TE gene territories in D seedlings. No sizeable differences in frequency were noticed in D to L seedlings.
Figure 3.
Distribution of Four Histone Modifications in Arabidopsis Genome and Their Relationships with Gene Expression.
(A) Frequencies of non-TE genes, TE-related genes, and intergenic regions in Arabidopsis genome based on the TAIR 8.0 release.
(B) Frequencies of non-TE genes, TE-related genes, and intergenic regions overlapping with H3K9ac, H3K9me9, H3K27ac, and H3K27me3 regions.
(C) Venn diagrams showing the overlap between growth conditions of non-TE genes targeted by H3K9ac, H3K9me9, H3K27ac, and H3K27me3. The percentage values in blue indicate the portion of modified non-TE genes in D seedlings also modified in D to L seedlings.
(D) Venn diagrams showing the overlap between growth conditions of TE genes targeted by H3K9ac, H3K9me9, H3K27ac, and H3K27me3. The percentage values in blue indicate the portion of modified TE genes in D seedlings also modified in D to L seedlings.
(E) Distribution along genes aligned at their TSSs of Spearman coefficients for correlation of gene expression data and the presence of a modification. The correlation coefficients were calculated in a 200-bp window sliding along the genome with a statistical significance of P < 0.0001.
Our ChIP-chip analysis revealed a total of 9325, 3977, 9997, and 8569 nonoverlapping non-TE genes targeted by H3K9ac, H3K9me3, H3K27ac, and H3K27me3, respectively, in the Arabidopsis genome across the two growth conditions (see Supplemental Data Sets 2 to 5 online). Comparisons of non-TE genes targeted by each modification in D and in D to L seedlings revealed a considerable degree of overlap (except H3K9me3; Figure 3C). Specifically, 83, 28, 67, and 62% of non-TE genes targeted by H3K9ac, H3K9me3, H3K27ac, and H3K27me3, respectively, in D seedlings were also targeted in D to L seedlings. Comparison of TE genes targeted by each modification in D and in D to L seedlings revealed little overlap (Figure 3D).
To correlate the density of H3K9ac, H3K9me3, H3K27ac, and H3K27me3 coverage with gene expression levels, we performed a Spearman correlation analysis (Spearman, 1904). We divided each gene into 20 intervals (5% each interval), and the 1-kb regions upstream and downstream of each gene were divided into 50-bp intervals. The correlation coefficients for modification and gene expression levels were plotted for each bin for each growth condition (Figure 3E). This analysis revealed a positive correlation between H3K9ac and H3K27ac levels and gene expression levels, indicating that actively transcribed genes are likely to be acetylated. The highest correlation coefficients were observed ∼100 bp downstream of the TSS. By contrast, we observed a weak positive correlation for H3K9me3 and a negative correlation between the levels of H3K27me3 and gene expression (Figure 3E). Moreover, non-TE genes showing low expression levels (0 to 20%) had the lowest levels of acetylation but the highest levels of H3K27me3 (see Supplemental Figure 2 online). On the other hand, highly expressed non-TE genes had the highest levels of acetylation but were virtually devoid of H3K27me3 (see Supplemental Figure 2 online). Interestingly, H3K27ac and H3K27me3 exhibited variations in correlation strength depending on the growth condition. H3K27ac displayed high correlation coefficients in D seedlings, while lower correlation coefficients were found in D to L seedlings (Figure 3E). This can be explained by the observation that highly expressed genes in D to L seedlings exhibited considerably lower H3K27ac levels compared with higher levels in D seedlings (see Supplemental Figure 2 online).
Taken together, these results indicate that the selected histone modifications preferentially target a large number of non-TE genes and that even though a substantial overlap is found between the different growth conditions, a large number of non-TE genes are specific to each condition, suggesting that light modulates chromatin changes. Moreover, these results strongly suggest that light affects the overall correlation strength between the presence of specific histone modifications like H3K27ac and transcription.
H3K9ac and H3K27ac Co-Occur and Are Mutually Exclusive with H3K27me3, Which Targets Genes Expressed in a Highly Tissue-Specific Manner
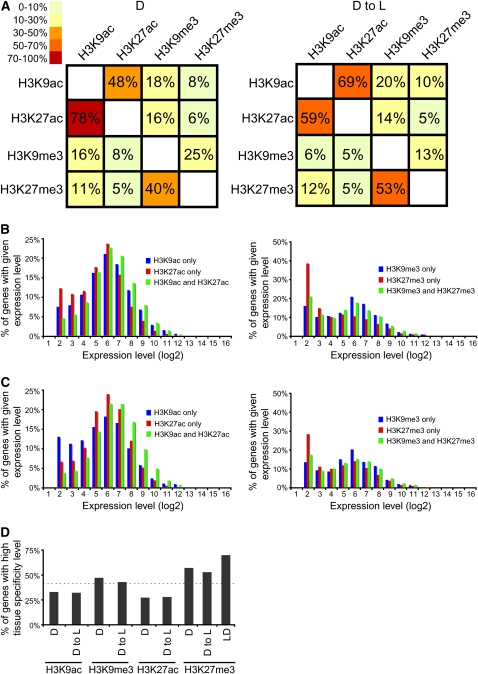
To gain an understanding of the combinatorial co-occurrence of histone modifications in response to light, we analyzed how often genomic locations were enriched for modification pairs. For example, H3K9ac and H3K27ac tended to colocalize under all growth conditions (Figure 4A). On the other hand, H3K9me3 occurred mainly in conjunction with H3K27me3 in wild-type seedlings (D and D to L).
Figure 4.
Co-Occurrence and Tissue Specificity Analyses of Histone Modifications.
(A) Contingency tables of the occurrence of histone modifications for D seedlings (left panel) and D to L seedlings (right panel).
(B) and (C) Association between gene expression levels and the presence of a specified combination of modifications in D (B) or D to L (C) seedlings. The x axes show the expression level (log2) calculated from the analysis presented in Figure 1. The y axes show the percentage of genes in the indicated category containing a specified combination of modifications.
(D) Tissue specificity of modified target genes. Tissue specificity measured by Shannon entropy values of genes modified by H3K9ac, H3K9me3, H3K27ac, and H3K27me3 in different growth conditions. Entropy values were calculated on a scale from 0 to 12 (low entropy values = high tissue specificity). Values from 0 to 6 were grouped as genes with high tissue specificity. The dotted line represents a high tissue specificity distribution of all Arabidopsis genes. LD, H3K27me3 target genes previously identified in wild-type plants grown under long-day conditions by Zhang et al. (2007). y axis: percentage of genes with high tissue specificity.
