Abstract
The accuracy of metabolic quantification in MR spectroscopy (MRS) is limited by the unknown radio-frequency field (B1) and longitudinal relaxation time (T1). To address both issues in proton (1H) MRS we obtained B1-corrected T1 maps of N-acetylaspartate (NAA), choline (Cho) and creatine (Cr) in five healthy rhesus macaques at 3 T. For efficient use of the 4 hour experiment, we used a new three-point protocol that optimizes the precision of T1 in 3D 1H-MRS localization for extensive, ~30%, brain coverage at (0.6×0.6×0.5 cm)3=180 μL spatial resolution. The resulting mean±standard error (SEM) T1s in 700 voxels were: NAA=1232±44, Cr=1238±23 and Cho=1107±56 ms. Their histograms from all 140 voxels in each animal were similar in position and shape - characterized by SEMs of the full width at half maximum divided by their means, of better than 8%. Regional gray matter NAA, Cho and Cr T1s: 1333±43, 1265±52 and 1131±28 ms were 5–10% longer than white matter: 1188±34, 1201±24 and 1082±50 ms (statistically significant for the NAA only), all within 10% of the corresponding published values in the human brain.
Keywords: Brain MR Spectroscopy, Brain Metabolites, B1 mapping, Longitudinal Relaxation Time, Rhesus Macaque, T1
Introduction
The similarities between the rhesus macaque brain and its human counterpart have lead to use of the former in studies of neurological disorders and their treatment (1–4). Due to the serial nature and high cost of these studies, non-invasive modalities: MRI for morphology or function and proton MR spectroscopy (1H-MRS) for metabolism, are often favored. The latter can be used to monitor neuronal cells, cell energetics and membrane turnover by quantification of their surrogate metabolic markers (5,6): N-acetylaspartate (NAA), creatine (Cr) and choline (Cho).
Metabolic quantification requires knowledge of several parameters that influence the 1H-MRS signal. Some, e.g., the echo- and recycle times (TE, TR) or voxel size are known. Instrumental factors such as the static (B0) and radio-frequency (RF, B1) field inhomogeneity can be handled by field mapping and internal water referencing (7–9). Accounting for the molecular environment, however, requires knowledge of the local transverse (T2) and longitudinal (T1) relaxation times (5). Although T1- and T2-weighting can be minimized at TE «T2 and TR»T1, the latter reduces both coverage and the signal-to-noise-ratio (SNR), thereby limiting the spatial resolution (10,11). Given the small, ~80 cm3, average macaque brain(12), either limitations can become prohibitive.
To optimize SNR/unit-time for the net α=90° nutation, 1H-MRS is often done at TR≈1.2·T1 (11), that also subjects the metabolites’ signals to unknown T1-weighting. Furthermore, the RF field itself can suffer regional variations that worsen with increasing B0 (13,14). Consequently, the observed steady-state MR signal, S, exhibits a complex dependence on B1 and T1 (15):
| [1] |
where α is the target nutation and B1, the ratio between the actual local and nominal B1 value, can exceed 20% at 4 T and 40% at 7 T over regions the size of a human head (13). Unknown B1 and T1 combine to confound determining ρ0, the voxel spin density, i.e., metabolic quantification.
In this paper we mapped the B1-corrected T1s of NAA, Cr and Cho at 3 T over ~30% of the macaque brain with three dimensional (3D) 1H-MRS at 180 μL spatial resolution in a three-point protocol that optimizes their precision per unit time (15). This also enabled us to test two implicit hypotheses often made in 1H-MRS (5,16): First, that inter-animal metabolite T1 distributions are sufficiently similar that obtaining them once is enough. Second, that regional T1s are similar enough that one value per metabolite suffices for reliable quantification using Eq. [1].
Materials and Methods
Non Human Primates
One male and four female healthy adult (3 – 5 years old, 3.9 – 6 kg) rhesus macaques (Macaca mulatta) were studied. Each was tranquilized with 15 – 20 mg/kg intra-muscular ketamine hydrochloride and intubated to ensure a patent airway during the experiment (no mechanical ventilation was needed). Intravenous injection of 0.4 mg/kg atropine was administered to prevent bradycardia and continuous infusion of 0.25 mg/kg/min propofol was maintained throughout via catheter in a saphenous vein. Heart and respiratory rates, oxygen saturation and end-tidal CO2 were monitored continuously and a water blanket used to prevent hypothermia. The animals were under constant veterinary supervision and the protocol was approved by the Harvard Medical School and Massachusetts General Hospital IACUCs.
