Abstract
Background
Nucleic acid amplification techniques have improved the diagnostic possibilities in respiratory tract infections, although their clinical applicability is not yet fully defined. We have evaluated a multiplex real-time PCR method for the detection of 13 respiratory viruses and 2 bacteria (Mycoplasma and Chlamydophila) in a clinical setting.
Objectives
The aim of the present study was to evaluate the diagnostic performance and clinical use of a novel multiplex PCR method in adults with community-acquired respiratory viral infection, and the impact of duration of symptoms on detection rates.
Study design
Nasopharyngeal swab samples were prospectively collected from 209 adult outpatients with respiratory infections and 100 asymptomatic controls.
Results
An infectious agent was identified in 43% of samples from patients and 2% of asymptomatic controls. The detection rate was significantly higher in samples from patients with a duration of symptoms of 6 days or less (51%) than in samples from patients with a duration of symptoms of 7 days or more (30%, p < 0.01). For human corona viruses, and influenza virus A and B there was a correlation between the amount of virus in each patient sample as measured Ct values and duration of symptoms.
Conclusions
Duration of symptoms significantly affects the detection rate of respiratory pathogens by multiplex real-time PCR in nasopharyngeal swab samples from adult patients with respiratory infections. Our finding should be taken into account when using these tests in clinical practise.
Keywords: Multiplex PCR, Respiratory viruses, Respiratory tract infection
1. Background
Real-time polymerase chain reaction (PCR) techniques are gaining increasing acceptance for diagnosis of viral respiratory tract infections (RTI). High sensitivity and short analysis time along with the ability to detect several pathogens in a single sample are advantages compared with serology, viral culture and antigen detection.1, 2, 3 PCR assays may enhance identification of viral respiratory pathogens by at least 50% compared with traditional diagnostic methods,4, 5, 6 partly by detecting viruses for which no conventional methods exist, such as human rhinovirus (hRV), human coronavirus (hCoV) and human metapneumovirus (hMPV). Kaye et al.7 found a respiratory virus as the only causative agent in 15% of adult patients admitted to hospital with acute RTI. In patients with severe pneumonia, requiring intensive care treatment, who had negative bacterial cultures from bronchoalveolar lavage (BAL) samples, Legoff et al.8 reported detection of a respiratory virus in as much as 63% of cases. Viruses believed to cause only mild upper respiratory tract infections (URTI), for example hRV and hCoV have been detected in the lower respiratory tract in patients with severe disease.9, 10, 11 On the other hand, it is not known if asymptomatic shedding occurs following clinical recovery or during subclinical infection, and results of molecular methods with high sensitivity must be assessed with caution regarding the clinical relevance of the finding.
Detection of viral agents in respiratory tract samples using multiplex real-time PCR is both quick and sensitive12, 13 and will likely replace traditional means of diagnosing RTI. The diagnostic performance of these methods in adult immunocompetent individuals and the optimal time point for specimen collection is, however, poorly defined.
2. Objectives
The aim of the present study was to evaluate the diagnostic performance and clinical use of a novel multiplex PCR method in adults with community-acquired respiratory viral infection, and the impact of duration of symptoms on detection rates.
3. Study design
We conducted a prospective study in Western Sweden during two consecutive winter seasons, October through April 2006–2008. Adult patients (≥18 years old) with symptoms of acute RTI with duration of less than 2 weeks were included in the study, by the treating physician. An acute RTI was defined as having at least two of the following symptoms; coryza, congestion, sneezing, soar throat, odynophagia, cough, chest pain, shortness of breath or fever, for which the physician found no other explanation. Patients with suspected or confirmed bacterial infection were excluded. Patients were asked to return for a follow-up visit 10 days (±2 days) after the initial visit. Signs and symptoms were recorded in a web-based form. The same protocol was used at initial and follow-up visit. A control group of 100 healthy adults without history of fever or symptoms of RTI during the preceding 14 days were also included. Control subjects were contacted for a telephone interview 2 days after testing. Individuals who had developed symptoms of RTI were excluded, to avoid detecting virus that might be shed in high levels prior to onset of symptoms.