To analyze further the combinatorial occurrence of modifications, we calculated the average expression levels of non-TE genes targeted by different combinations of histone modifications (Figures 4B and 4C). In non-TE genes simultaneously targeted by both H3K9ac and H3K27ac, we observed a higher level of gene expression compared with genes targeted only by H3K9ac or H3K27ac. However, this increase in transcription was light independent since it was detected in all growth conditions. In wild-type seedlings (D and D to L), where H3K9me3 occurred mainly in conjunction with H3K27me3, genes that were modified only by H3K9me3 were expressed at higher levels than those that contained H3K27me3.
Recently, genes targeted by H3K27me3 were reported to be expressed in a tissue-specific manner in Arabidopsis (Zhang et al., 2007). To determine if light affects the tissue specificity of the genes targeted by H3K9ac, H3K9me3, H3K27ac, and H3K27me3, we introduced a definition of tissue specificity based on Shannon entropy (Schug et al., 2005) to rank modified genes according to their overall tissue specificity (Figure 4D). As expected, a significant portion of H3K27me3 targets were expressed in a highly tissue-specific manner. However, genes targeted by H3K9ac, H3K9me3, and H3K27ac were preferentially expressed in a non-tissue-specific manner, suggesting that these modifications target genes expressed in a multitude of tissues and organs and that they are likely to co-occur.
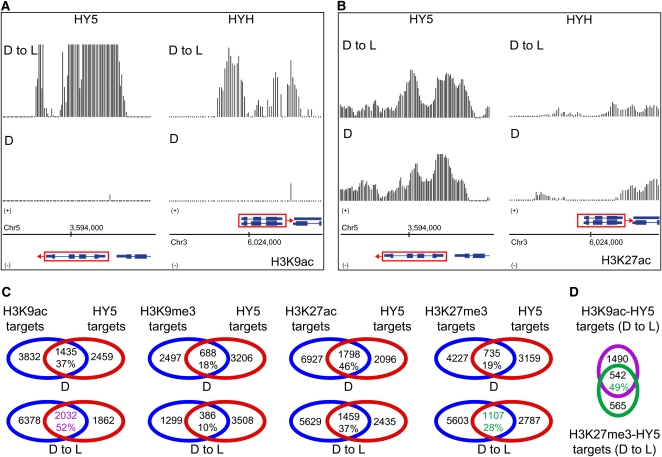
H3K9ac Marks HY5 and HYH in a Light-Dependent Manner
To explore the relationships between photomorphogenesis and activating or silencing histone marks, we analyzed the differences in chromatin profiles over higher-level hierarchical regulators (HY5 and HYH) of the transcriptional cascades for photomorphogenesis. The transcription of the HY5 and HYH genes is known to be dramatically higher in light-grown seedlings compared with seedlings grown in darkness (Holm et al., 2002). Inspection of the HY5 and HYH loci revealed considerable differences in modification patterns between D and D to L seedlings. In D to L seedlings, H3K9ac showed a massive peak through the transcribed region of HY5 (Figure 5A), while no peak was detected in D seedlings. A similar situation could be observed for HYH, in which substantial H3K9ac was found at the promoter region with lower, but significant, levels throughout the coding sequence in the D to L condition. No signal differences or significant signals were found for H3K27ac (Figure 5B), H3K9me3, and H3K27me3 (data not shown) over the HY5 and HYH loci. The differential accumulation of H3K9ac at the HY5 locus (as well as at additional loci) in response to light was validated by ChIP-PCR using independently prepared ChIP samples (see Supplemental Figures 3 to 6 online).
Figure 5.
Distribution of Histone Modifications within Transcription Factors Involved in Photomorphogenesis.
(A) and (B) Signals for H3K9ac (A) and H3K27ac (B) are shown within regions encompassing HY5 and HYH in D seedlings (bottom panel) and D to L seedlings (top panel). A schematic representation of each gene (rectangles = exons and lines = introns) within these regions is shown at the bottom. Red boxes with arrows indicate direction of transcription. Data shown in this figure were corrected for total H3 (nucleosomal DNA). Chromosome coordinates are indicated in base pairs at the bottom.
(C) Venn diagrams showing the overlap of non-TE genes targeted by H3K9ac, H3K9me9, H3K27ac, and H3K27me3 with putative HY5 binding sites (Lee et al., 2007) in D and D to L seedlings. The percentage values indicate the portion of modified target genes also modified by HY5 in D and D to L seedlings. The target groups in green and purple are compared in (D).
(D) Venn diagrams showing the overlap of H3K9ac-HY5 and H3K27me3-HY5 targets in D to L seedlings. The percentage value indicates the portion of H3K27me3-HY5 target genes also targeted by H3K9ac in D to L seedlings.
To investigate further the impact of histone modifications on photomorphogenic development, we analyzed the overlap between modified genes and putative HY5 binding sites identified by Lee et al. (2007). Thirty-seven percent of the putative HY5 target genes were also targeted by H3K9ac in D seedlings, while the overlap increased to 52% in D to L seedlings (Figure 5C). A similar situation was noticed for H3K27me3. Interestingly, 49% of genes targeted by both H3K27me3 and HY5 were also modified by H3K9ac in D to L seedlings (Figure 5D). This is considerably higher than the 10% co-occurrence frequency previously observed between these two modifications (Figure 4A). Taken together, these data suggest that acetylation, in particular H3K9ac, is an important contributor to light-regulated genome expression not only through its activating action on HY5 and HYH but also on their downstream targets.