MRI and 1H-MRS
All experiments were done in a 3 T whole-body MR imager (Magnetom TIM Trio, Siemens AG, Erlangen, Germany) with a circularly-polarized transmit-receive human knee coil. To guide the 1H-MRS volume of interest (VOI) we acquired sagittal, coronal and axial turbo spin echo MRI at TE/TR=15/10000 ms, 512×512 matrix, 140×140 mm2 field of view (FOV) and 1 mm slice thickness. A 4.2×3.0×2.0 cm3 anterior-posterior × left-right × inferior-superior (AP×LR×IS) VOI was then image-guided along the splenium-genu axis of the corpus callosum, as shown in Fig. 1a and b. The manufacturer’s automatic shim procedure then adjusted its water linewidth to a consistent 26±1 Hz full-width-at-half-maximum (FWHM).
Fig. 1.
Top: Sagittal (a) and axial (b) T2-weighted MRI from a macaque head showing VOI and FOV (solid and dashed white frames) placement and their relative size.
Bottom: Real part of the axial 5×7 (LR×AP) spectra matrices from the VOI on slice b for each of the TRi, averages-Ni and flip-angles, αi (i=1..3), on common 3.6 – 1.6 ppm and intensity scales. Note the spectral resolution and SNRs at each of the three TRs.
The VOI was selectively excited using TE=42 ms PRESS with the first 5.12 ms RF pulse also performing 4th-order transverse Hadamard encoding in the IS direction (17,18). Its 4.5 mT/m slice-select gradient applied kept the chemical shift displacement between NAA and Cho under 0.08 cm, ~16% of the slices thickness (18). The AP and LR aspects of the VOI were defined by numerically optimized 15.4 ms 180°s pulsed under 1.9 and 2.7 mT/m, sustaining 0.18 and 0.13 cm (30% and 22%) relative displacement between NAA and Cho at the VOI edges. The slices’ LR×AP planes were partitioned with 16×16 chemical shift imaging (CSI) over a 9.6×9.6 cm2 FOV into 0.6×0.6 ×0.5 cm3 (180 μL) voxels. With no k-space filters applied the effective resolution, accounting for full width at half maximum of the point spread function for the uniform 2D phase encoding, was 226 μL (19). [In the Hadamard direction the nominal equals the actual voxel size (20)]. The signal was acquired for 256 ms at ±1 kHz bandwidth.
Choice of acquisition parameters
Since T1 precision is a function of the source data SNR (15,21), noisy 1H-MRS experiments at high spatial resolution lead to long acquisition. For its efficient use we combined two approaches: (i) TriTone - a three-point scheme optimizing B1 and T1 precision by manipulating the number of averages: N1, N2, N3 and nominal flip angles: α1, α2, α3, at TR1, TR2 and TR3 (15). (ii) 3D 1H-MRS to cover extensive brain volume at the same SNR as single-voxel methods (11,22).
Since the uncertainty in TriTone-estimated T1s varies by less than 10% from ×0.8 to ×1.6 of the T1tune for which the protocol is adjusted, we used 800 ms for that parameter to keep the total experiment duration to under 4 hours. This led to the following three-point protocol (15): TR1=600 ms, α1=85°, N1=8 (for 82 min.); TR2=1280 ms; α2=40°, N2=1 (22 min.); and TR3=3600 ms, α3=115°, N3=2 (123 minutes). To keep the SAR within the FDA approved limits in human subjects for the shortest TR=600 ms acquisition we extended each of the two PRESS 180° pulses to 15.4 ms leading to a TE of 42 ms.
Post-processing and T1 determination
The 1H-MRS data were processed off-line with in-house software. Residual water signals were removed in the time domain (23), voxel-shift aligned the CSI grid with the NAA VOI and the data was zero-filled to 2048 and 32×32 in the time and axial (CSI) directions. [Although it does not add new information to the raw data, Zero-filling can increase the effective spatial resolution by providing overlapping voxels that can reduce partial volume effects (24,25)]. Fourier and Hadamard transforms in the time and spatial dimensions followed with frequency and zero-order phase shifts done automatically in reference to the NAA peak in each voxel.