Nasopharyngeal and throat swab specimens were collected from each patient and control subject. The swabs were jointly placed in a sterile container with 1 ml of sodium-chloride solution, and sent to the virology laboratory the same day. At the laboratory, specimens were either analysed directly or frozen at −70 °C for later analysis.
The research ethics committee at Gothenburg University, Gothenburg, Sweden, approved the study and all patients gave their informed consent. We have focused on the analysis of viruses found with our PCR assay, although two bacteria were also included in the panel.
3.1. Nucleic acid extraction and RT-PCR
We utilized a real-time PCR procedure, based on automated specimen extraction and multiplex amplification. Primers and hydrolysis probes were obtained from the literature or developed in our laboratory. Nucleic acid from 100 μL of specimen was extracted into an elution volume of 100 μL by a Magnapure LC robot (Roche Molecular Systems, Mannheim, Germany), using the Total Nucleic Acid protocol, and was amplified in an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA). Amplification was then carried out in 50 μL reaction volumes. After a reverse transcription step, 45 cycles of two-step PCR was performed. Each sample was amplified in 5 parallel reactions, each containing primers and probes specific for 3 targets, as previously described,14 targeting 15 respiratory agents. Included in the panel were parainfluenza virus 1–3 (PIV), influenza virus A (IfA) and B (IfB), human metapneumovirus (hMPV), respiratory syncytial virus (RSV), human rhinovirus (hRV), enterovirus (EV), adenovirus (AdV), human corona viruses 229E, OC43 and NL63, M. pneumoniae and C. pneumoniae.
Ct values (cycle threshold) for each patient sample positive by real-time PCR were recorded for semi-quantitative estimation of the amount of DNA/RNA in each specimen.
3.2. Statistical analysis
Chi-square test was used to compare proportions and Spearman's rank correlation coefficient to analyse correlations. A p-value of <0.05 was considered as significant.
4. Results
Two hundred and nineteen patients with symptoms of RTI and 100 asymptomatic controls were included in the study. Ten patients were excluded (Table 1 ), leaving 209 for the final analysis.
Table 1.
Demographic characteristics and detected agents with real-time PCR analysis of nasopharyngeal/throat swab specimens from patients and controls.
| Variables | Patients | Controls |
|---|---|---|
| Included, n | 219 | 100 |
| Excluded, n | 10 | 0 |
| Duration of symptoms > 2 weeks | 3 | |
| Confirmed bacterial infectiona | 2 | |
| Incorrect sampling | 3 | |
| Patient withdrawal from study | 2 | |
| Age, median (range) | 41 (19–87) | 43 (22–70) |
| Female sex, n (%) | 125 (57) | 79 (79) |
| No of samples tested, n | 209 | 100 |
| Positive outcomes, n (%) | ||
| Influenza A virus | 28 (30) | 0 |
| Rhinovirus | 19 (20) | 2 (2) |
| Human Coronavirus NL63 | 6 (6) | 0 |
| Human Coronavirus OC43 | 7 (7) | 0 |
| Influenza B virus | 10 (11) | 0 |
| Mycoplasma pneumoniae | 7 (7) | 0 |
| Respiratory syncytial virus | 6 (6) | 0 |
| Human metapneumovirus | 6 (6) | 0 |
| Adenovirus | 3 (3) | 0 |
| Parainfluenza virus 1–3 | 2 (2) | 0 |
| No of samples positive for one microbial agent or more, n (%) | 94 (43) | 2 (2) |
One patient was excluded prospectively due to bacterial tonsillitis and one patient was excluded retrospectively due to pneumococcal sepsis.
An infectious agent was identified in 94 patients (43%) and in 2 controls (2%). Demographic characteristics and the agents detected are presented in Table 1. No cases of Enteroviruses or Chlamydia pneumoniae were identified. In 7% of positive outcomes (n = 7) more than one agent was identified (data not shown).