Acetylation Positively Correlates with Activation of Photosynthetic Genes, While H3K27me3 Marks Gibberellin Biosynthesis and Inactivation
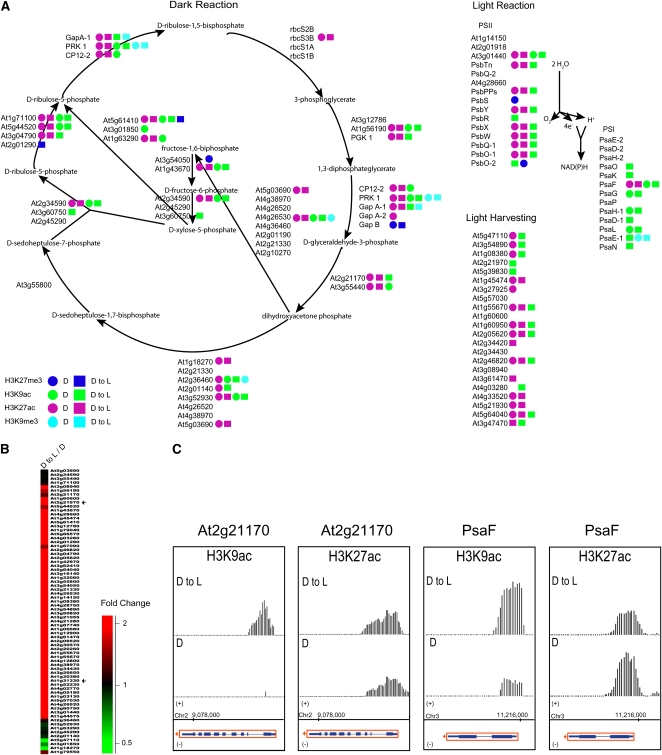
It has been previously shown that light induces expression of genes in most major metabolic pathways in Arabidopsis (Jiao et al., 2005). Therefore, to verify if histone modifications target specific metabolic pathways, we employed the AraCyc tool to visualize biochemical pathways of Arabidopsis (Mueller et al., 2003). AraCyc currently includes 288 pathways and 5847 Arabidopsis enzyme-coding genes. As query, we used non-TE genes targeted by each modification in different growth conditions. Interestingly, we found metabolic pathways that were mostly modified by activating modifications (H3K9ac and H3K27ac; i.e., photosynthesis), while other pathways were almost exclusively modified by repressive modifications (H3K27me3; i.e., gibberellin metabolism). To examine the influence of light on the extent of chromatin-based regulation of these metabolic pathways, we determined the presence or absence of a specific modification in these pathways in D, and D to L seedlings (Figures 6A and 7A).
Figure 6.
Photosynthetic Genes Are Acetylated.
(A) Diagram of the photosynthesis pathway. Arrows represent enzymatic reactions. Colored circles mark genes targeted by a specific modification in D seedlings. Colored squares mark genes targeted by a specific modification in D to L seedlings. Green = H3K9ac; turquoise = H3K9me3; purple = H3K27ac; blue = H3K27me3.
(B) Overview of photosynthesis gene expression by cluster display. D to L/D, dark to light versus dark. The color scale is shown at the bottom. A magnified view of the display is presented in Supplemental Figure 11 online. Arrows indicate the position of the At2g21170 and PsaF genes in the cluster display.
(C) Signals for H3K9ac and H3K27ac are shown within regions encompassing At2g21170 and PsaF in D seedlings (bottom panel) and D to L seedlings (top panel). A depiction of each gene (rectangles = exons and lines = introns) within these regions is shown at the bottom. Red boxes with arrows indicate direction of transcription. Data shown in this figure were corrected for total H3 (nucleosomal DNA). Chromosome coordinates are indicated in base pairs at the bottom.
Figure 7.
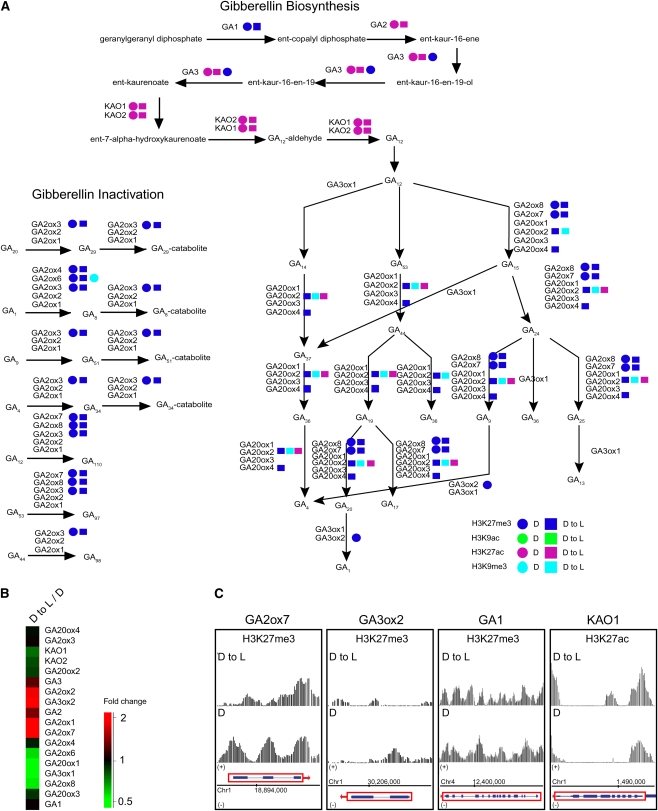
H3K27me3 Marks the Gibberellin Biosynthesis and Inactivation Pathways.
(A) Diagram of the gibberellin biosynthesis and inactivation pathways. Arrows represent enzymatic reactions. Colored circles mark genes targeted by a specific modification in D seedlings. Colored squares mark genes targeted by a specific modification in D to L seedlings. Green = H3K9ac; turquoise = H3K9me3; purple = H3K27ac; blue = H3K27me3.
(B) Cluster display of expression profiles of genes involved in gibberellin metabolism. D to L/D, dark to light versus dark. The color scale is shown at the bottom.
(C) Signals for H3K27me3 and H3K27ac are shown within regions encompassing GA2ox7, GA3ox2, and GA1, and KAO1, respectively, in D seedlings (bottom panel) and D to L seedlings (top panel). A schematic representation of each gene (rectangles = exons and lines = introns) within these regions is shown at the bottom. Red boxes with arrows indicate direction of transcription. Data shown in this figure were corrected for total H3 (nucleosomal DNA). Chromosome coordinates are indicated in base pairs at the bottom.
One of the most striking observations was that photosynthetic genes were targeted by H3K9 and H3K27 acetylation in D, and D to L seedlings, but not by H3K9me3 or H3K27me3 (Figure 6A). Our analysis of transcriptomic data using hierarchical clustering showed that the majority of photosynthetic genes were upregulated by light under our experimental conditions (Figure 6B), confirming previous observations (Eberhard et al., 2008). Thus, the presence of acetylation could be a prerequisite to the activation of photosynthetic genes at this developmental stage. In general, photosynthetic genes had stronger H3K9ac signals in D to L seedlings than other genes. On the other hand, the H3K27ac signals were either equal among all conditions or more pronounced in D seedlings. Several examples illustrating these dynamic changes in response to light are presented in Figure 6C.