Relative level of the i-th metabolite (i=NAA, Cr or Cho) at each point, S1i, S2i, and S3i, were estimated from their peak area with the Spectroscopic Imaging Toolkit – Fitting (SITools-FITT) parametric spectral modeling and least-squares optimization package of Soher et al. (8) with NAA, Cho, Cr, glutamate, glutamine, taurine and myo-inositol included in the model. The relative level were converted into spherical angles:
| [2] |
and polled against a look-up table T1(φ, θ) constructed based on the sequence parameters (15). Their inter-animal reproducibility was assessed for each metabolite in terms of the coefficient of variation (CV=standard deviation/mean) using as data for each animal the mean over all voxels.
Statistical Analyses
Two-way analysis of variance based on ranks was used to compare brain regions with respect to the mean of each metabolite. Specifically, the data for a given metabolite were first converted to ranks which were then used as the dependent variable in order to satisfy underlying distribution assumptions. The model included animal ID as a random classification factor to allow comparisons to be made within each animal and to account for the lack of statistical independence among observations made for the same animal. Statistical significance was defined as a two-sided p≤0.05. SAS version 9.0 (SAS Institute, Cary, NC) was used for all analyses.
Results
Spectra from one slice at each of the three TriTone acquisition points, shown in Fig. 1, demonstrate the SNRs and spectral resolution. Specifically, metabolite linewidth in each voxel, also estimated by the modeling software, averaged 8.7±3.1 Hz FWHM corresponding to T2*s (=1/πΔω assuming Lorentzian shape) of 37±13 ms. The SNRs from the 700 voxels and the inter-animal coefficient of variation in the five macaques, are compiled in Table 1.
Table 1.
The SNR of 700 (35 voxels per slice × 4 slices per animal × 5 animals) voxels (mean±standard deviation) of NAA, Cr and Cho at the three different TRs, and the inter-animal T1 coefficient of variation (CV) for each of the three metabolites.
| Metabolite | SNR |
Inter-animal T1 CV | ||
|---|---|---|---|---|
| TR1 | TR2 | TR3 | ||
| NAA | 31.7±10.1 | 21.7±7.0 | 26.4±8.8 | 3.5% |
| Cr | 22.7±9.3 | 14.9±5.6 | 23.7±8.9 | 1.9% |
| Cho | 14.1±6.8 | 13.8±6.8 | 20.9±9.7 | 4.4% |
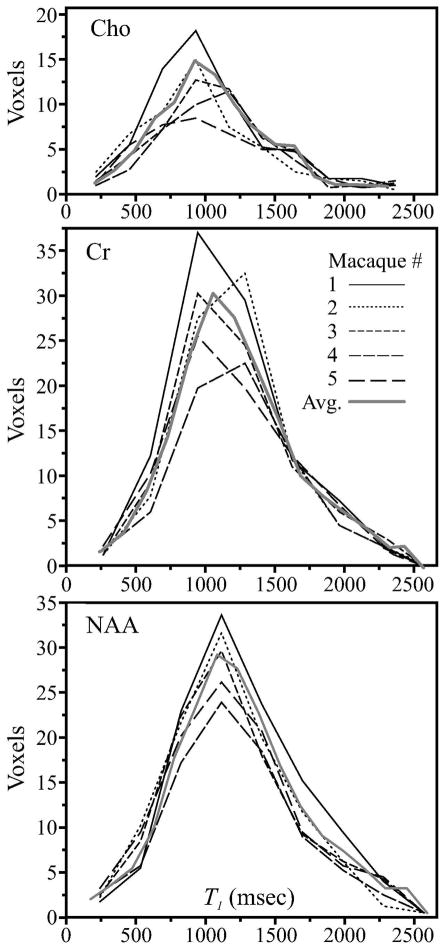
The B1-corrected T1 histograms for each metabolite in the 140 voxels from every monkey together with a normalized histogram for each metabolite from the 700 voxels in all the animals, are shown in Fig. 2. Their distributions exhibit a gratifying similarity in: (a) peak position, also reflected by less than 5% inter-animal CV (see Table 1); and (b) width, characterize by the CV of histograms FWHM: NAA=11%, Cr=16% and Cho=19%. The mean ± standard error (SEM) of the 140 T1s in each of the 5 animals were: NAA=1232±44, Cr=1238±23 and Cho=1107±56 ms. Note that all T1 values in Table 2 are in the ×0.8 to ×1.6 range about the 800 ms T1tune for which the protocol is adjusted, i.e., within 10% of the optimal value obtainable for the acquisition time.