4.1. Duration of symptoms
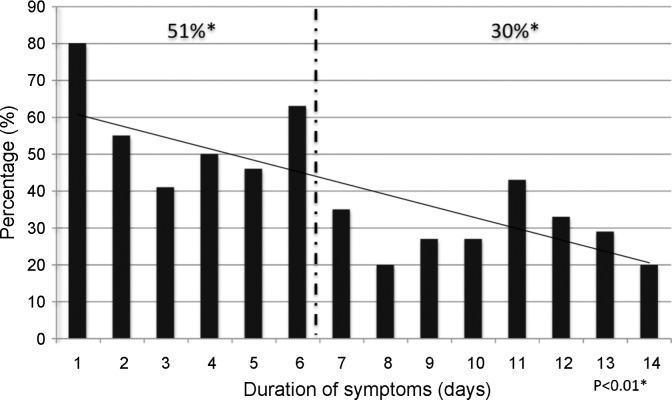
The duration of symptoms ranged from 0 to 14 days. The majority of patients (63%) were included within the first 7 days of disease. The detection rate was significantly higher in samples from patients with a duration of symptoms of 6 days or less (51%) than in samples from patients with a duration of symptoms of 7 days or more (30%, p < 0.01) (Fig. 1 ).
Fig. 1.
Distribution of real-time PCR positive samples (n = 94) according to duration of symptoms of patients (n = 219). * 51% vs. 30%, Chi-square test.
4.2. Relative viral load
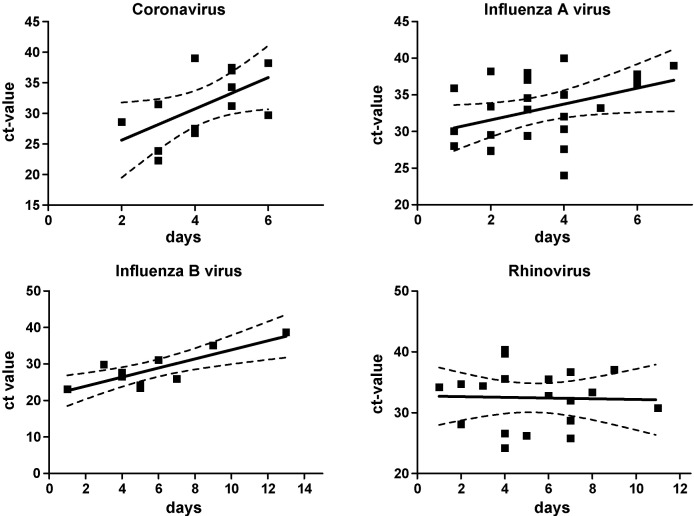
For HCoV, IfA and IfB there was a correlation between Ct values and duration of symptoms (n = 13, rs = 0.33, p < 0.05; n = 24, rs = 0.17, p < 0.05; n = 10, rs = 0.65, p < 0.01) For hRV no such correlation was found (Fig. 2 ). Two patients with influenza A were omitted because of results incongruent with patient history (data not shown).
Fig. 2.
Cycle threshold values (Ct-values) in relation to duration of symptoms in patients with acute respiratory tract infections. Human Corona virus OC43 and NL63, influenza A virus, influenza B virus and rhinovirus.
4.3. Follow-up testing
In total, 63% (n = 138) of the patients returned for a follow-up visit 10 ± 2 days after the initial visit. Nasopharynx/throat swab samples were collected. Out of 94 patients with a positive sample at initial visit, 57 patients (61%) showed up for follow-up testing. The same pathogen was detected in 13 of these patients (23%) (see Table 2 ).
Table 2.
Follow-up (10 ± 2 days after initial visit) test result from analysis with real-time PCR of nasopharyngeal/throat swab specimens.
| Patients and samples | N (%) |
|---|---|
| Patients at follow-up visit | 138 (63.0) |
| Patients with positive PCR analysis at initial visit | 94 (42.9) |
| Nasopharyngeal and throat swab samples collected at follow-up | 57 (60.6) |
| Patients with positive PCR analysis at initial visit and follow-up | 18 (31.6) |
| Patients with same pathogen at initial visit and at follow-up | 13 (22.8) |
| Pathogens (in patients with same pathogen at initial visit and follow-up, n = 13) | |
| Rhinovirus | 8 (61.5) |
| Human Corona virus NL63 | 2 (15.4) |
| Human Corona virus OC43 | 1 (7.7) |
| Mycoplasma pneumoniae | 1 (7.7) |
| Respiratory syncytial virus | 1 (7.7) |
Eight of 13 (62%) of samples positive for the same virus at initial and follow-up visit were positive for hRV. A new microbial agent was found in five subjects, all of who still had respiratory tract symptoms at follow-up. All patients still positive for the same agent on follow-up had a higher Ct-value (corresponding to a lower amount of viral RNA in the specimen) 10 days later except one patient with hRV on both occasions (data not shown).