At1g21170 is a gene coding for a triosephosphate isomerase involved in the conversion of d-glyceraldehyde-3-phosphate into dihydroxyacetone phosphate in the photosynthetic dark reactions or Calvin cycle. In D seedlings, this gene was unmodified by H3K9ac, while clear signal peaks were present in D to L seedlings, suggesting gene activation under these two conditions. No difference in H3K27ac levels was noticed between D and D to L seedlings for At1g21170.
The photosystem I subunit F (PsaF) is involved in the transfer of electrons from plastocyanin to ferredoxin in higher plants. The PsaF gene showed a clear increase in its H3K9ac signal at its 5′ end in wild-type seedlings upon transfer to light conditions, which suggests gene activation. However, a lower H3K27ac signal in D to L seedlings was detected compared with D seedlings. These two examples clearly demonstrate that dynamic acetylation changes in response to light have a major role in the dynamic regulation in the expression of photosynthetic genes.
A relationship between light-regulated plant development and gibberellin metabolism has long been proposed, and molecular evidence has recently been reported to support this model (Kamiya and García-Martínez, 1999; Feng et al., 2008). The superpathway of gibberellic acid (GA) biosynthesis and inactivation in higher plants is compartmentalized between plastids, the endomembrane system, and the cytoplasm (Hedden and Phillips, 2000). Our analysis demonstrates that the genes coding for enzymes acting in the plastids and the endomembrane system, such as GA REQUIRING1 (GA1), GA2, and GA3 and ENT-KAURENOIC ACID HYDROXYLASE1 (KAO1) and KAO2, were mainly modified by H3K27ac, while components of the cytoplasmic portion of the pathway were under the regulation of H3K27me3 in D and D to L, seedlings (Figure 7A). It has been reported that dioxygenase genes are major targets for light regulation of the GA metabolism (Kamiya and García-Martínez, 1999). Consistent with these findings, we observed that most genes in the GA pathway were differentially regulated in D and D to L seedlings (Figure 7B). To some degree, this regulation can be explained by the modulation of the levels of H3K27me3 at these loci. Examples are presented in Figure 7C. The two dioxygenases genes GA2ox7 and GA3ox2 were upregulated in D to L seedlings (Figure 7B). Analysis of the H3K27me3 landscape at these loci revealed a clear signal reduction over the promoter regions and first exons of dioxygenase genes in the D to L seedlings, suggesting that the repressive effect of this modification was removed.
Furthermore, we observed that genes that are not affected by the GA pathway did not show any difference in H3K27me3 levels under our experimental conditions. Highly repressed genes (GA2ox6, GA20ox1, and GA3ox1) were not modified by H3K27me3, suggesting that other repressive mechanisms might also be involved. Taken together, these results suggest that changes in the levels of H3K27ac and H3K27me3 are major contributors to light regulation of the gibberellin metabolism.
DISCUSSION
In this study, we systematically investigated the dynamic landscape of four histone modifications in response to light. This exhaustive analysis revealed a combinatorial interplay between histone modifications and light-regulated gene expression and delivers new insights into the chromatin-based regulation of photomorphogenesis.
Characteristic Patterns of H3K9ac, H3K9me3, H3K27ac, and H3K27me3 in Arabidopsis Genes
The coverage provided by our analysis in a specific growth condition is consistent with results from other studies using similar tiling array platforms (Zhang et al., 2006, 2007). By studying the effect of the light signal on wild-type seedlings grown under different light conditions, we were able to uncover a significant number of genes targeted by selected histone modifications (9325 for H3K9ac, 3977 for H3K9me3, 9997 for H3K27ac, and 8569 for H3K27me3; Figure 3). However, these numbers are in all likelihood conservative estimates of all possible targets due to the limited number of developmental stages studied, as well as due to technical and computational limitations.
All four histone modifications examined in this study (H3K9ac, H3K9me3, H3K27ac, and H3K27me3) preferentially targeted genes (Figure 3C). This was an expected result because cytological observations established that these modifications (except H3K27ac) target euchromatin in Arabidopsis (Naumann et al., 2005). Enrichment of histone acetylation is regarded as a positive marker of histone modification associated with gene activation in yeast, mammals, and plants (Bernstein et al., 2002, 2005; Benhamed et al., 2006; Millar and Grunstein, 2006). Several studies in human and yeast demonstrated that histone acetylation is enriched in the promoter region of activated genes and that the level of acetylation in the promoter strongly correlates with gene transcription (Bernstein et al., 2002, 2005; Roh et al., 2005; Nishida et al., 2006; Heintzman et al., 2007). Even though this phenomenon has been observed in plants in the past, for example, the light induction of H3K9ac at the distal promoter of the maize (Zea mays) Pepc gene (Offermann et al., 2008), a different image is starting to emerge. Global mapping of H3K9ac in maize shoots and roots found this mark to be mostly enriched at the translation start site of non-TE genes (Wang et al., 2009). Accordingly, we observed that in Arabidopsis the presence of H3K9ac and H3K27ac correlates better with transcription when located at the beginning of the gene body region just downstream of the TSS (Figure 3E; see Supplemental Figures 7 to 9 online). Similar enrichment profiles for H3K9ac, H3K27ac, and H3K14ac at several drought-responsive loci have been observed in both stressed and unstressed Arabidopsis plants (Kim et al., 2008). Moreover, this pattern has also been reported in response to light for the LHB1B2 gene in Arabidopsis (Guo et al., 2008). These observations clearly suggest a transcriptional regulatory mechanism that might be specific to plant genes.
Our results identified H3K9me3 as an activating mark that precisely targets TSSs in Arabidopsis (Figure 3E; see Supplemental Figure 7 online). This finding agrees well with the observed distribution of H3K9me3 over the region located just upstream of the open reading frames of GLABRA2 and CAPRICE in Arabidopsis (Caro et al., 2007). However, when compared with acetylation, this modification was found to be a weak activator (Figure 3E). In the mammalian system, H3K9me3 has been associated with both constitutive and facultative heterochromatin and with both transcriptional repression and activation (Peters et al., 2003; Vakoc et al., 2005). In Arabidopsis, cytological observations presented clear evidence that H3K9me3 preferentially localizes within euchromatin (Naumann et al., 2005). Recently, ChIP-chip analysis of H3K9me3 demonstrated that this mark is detected mostly within the euchromatic parts of chromosome 4 in Arabidopsis (Turck et al., 2007). However, these studies did not present evidence regarding whether this modification is a repressive or an activating mark. Our results clearly point toward a mild activating action of this mark on gene transcription. Interestingly, almost half of the non-TE genes targeted by H3K9me3 in this study were also H3K27me3 targets (Figure 4A). This co-occurrence was unexpected since previous work demonstrated that only 6% of H3K9me3 target genes were marked by H3K27me3 (Turck et al., 2007). The reasons behind this disparity are unknown and do not solely involve gene set discrepancies since 40% of the H3K9me3 target genes are common between the Turck data set and the data generated here (see Supplemental Figure 10 online).