Fig. 2.
Cho, Cr and NAA B1-corrected T1s histograms from all 140 voxels in the VOI of each macaque (black lines), as well a normalized histogram for each metabolite formed from the 700 voxels in all the animals (thick solid gray line). Note the inter-animal histogram similarity reflected by the peak position and FWHM for each metabolite and proximity to the “global” histogram from all, suggesting that these distributions are characteristic.
Table 2.
Mean±SEM values of proton T1 relaxation times (milliseconds) at 3 T of the NAA, Cr and Cho in the various GM and WM brain regions and structures of the rhesus macaque brain shown in Fig. 3.
| T1 (ms) | ||||
|---|---|---|---|---|
| NAA | Cr | Cho | ||
| GM | Caudate head | 1376±61 | 1370±75 | 1072±121 |
| Thalamus | 1330±78 | 1272±30 | 1099±106 | |
| Lentiform nucleus | 1212±70 | 1293±40 | 1198±42 | |
| Cingulate gyrus | 1413±89 | 1123±99 | 1154±75 | |
| aMean±SEM | 1335±25 | 1263±22 | 1147±68 | |
| WM | Splenium of the b CC | 1222±33 | 1176±55 | 1132±27 |
| Centrum semiovale | 1154±51 | 1224±49 | 1032±76 | |
| aMean±SEM | 1166±19 | 1214±41 | 1047±44 | |
Mean±SEM of the average of the T1s in all the structures in that tissue type (GM or WM) in each of the 5 animals.
CC – corpus callosum.
Regional T1s were estimated in the six gray and white matter (GM, WM) brain regions shown in Fig. 3 with their average spectra at each TR overlaid with the SITools-FITT model functions. Each was outlined on the axial MRI and our software averaged the T1s in all voxels that fell entirely or partially within. No chemical shift displacement (at the VOI edges) or CSF partial volume correction were made since they cancel in the ratios of Eq. [2] and exert only minor influence on T1 precision in affected voxels (due to lower SNR) but not on its accuracy. The regional values compiled in Table 2 show GM T1s to be 5 - 10% longer than WM, although this difference was statistically significant only for the NAA.
Fig. 3.
Top: Axial MRI showing the 3×4.2 cm2 VOI (thick white frame), CSI grid (thin white lines) and the brain regions where the voxels’ T1 were averaged (thick white lines): a- cingulate gyrus, d- caudate head, e- lentiform nucleus, f- thalamus in the GM; and b- centrum semiovale and c- splenium of the corpus callosum in WM.
Bottom: Real part of the average spectra from each region and time point (black lines) overlaid with the model lineshapes (gray lines). All are on common 3.7 – 1.6 ppm and intensity scales. Note their improved SNRs versus single-voxel spectra in Fig. 1.
Discussion
Nutation angle variation due to B1 inhomogeneity (even in volume coils at low fields) is well known to be the main source of T1 estimation errors (26,27), that worsen at higher B0s (10,13,14). Unknown B1 and T1 combine to impede use of Eq. [1] in two ways. First, the assumption that the nutation equals its nominal value fails due to B1 variations of 20% and 40% at 4 and 7 T over VOIs the size of a human head (13,26) that can lead to 40% – 80% error in T1 (26). Second, if TR≈T1 is used for optimum SNR [even if B1 variations are addressed through usage adiabatic pulses (28–30)], the lack of knowledge of metabolite T1’s lowers quantification accuracy.
Although other B1 mapping methods are available, besides different RF power, imaging time, and post-processing that they require, they all need at a minimum two separate scans each done with different sequence parameters (typically different flip angles) in order to obtain B1 (26,31). At least two more scans with different acquisition parameters (typically different TRs) are subsequently needed to obtain T1 (32), rendering the portion of the time used to map B1 unavailable to measure T1 and vice versa. TriTone in comparison avails all the time for both. Since the precision of T1 and B1 is a function of the SNR of the data (15), this gives TriTone an advantage that increases at higher spatial resolutions that require longer acquisition.