5. Discussion
We have shown that the diagnostic yield increases significantly if a multiplex PCR assay for respiratory viruses is used within the initial 6 days of symptoms, in immunocompetent adults with RTI. This is in concordance with reports on single-plex PCR assays for influenza where the detection rate correlates to duration of symptoms.15
We detected an infectious agent in 43% of patients. Previously, detection rates ranging between 43% and 63% have been described,6, 12, 16, 17 with exceptions for very high rates among young children and infants.18, 19 In asymptomatic children, higher frequencies of positive results have been reported.20, 21 In particular detection of hRV and EV may represent a previous infection dating back 4–5 weeks, due to prolonged shedding of virus.22 Lack of pre-existing immunity, greater viral exposure as well as an immature immune system may cause higher levels of viral replication and prolonged viral shedding in children. Templeton et al. reported a detection rate of only 24%, but hRV, hCoV, AdV or hMPV were not included in their panel.23 Studies performed retrospectively on clinical samples will select for a short duration of symptoms as will studies including children, who tend to seek medical care early and who may shed more virus during a longer time period. Thus, the prospective approach and the adult population may explain the relatively low detection rate found in our sample. As the study period included the influenza season, we detected influenza A and B virus in a relatively high proportion of patients (41%), which is comparable to other studies of adults.9
We detected a pathogen in only 2% of control subjects, both hRV. Although most studies do not include controls, Creer et al.16 found a respiratory virus in 12% of controls, who reported no signs of respiratory tract infection during 2 months before sampling, predominantly hRV. Asymptomatic shedding of hRV, RSV and PIV has also been reported in immunocompromised patients.24, 25 Our results suggest that viral shedding exceeding 14 days is rare in immunocompetent adults. The rate of asymptomatic carriage of virus might be affected by epidemiological factors that vary over time. Our control samples were, however, collected throughout the entire study period.
For several viruses analysed in our study, the amount of viral DNA/RNA decreased with duration of symptoms, suggesting a gradual reduction of viral shedding from the epithelial surface over time. This is in concordance with the gradual reduction of RSV levels in nasopharyngeal aspirates described by Gerna et al.26 and Campanini et al.27
Dual viral infections are previously described in approximately 5–20% of infected patients, with a higher frequency among young children.17, 28, 29 In accordance with previous studies we found two viruses in 7% of the samples. The clinical impact of multiple infections remains to be determined. In one study of children with RSV infection, higher fever, longer hospital stays and more frequent use of antibiotics was associated with multiple infections.30 Semi-quantification of viral DNA, as measured by Ct values, may be of use when determining the clinical relevance of a positive test, particularly in multiple infections.
We found a correlation between Ct values and duration of symptoms for hCoV, IfA, and IfB but not for hRV. The high genetic variability of hRVs, which increase the probability of probe-target mismatch, may explain this. For other viruses, such as influenza B virus, with less genetic variability viral detection will be more stable. The lack of association between duration of illness and Ct values in hRV infection may also reflect variations in viral shedding and differences in pre-existing immunity between individuals as well as limited sample size. Interestingly, the majority of patients, 7/13 (62%) positive for any agent at the follow-up visit (day 10 ± 2 days) had hRV. Whether this translates into a longer period of infectivity remains to be determined. However, the semi-quantitative estimation of viral loads by Ct values must be interpreted with caution since relative viral loads were not compared to standard amounts of virus.
The clinical relevance of the outcome of multiplex PCR tests for respiratory viruses is not yet fully determined. For some agents, such as influenza A and B virus, a positive test may provide the basis for antiviral treatment. It has been suggested that a rapid etiologic diagnosis of viral RTI could reduce unnecessary prescription of antibiotics, but this remains to be shown.