Recent genome-wide mapping in Arabidopsis revealed that H3K27me3 is a major silencing mechanism in plants that targets a large number of genes (Turck et al., 2007; Zhang et al., 2007; Oh et al., 2008). Our H3K27me3 data agree well with the analyses of Zhang et al. (2007) and Turck et al. (2007). We found that ∼40% of the target genes identified under different light conditions overlapped with the Zhang and Turck data sets, respectively (see Supplemental Figure 10 online). These numbers are impressive considering the obvious differences in tissue composition of the plant material analyzed. While both the Zhang and Turck data sets, which overlap by 65%, were generated from whole seedlings presenting photomorphogenic phenotypes (light-grown for 10 d), our data set was generated from seedlings presenting mostly skotomorphogenic phenotypes (Figure 1A). Even if our data set is to some extent different, essential characteristics of H3K27me3 targets are retained. We observed that H3K27me3 was correlated with transcriptional repression and that genes targeted by this modification were expressed in a tissue-specific manner (Figures 3E and 4D). However, the H3K27me3 regions detected by our analysis were 43% longer (Table 1). Using the same analysis method, Zhang et al. (2007) found few regions longer than 1 kb (∼700 bp on average). Our analysis indicates that enriched regions in the different conditions have a length of at least 1100 bp. However, this average length is still considerably below the size of H3K27me3 regions observed in animals, which can span up to hundreds of kilobases and cover multiple genes, maintaining them in a transcriptionally repressed state at appropriate developmental stages (Bernstein et al., 2005; Nègre et al., 2006; Tolhuis et al., 2006). Nonetheless, this discrepancy in the length of H3K27me3-covered regions observed between these two Arabidopsis data sets might be the consequence of different developmental stages and growth conditions studied and most likely points toward dynamic changes in the deposition pattern of this modification during morphogenesis.
Differences in Histone Modifications and Transcription in Response to Light
One of the goals of this study was to analyze specifically the possibility of histone modifications being transcription regulators during light-regulated plant development. Recent studies in Arabidopsis showed that factors involved in the regulation of histone modifications, such as histone acetyltransferases and histone deacetylases, are light responsive and that acetylated histones are required for light-regulated gene expression (Chua et al., 2001; Bertrand et al., 2005; Benhamed et al., 2006). However, these studies only offer a glimpse of the global epigenetic landscape due to the limited number of genes studied. Our genome-wide approach offers the most comprehensive overview of histone modification changes in response to light to date. Furthermore, the results of this study also suggest the existence of a chromatin-based program regulating photomorphogenesis. We observed that more protein-coding genes contained H3K9ac and H3K27me3 in D to L seedlings than in D seedlings (Figure 3C). These results illustrate that each growth condition has its own epigenome, corroborating the hypothesis that dynamic changes occur in response to environmental stimuli. We demonstrated that differences in histone modifications are correlated with differences in transcript abundance (Figure 3E). Furthermore, the differences in correlation strength between the modification signal and transcription exhibited by specific histone marks in response to light suggest that specific histone modifications might have dominant roles in photomorphogenesis. Taken together, our observations support the general concept that differential modification of the epigenome is an important component of developmental regulation of gene transcription.
Light-Regulated H3K9ac Targets Photomorphogenesis-Promoting Components, Such as HY5 and HYH
Through an analysis of det1, cop1, and hy5 mutants, direct evidence has been provided that some key factors of the photomorphogenesis pathway also participate in the regulation of histone modifications by influencing the relative enrichment of H3K9ac and Pol II during gene transcription (Guo et al., 2008). Moreover, it has been shown that a significant proportion of light-regulated genes depend on both GCN5 histone acetyltransferase and HY5 for expression (Benhamed et al., 2008). Our findings that H3K9ac specifically targets HY5 and HYH in a light-dependent manner (Figure 5) further reinforce the concept that histone marks, such as acetylation, might be key players or switches to relay environmental stimuli like light to gene transcription in Arabidopsis. Moreover, the observation that a large number of HY5 targets are also targeted by acetylation provides evidence that histone marks might form a second layer of regulation for this subset of genes. Consequently, acetylation might act synergistically with HY5 and HYH to properly regulate the transcription of downstream effectors.
The presence of a number of bivalent genes, which simultaneously have activating and repressive histone modifications, is also an indication that this subset is composed of developmentally important genes. Bivalent genes were observed in embryonic stem cells where the co-occurrence of H3K27me3 and H3K4me3 was described for a group of key developmental regulators (Bernstein et al., 2006). Bivalent domains were proposed to silence developmental genes while keeping them poised for activation. Since we observed that a significant number of HY5 targets are bivalent genes, it is possible that a similar situation occurs during light-regulated gene transcription, in which the expression of key effectors is tightly regulated by the simultaneous presence of activating and repressing marks. However, we cannot rule out allelic differences in chromatin modification and the complexity of the tissues used as starting material as alternative explanations for the apparent coexistence of both activating and repressive histone marks.
Photosynthesis-Related Genes Are Marked by Acetylation
Photosynthesis-related genes were the most highly enriched functional group among HY5 targets (Lee et al., 2007). This observation supports our findings that two acetylation marks studied here specifically targeted photosynthetic genes (Figure 6). Furthermore, several examples of photosynthetic genes modified by acetylation in response to light conditions have been published. It has been demonstrated that light-regulated expression of the pea plastocyanin gene (PetE) was specifically associated with the acetylation of histones H3 and H4 (Chua et al., 2001, 2003). Offermann et al. (2006) previously reported that the chromatin of the phosphoenolpyruvate carboxylase (Pepc) gene in maize is acetylated in response to light. Furthermore, it has been shown that light selectively induced the acetylation of H4K5 and H3K9 in both the promoter and the transcribed region of the maize Pepc gene and that the induction was fully reversible in the dark (Offermann et al., 2008). Our results extend the observations provided by these reports by showing that the vast majority of photosynthetic genes are targeted by acetylation in response to light. Furthermore, this modulation of the acetylation signal is consistent with the activation level of these genes during the dark-to-light transition (Figure 6). These findings support our conclusion that acetylation directly regulates the transcription of photosynthesis-related genes during seedling photomorphogenesis.
GA-Related Genes Are Marked by H3K27me3
The manifold roles of GAs in plant development suggest a complex regulation of GA-related genes. Recently, GA and light signaling pathways were shown to interact to regulate the deetiolation process by modulating the activity of the HY5 and PHYTOCHROME INTERACTING FACTOR light signaling elements, which represent integration nodes for both pathways (Alabadi et al., 2008; de Lucas et al., 2008; Feng et al., 2008). Moreover, several genes involved in GA metabolism were found among the HY5 binding targets (Lee et al., 2006) and many dioxygenase genes are light regulated (Hedden and Phillips, 2000). Our analysis clearly demonstrated that dioxygenase genes were heavily targeted by H3K27me3 and that the level of this modification was correlated with expression changes, suggesting that H3K27me3 is a major player in the regulation of this pathway (Figure 7). Previous reports have linked the repressive histone mark H3K27me3 to GA through the regulator PICKLE (PKL; Zhang et al., 2008). pkl mutants exhibit the phenotypic hallmarks of a plant that is defective in the ability to respond to GA, including reduced responsiveness to GA and elevated levels of bioactive GAs (Ogas et al., 1997). PKL codes for an ATP-dependent chromatin remodeling factor in the CHD3 family (Ogas et al., 1999). In animal systems, CHD3 proteins are a component of the Mi-2/NuRD complex that contributes to transcriptional repression (Denslow and Wade, 2007). In plants, it has recently been suggested that PKL plays a role in deposition and/or maintenance of H3K27me3 since levels of this mark were reduced at several loci in pkl (Zhang et al., 2008). Furthermore, a substantial overlap between PKL-dependent genes and genes that are enriched for trimethylation of histone H3K27me3 was found (Zhang et al., 2007, 2008). Interestingly, PKL and GA were found to act together or synergistically during specific developmental windows during which PKL and GA act to repress expression of seed-associated developmental programs (Rider et al., 2003; Zhang et al., 2008). Therefore, it is possible that the modulation of H3K27me3 levels in dioxygenase genes in response to light might represent a yet undiscovered integration node between the GA and light pathways.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was the genetic background used for this study. Surface-sterilized seeds (250 mg) were sown on sterile half-strength Murashige and Skoog medium (Sigma-Aldrich) containing 1% sucrose. Seeds were stratified for 2 d at 4°C and then germinated and grown for 5 d at 22°C in continuous darkness (D). Six hours before harvest, a number of plates were transferred to continuous white light (D to L). Seedlings from all conditions (D and D to L) were harvested at the same time of day. A total of eight independent biological replicates for each condition were collected for further analysis (four for array hybridization and four for validation).
ChIP and Microarray Hybridization
Chromatin isolation was performed with 10 g of whole seedlings tissue according to Bowler et al. (2004). The resuspended chromatin pellet was sonicated at 4°C with a Diagenode Bioruptor set at high intensity for 10 min (30 s on, 30 s off intervals). The DNA was sheared to ∼0.3- to 0.8-kb fragments. Chromatin was immunoprecipitated, washed, reverse cross-linked, amplified, and hybridized according to the Affymetrix Chromatin Immunoprecipitation Assay Protocol Rev.3. Three microliters of the following antibodies were used: anti-H3K9ac (Upstate 06-942), anti-H3K9me3 (Upstate 07-442), anti-H3K27ac (Upstate 07-360), anti-H3K27me3 (Upstate 07-449), and anti-H3 (Upstate 07-690). An aliquot of untreated sonicated chromatin was reverse cross-linked and used as a total input DNA control for ChIP-PCR experiments. Four biological replicates for each condition were hybridized to tiling arrays.
Microarray Data Analysis
Data were analyzed with the integrated tools of CisGenome (http://www.biostat.jhsph.edu/∼hji/cisgenome/) (Ji et al., 2008). Raw microarray data were quantile normalized and modified regions were detected with TileMap using the Hidden Markov model option and the Unbalanced Mixture Subtraction method (Ji and Wong, 2005) to provide an approximate estimate of model parameters for HMM (false discovery rate). Analysis parameters were set according to published studies (Zhang et al., 2006, 2007). Briefly, neighboring probes yielding posterior probabilities of 0.5 or higher were joined into regions by requiring a minimal run of 100 bp and allowing a maximal gap of 200 bp. Default settings were used for the remaining parameters. Genomic regions associated with each modification were identified as those presenting significantly higher hybridization signals when probed with ChIP samples than with nucleosomal DNA (DNA pulled down with the anti-H3 antibody). All raw microarray data (CEL files) have been deposited in the Gene Expression Omnibus (GSE15597). Arabidopsis full-length cDNAs were mapped onto the Arabidopsis genome to identify TSSs as described by Tanaka et al. (2009). If two or more transcript variants were derived from the same locus, the most upstream TSS was selected. No clustering analysis was performed to assign TSSs. Tissue specificity analyses were performed as described (Schug et al., 2005). Shannon entropy values were calculated on a scale from 0 to 12 (low entropy values = high tissue specificity). Gene expression data used for the analysis shown in Figure 4 were from a previous publication reporting the transcriptional profiling of Arabidopsis genes across various developmental stages (Schmid et al., 2005).
Gene Expression
Four independent biological replicates were grown as detailed above and harvested for RNA isolation (RNeasy; Qiagen). RNA quality was assessed with an Agilent 2100 Bioanalyzer, and hybridization to the Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays was performed according to the manufacturer's instructions. For assessing differential expression (D to L versus D), raw microarray data were normalized using the robust multi-array average analysis (Irizarry et al., 2003) and subsequently analyzed with an Empirical Bayes model (Wright and Simon, 2003) and a false discovery rate controlling procedure (Benjamini and Hochberg, 1995) included in the Flexarray software package (http://genomequebec.mcgill.ca/FlexArray/).
Validation of ChIP-Chip Results
Modified and unmodified regions were validated using four independently prepared ChIP samples for each condition. The ChIP-DNA samples were resuspended in 30 μL water, and 1 μL was used for PCR amplification with specific primers listed in Supplemental Table 1 online. PCR conditions were as follows: 95°C for 2 min, 28 to 30 cycles at 95°C for 30 s, 60°C for 30 s, 72°C for 1 min, followed by 72°C for 10 min. Amplified DNA products were separated on a 1.5% agarose gel subsequently stained with SYBR Green I Nucleic Acid Gel Stain (Lonza). Images were digitally captured with a GelDoc 2000 system (Bio-Rad) and quantified using ImageJ version 1.34s (http://rsbweb.nih.gov/ij/). The enrichment of a modified region was determined as the fold change of modification over nucleosomal DNA and normalized by the first negative locus in each region as previously described (Zhang et al., 2007). Quantum RNA 18S internal standards (Ambion; AM1718) were used as internal controls to compare D and D to L samples.