We conclude that duration of symptoms affects detection rate by real-time multiplex PCR in adult patients with RTI. Duration of symptoms should be taken into account when using these tests in clinical practise. For some viruses, with relatively low genetic variability, semi-quantification might be of value when interpreting results where more than one respiratory virus is found.
Conflict of interest
None declared.
Acknowledgements
The authors would like to acknowledge the staff at the clinical virology laboratory, department of virus detection, at Sahlgrenska University Hospital for technical expertise and patients and staff at the following centres in the Västra Götaland Region; The Infectious disease clinics in Uddevalla, Skövde, Borås, and Göteborg, and Primary health care centres in Stenungsund, Sollebrunn, Floda, Kungshöjd, Kungsten, Olskroken, Carlanderska, and Capio Axess.
Funding: The study was partly funded by The Swedish strategic programme against antibiotic resistance (STRAMA), Capio Research foundation, grant # 2006-1166 and the Västra Götaland Region research funds, grant # VGFOUREG-8402.
Ethical approval: Granted by the research ethics committee at Gothenburg University, # 403-06.
References
- 1.Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 2005 Jun;126(1–2):53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiveljung-Lindell A., Rotzen-Ostlund M., Gupta S., Ullstrand R., Grillner L., Zweygberg-Wirgart B. Development and implementation of a molecular diagnostic platform for daily rapid detection of 15 respiratory viruses. J Med Virol. 2009;81(January (1)):167–175. doi: 10.1002/jmv.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunson R.N., Collins T.C., Carman W.F. Practical experience of high throughput real time PCR in the routine diagnostic virology setting. J Clin Virol. 2006;35(April (4)):355–367. doi: 10.1016/j.jcv.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbino J., Gerbase M.W., Wunderli W., Kolarova L., Nicod L.P., Rochat T. Respiratory viruses and severe lower respiratory tract complications in hospitalized patients. Chest. 2004;125(March (3)):1033–1039. doi: 10.1378/chest.125.3.1033. [DOI] [PubMed] [Google Scholar]
- 5.Lee B.E., Robinson J.L., Khurana V., Pang X.L., Preiksaitis J.K., Fox J.D. Enhanced identification of viral and atypical bacterial pathogens in lower respiratory tract samples with nucleic acid amplification tests. J Med Virol. 2006;78(May (5)):702–710. doi: 10.1002/jmv.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oosterheert J.J., van Loon A.M., Schuurman R., Hoepelman A.I., Hak E., Thijsen S. Impact of rapid detection of viral and atypical bacterial pathogens by real-time polymerase chain reaction for patients with lower respiratory tract infection. Clin Infect Dis. 2005;41(November (10)):1438–1444. doi: 10.1086/497134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaye M., Skidmore S., Osman H., Weinbren M., Warren R. Surveillance of respiratory virus infections in adult hospital admissions using rapid methods. Epidemiol Infect. 2006;134(August (4)):792–798. doi: 10.1017/S0950268805005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legoff J., Guerot E., Ndjoyi-Mbiguino A., Matta M., Si-Mohamed A., Gutmann L. High prevalence of respiratory viral infections in patients hospitalized in an intensive care unit for acute respiratory infections as detected by nucleic acid-based assays. J Clin Microbiol. 2005;43(January (1)):455–457. doi: 10.1128/JCM.43.1.455-457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(October (4)):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renwick N., Schweiger B., Kapoor V., Liu Z., Villari J., Bullmann R. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J Infect Dis. 2007;196(December (12)):1754–1760. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louie J.K., Roy-Burman A., Guardia-Labar L., Boston E.J., Kiang D., Padilla T. Rhinovirus associated with severe lower respiratory tract infections in children. Pediatr Infect Dis J. 2009;28(April (4)):337–339. doi: 10.1097/INF.0b013e31818ffc1b. [DOI] [PubMed] [Google Scholar]
- 12.Lam W.Y., Yeung A.C., Tang J.W., Ip M., Chan E.W., Hui M. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007;45(November (11)):3631–3640. doi: 10.1128/JCM.00280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace L.A., McAulay K.A., Douglas J.D., Elder A.G., Stott D.J., Carman W.F. Influenza diagnosis: from dark isolation into the molecular light West of Scotland Respiratory Virus Study Group. J Infect. 1999;39(November (3)):221–226. doi: 10.1016/s0163-4453(99)90053-1. [DOI] [PubMed] [Google Scholar]