Histone Extraction and Protein Gel Blot Analysis
Histones extraction was performed according to Tariq et al. (2003) with modifications. Four grams of seedlings were homogenized in extraction buffer 1 (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 5 mM 2-mercaptoethanol, 0.1 mM PMSF, and protease inhibitors), filtered through four layers of miracloth and centrifuged for 20 min at 2880g at 4°C. Pellets were resuspended in 1 mL of histone extraction buffer (10 mM Tris-HCl, 2 mM EDTA, 0.25 M HCl, 5 mM 2-mercaptoethanol, 0.2 mM PMSF, and protease inhibitors), and soluble proteins were isolated by centrifugation, precipitated with 25% trichloroacetic acid, and repelleted by centrifugation at 17,000g for 30 min. Pellets were washed twice with ice-cold acetone, resuspended in Laemmli buffer, separated electrophoretically on Novex 4 to 20% Tris-glycine gel (Invitrogen), and transferred to Immobilon PVDF membranes (Millipore). Histone modifications were detected with the antibodies used for ChIP experiments (see above) and visualized with film using a horseradish peroxidase–conjugated anti-rabbit IgG secondary antibody (Sigma-Aldrich) and the ECL detection system (GE healthcare).
Accession Numbers
The Arabidopsis Genome Initiative locus number for the major genes discussed in this article are At1g02400 for GA2ox6, At1g05160 for KAO1, At1g15550 for GA3ox1, At1g21170 for triosephosphate isomerase, At1g31330 for PsaF, At1g50960 for GA2ox7, At1g79460 for GA2, At1g80340 for GA3ox2, At2g25070 for PP2C, At2g32440 for KAO2, At3g15095 for hypothetical protein, At3g17609 for HYH, At3g47470 for CAB4, At3g57010 for strictosidine synthases 1, At3g57020 for strictosidine synthases 2, At3g57030 for strictosidine synthases 3, At4g02780 for GA1, At4g25420 for GA20ox1, At5g11260 for HY5, At5g25900 for GA3, At5g66400 for RAB18, and At5g67630 for putative DNA helicase. All raw microarray data (CEL files) have been deposited in the Gene Expression Omnibus (GSE15597).
Author Contributions
X.W.D. and J.-B.F.C conceived the project. J.-B.F.C. performed the experiments. H.H. and J.-B.F.C. conducted bioinformatics analyses. J.-B.F.C., A.A.E, and X.W.D. prepared the manuscript.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunodetection of Acetylated and Trimethylated Histone H3.
Supplemental Figure 2. H3K9ac, H3K9me3, H3K27ac, and H3K27me3 Mark Different Levels of Gene Expression.
Supplemental Figure 3. Validation of H3K9ac ChIP-Chip Results by ChIP-PCR from Four Independently Prepared Biological ChIP Replicates.
Supplemental Figure 4. Additional Validation of H3K9ac ChIP-Chip Results by ChIP-PCR from Four Independently Prepared Biological ChIP Replicates.
Supplemental Figure 5. Validation of H3K27ac ChIP-Chip Results by ChIP-PCR from Four Independently Prepared Biological ChIP Replicates.
Supplemental Figure 6. Validation of H3K27me3 ChIP-Chip Results by ChIP-PCR from Four Independently Prepared Biological ChIP Replicates.
Supplemental Figure 7. Distribution of Four Histone Modifications within Arabidopsis Genes.
Supplemental Figure 8. Histone Modifications in Gene Promoter and Body Regions.
Supplemental Figure 9. Acetylation of Promoter Regions Does Not Increase Gene Expression Levels.
Supplemental Figure 10. Comparison of Identified Target Genes with a Previously Published Data Set.
Supplemental Figure 11. Magnification of Figure 6B.
Supplemental Table 1. PCR Primers Used for Validation of Modified Regions.
Supplemental Data Set 1. Expression of ∼24,000 Arabidopsis Genes in Response to Light Exposure.
Supplemental Data Set 2. List of H3K9ac Target Genes in Arabidopsis.
Supplemental Data Set 3. List of H3K9me3 Target Genes in Arabidopsis.
Supplemental Data Set 4. List of H3K27ac Target Genes in Arabidopsis.
Supplemental Data Set 5. List of H3K27me3 Target Genes in Arabidopsis.
Acknowledgments
This research was supported by funds from the National Institutes of Health (GM47850) to X.W.D. J.-B.F.C. was in part supported by postdoctoral fellowships from Le Fonds Québécois de la Recherche sur la Nature et les Technologies, Natural Sciences and Engineering Research Council of Canada, and Yale University. We thank Kenneth Nelson for assistance with microarray hybridization.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
References
- Alabadi, D., Gallego-Bartolome, J., Orlando, L., Garcia-Carcel, L., Rubio, V., Martinez, C., Frigerio, M., Iglesias-Pedraz, J.M., Espinosa, A., Deng, X.W., and Blazquez, M.A. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53 324–335. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., et al. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed, M., Bertrand, C., Servet, C., and Zhou, D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed, M., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56 493–504. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57 289–300. [Google Scholar]
- Berger, S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., Humphrey, E.L., Erlich, R.L., Schneider, R., Bouman, P., Liu, J.S., Kouzarides, T., and Schreiber, S.L. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99 8695–8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B.E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D.K., Huebert, D.J., McMahon, S., Karlsson, E.K., Kulbokas III, E.J., Gingeras, T.R., Schreiber, S.L., and Lander, E.S. (2005). Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120 169–181. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 315–326. [DOI] [PubMed] [Google Scholar]
- Bertrand, C., Benhamed, M., Li, Y.F., Ayadi, M., Lemonnier, G., Renou, J.P., Delarue, M., and Zhou, D.X. (2005). Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 280 1465–1473. [DOI] [PubMed] [Google Scholar]
- Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A.V., Tariq, M., and Paszkowski, J. (2004). Chromatin techniques for plant cells. Plant J. 39 776–789. [DOI] [PubMed] [Google Scholar]
- Caro, E., Castellano, M.M., and Gutierrez, C. (2007). A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447 213–217. [DOI] [PubMed] [Google Scholar]
- Chua, Y.L., Brown, A.P., and Gray, J.C. (2001). Targeted histone acetylation and altered nuclease accessibility over short regions of the pea plastocyanin gene. Plant Cell 13 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, Y.L., Watson, L.A., and Gray, J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas, M., Davière, J.M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blázquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Denslow, S.A., and Wade, P.A. (2007). The human Mi-2/NuRD complex and gene regulation. Oncogene 26 5433–5438. [DOI] [PubMed] [Google Scholar]
- Eberhard, S., Finazzi, G., and Wollman, F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42 463–515. [DOI] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L., Zhou, J., Elling, A.A., Charron, J.B., and Deng, X.W. (2008). Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Heintzman, N.D., et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39 311–318. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9 15–26. [DOI] [PubMed] [Google Scholar]