- 14.Brittain-Long R., Nord S., Olofsson S., Westin J., Anderson L.M., Lindh M. Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol. 2008;41(January (1)):53–56. doi: 10.1016/j.jcv.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leekha S., Zitterkopf N.L., Espy M.J., Smith T.F., Thompson R.L., Sampathkumar P. Duration of influenza A virus shedding in hospitalized patients and implications for infection control. Infect Cntrl Hosp Epidemiol. 2007;28(September (9)):1071–1076. doi: 10.1086/520101. [DOI] [PubMed] [Google Scholar]
- 16.Creer D.D., Dilworth J.P., Gillespie S.H., Johnston A.R., Johnston S.L., Ling C. Aetiological role of viral and bacterial infections in acute adult lower respiratory tract infection (LRTI) in primary care. Thorax. 2006;61(January (1)):75–79. doi: 10.1136/thx.2004.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahony J., Chong S., Merante F., Yaghoubian S., Sinha T., Lisle C. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J Clin Microbiol. 2007;45(September (9)):2965–2970. doi: 10.1128/JCM.02436-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruohola A., Waris M., Allander T., Ziegler T., Heikkinen T., Ruuskanen O. Viral etiology of common cold in children, Finland. Emerg Infect Dis. 2009;15(February (2)):344–346. doi: 10.3201/eid1502.081468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freymuth F., Vabret A., Cuvillon-Nimal D., Simon S., Dina J., Legrand L. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006;78(November (11)):1498–1504. doi: 10.1002/jmv.20725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nokso-Koivisto J., Kinnari T.J., Lindahl P., Hovi T., Pitkaranta A. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J Med Virol. 2002;66(March (3)):417–420. doi: 10.1002/jmv.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Benten I., Koopman L., Niesters B., Hop W., van Middelkoop B., de Waal L. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14(October (5)):363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jartti T., Lehtinen P., Vuorinen T., Koskenvuo M., Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72(April (4)):695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 23.Templeton K.E., Scheltinga S.A., Beersma M.F., Kroes A.C., Claas E.C. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42(April (4)):1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ison M.G. Respiratory viral infections in transplant recipients. Antivir Ther. 2007;(4 Pt. B):627–638. [PubMed] [Google Scholar]
- 25.Peck A.J., Englund J.A., Kuypers J., Guthrie K.A., Corey L., Morrow R. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood. 2007;110(September (5)):1681–1688. doi: 10.1182/blood-2006-12-060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerna G., Campanini G., Rognoni V., Marchi A., Rovida F., Piralla A. Correlation of viral load as determined by real-time RT-PCR and clinical characteristics of respiratory syncytial virus lower respiratory tract infections in early infancy. J Clin Virol. 2008;41(January (1)):45–48. doi: 10.1016/j.jcv.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Campanini G., Percivalle E., Baldanti F., Rovida F., Bertaina A., Marchi A. Human respiratory syncytial virus (hRSV) RNA quantification in nasopharyngeal secretions identifies the hRSV etiologic role in acute respiratory tract infections of hospitalized infants. J Clin Virol. 2007;39(June (2)):119–124. doi: 10.1016/j.jcv.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung J.Y., Han T.H., Kim S.W., Hwang E.S. Respiratory picornavirus infections in Korean children with lower respiratory tract infections. Scand J Infect Dis. 2007;39(3):250–254. doi: 10.1080/00365540600999126. [DOI] [PubMed] [Google Scholar]
- 29.Pierangeli A., Gentile M., Di Marco P., Pagnotti P., Scagnolari C., Trombetti S. Detection and typing by molecular techniques of respiratory viruses in children hospitalized for acute respiratory infection in Rome, Italy. J Med Virol. 2007;79(April (4)):463–468. doi: 10.1002/jmv.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvo C., Garcia-Garcia M.L., Blanco C., Vazquez M.C., Frias M.E., Perez-Brena P. Multiple simultaneous viral infections in infants with acute respiratory tract infections in Spain. J Clin Virol. 2008;42(July (3)):268–272. doi: 10.1016/j.jcv.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]