- Holm, M., Ma, L.G., Qu, L.J., and Deng, X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Ji, H., Jiang, H., Ma, W., Johnson, D.S., Myers, R.M., and Wong, W.H. (2008). An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H., and Wong, W.H. (2005). TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics 21 3629–3636. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., Lau, O.S., and Deng, X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 217–230. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., Ma, L., Strickland, E., and Deng, X.W. (2005). Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17 3239–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, Y., and García-Martínez, J.L. (1999). Regulation of gibberellin biosynthesis by light. Curr. Opin. Plant Biol. 2 398–403. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kim, J.M., To, T.K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., Toyoda, T., Kimura, H., Shinozaki, K., and Seki, M. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49 1580–1588. [DOI] [PubMed] [Google Scholar]
- Lee, J., He, K., Stolc, V., Lee, H., Figueroa, P., Gao, Y., Tongprasit, W., Zhao, H., Lee, I., and Deng, X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., et al. (2008). High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20 259–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Gao, Y., Qu, L., Chen, Z., Li, J., Zhao, H., and Deng, X.W. (2002). Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron, R., Trojer, P., and Reinberg, D. (2005). The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15 163–176. [DOI] [PubMed] [Google Scholar]
- Millar, C.B., and Grunstein, M. (2006). Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7 657–666. [DOI] [PubMed] [Google Scholar]
- Mueller, L.A., Zhang, P., and Rhee, S.Y. (2003). AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann, K., Fischer, A., Hofmann, I., Krauss, V., Phalke, S., Irmler, K., Hause, G., Aurich, A.C., Dorn, R., Jenuwein, T., and Reuter, G. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre, N., Hennetin, J., Sun, L.V., Lavrov, S., Bellis, M., White, K.P., and Cavalli, G. (2006). Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4 e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, H., Suzuki, T., Kondo, S., Miura, H., Fujimura, Y., and Hayashizaki, Y. (2006). Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res. 14 203–211. [DOI] [PubMed] [Google Scholar]
- Offermann, S., Danker, T., Dreymüller, D., Kalamajka, R., Töpsch, S., Weyand, K., and Peterhänsel, C. (2006). Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phosphoenolpyruvate carboxylase promoter in maize. Plant Physiol. 141 1078–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann, S., Dreesen, B., Horst, I., Danker, T., Jaskiewicz, M., and Peterhänsel, C. (2008). Developmental and environmental signals induce distinct histone acetylation profiles on distal and proximal promoter elements of the C4-Pepc gene in maize. Genetics 179 1891–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Cheng, J.C., Sung, Z.R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S., Park, S., and van Nocker, S. (2008). Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 4 e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., Kubicek, S., Mechtler, K., O'Sullivan, R.J., Derijck, A.A., Perez-Burgos, L., Kohlmaier, A., Opravil, S., Tachibana, M., Shinkai, Y., Martens, J.H., and Jenuwein, T. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12 1577–1589. [DOI] [PubMed] [Google Scholar]
- Puente, P., Wei, N., and Deng, X.W. (1996). Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Rice, J.C., and Allis, C.D. (2001). Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr. Opin. Cell Biol. 13 263–273. [DOI] [PubMed] [Google Scholar]
- Rider, S.D., Henderson, J.T., Jerome, R.E., Edenberg, H.J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh, T.Y., Cuddapah, S., and Zhao, K. (2005). Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schones, D.E., Cui, K., Cuddapah, S., Roh, T.Y., Barski, A., Wang, Z., Wei, G., and Zhao, K. (2008). Dynamic regulation of nucleosome positioning in the human genome. Cell 132 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug, J., Schuller, W.P., Kappen, C., Salbaum, J.M., Bucan, M., and Stoeckert, C.J., Jr. (2005). Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 6 R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman, C. (1904). The proof and measurement of association between two things. Am. J. Psychol. 15 72–101. [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403 41–45. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Koyanagi, K.O., and Itoh, T. (2009). Highly diversified molecular evolution of downstream transcription start sites in rice and Arabidopsis. Plant Physiol. 149 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi, W.B., and Cashmore, A.R. (1995). Photomorphogenesis. Seeing the light in plant development. Curr. Biol. 5 466–468. [DOI] [PubMed] [Google Scholar]
- Tolhuis, B., de Wit, E., Muijrers, I., Teunissen, H., Talhout, W., van Steensel, B., and van Lohuizen, M. (2006). Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38 694–699. [DOI] [PubMed] [Google Scholar]
- Turck, F., Roudier, F., Farrona, S., Martin-Magniette, M.L., Guillaume, E., Buisine, N., Gagnot, S., Martienssen, R.A., Coupland, G., and Colot, V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3 e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc, C.R., Mandat, S.A., Olenchock, B.A., and Blobel, G.A. (2005). Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19 381–391. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045. [DOI] [PubMed] [Google Scholar]
- Wang, X., Elling, A.A., Li, X., Li, N., Peng, Z., He, G., Sun, H., Qi, Y., Liu, X.S., and Deng, X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Zang, C., Rosenfeld, J.A., Schones, D.E., Barski, A., Cuddapah, S., Cui, K., Roh, T.Y., Peng, W., Zhang, M.Q., and Zhao, K. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, G.W., and Simon, R.M. (2003). A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19 2448–2455. [DOI] [PubMed] [Google Scholar]
- Yamada, K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846. [DOI] [PubMed] [Google Scholar]
- Ye, J., Fang, L., Zheng, H., Zhang, Y., Chen, J., Zhang, Z., Wang, J., Li, S., Li, R., Bolund, L., and Wang, J. (2006). WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 34 W293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y.V., Pellegrini, M., Goodrich, J., and Jacobsen, S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Rider, S.D., Jr., Henderson, J.T., Fountain, M., Chuang, K., Kandachar, V., Simons, A., Edenberg, H.J., Romero-Severson, J., Muir, W.M., and Ogas, J. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283 22637–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W., Chen, H., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]