Combining the coverage of 3D 1H-MRS with the efficiency of TriTone, therefore, enabled us to map the B1-corrected T1s of the three main metabolites over 25.2 cm3 (≈30%) of the ~80 cm3 macaque brain at 180 μL spatial resolution and best precision for the 4 hour experiment (15,33). This in turn yielded global and regional T1s in 5 animals in order to ascertain how characteristic they are (i) across rhesus macaques; and (ii) among different brain regions and tissue types.
Indeed, the inter-animal T1s distribution reproducibility reflected by the similar metabolite T1 histograms’ peak-position and shapes (under 10% SEM of their widths - see Fig. 2) support the first hypothesis: that these T1 distributions are characteristic of the healthy rhesus macaque brain. Consequently, additional measurements even if they were feasible given the time and cost, are unnecessary since they are unlikely to yield significantly different results.
Furthermore, the less than 5% T1 SEMs for the GM and WM for all metabolites (see Table 2) reflect inter-animal similarity that substantiates the second hypothesis: that for quantification purposes a global mean T1 for each metabolite is sufficient. Specifically, the GM 5 - 10% longer T1s will lead to minimal (under 5%) difference in relative T1-weighting compared with the WM at TR=1.2·T1≈1450 ms in these regions and metabolites (11). The practical implication is that around this TR use of the mean T1 for each metabolite suffices to reduce that metabolite’s T1-weighting quantification errors to below typical voxel noise levels, rendering them insignificant.
The metabolites’ T1s obtained are all within 10% of the 1.4, 1.2 and 1.3 s reported for NAA, Cr and Cho in the human brain at 3 T (34,35). The minor discrepancies may be due to relatively low, 8–12 cm3, spatial resolution single-voxels without B1 correction [long recognized as the main source of T1 estimation errors (27)] of the latter. These may have combined to confound tissue-specific T1 estimates due to partial WM volume in GM voxels and vice versa, an issue both studies acknowledge but only estimated its impact. Our use of 3D localization allowed us to reduce the voxel-volume nearly 50 fold and to benefit from zero-filling and regional averaging to better resolve GM from WM T1s by greatly reducing (although not entirely eliminating) partial volume. The similar T1s together with the validation of the two hypotheses ( vide supra) establish the rhesus macaque brain as a good model for its human counterpart with respect to this molecular environment parameter and also yield two additional encouraging ramifications for 1H-MRS in this model system. First, only one set of T1s need be retained for quantification in humans and macaques using Eq. [1]. Second (alternatively), at common TR all the metabolites will incur the same T1-weighting in the macaque as in the human brain.
Averaging adjacent voxels in the same structure reduces their the CV of the T1s (from the ~38%, 33% and 40% for NAA, Cr and Cho, estimated from the half-width at half height of the histograms in Fig. 2) via two mechanisms: Primarily due to smaller T1 variations in similar tissue that lower the biological variations; and secondly, by increasing the SNR (over a single voxel’s - compare the spectra in Fig. 1 with Fig. 3) to reduce the instrumental noise as (the number of voxels)½. Together they yield T1s at 5–10% regional CVs (see Table 2), suggesting that additional measurement in these structures in more animals (at similar spatial resolution and time) is unlikely to yield significantly different values.
Finally, it is noteworthy that although several minor metabolites, most notably myo-inositol, could also be detected at the TE=42 ms used (cf. Fig. 3), we focused only on the three major singlets. The motivation was to balance the dependence of the precision at which B1 and T1 are estimated on the voxel SNR (15), with the spatial resolution sought and total experiment duration. Specifically, the minor metabolites’ SNRs are fivefold to an order of magnitude lower than the main singlets. Even if their T1s are fortuitously close to T1tune of NAA, Cho and Cr; at 180 μL resolution and ~4 hours of acquisition their inferior SNRs would lead to unsatisfactory precision.
Conclusion
Combining 3D 1H-MRS with an optimized acquisition protocol makes for efficient use of four hours to map regional brain metabolites’ T1s. The results reveal that these T1s are sufficiently reproducible and that their variations among WM and GM structures in healthy rhesus macaques are small enough that use of a global average T1 for each metabolite is justifiable for metabolic quantification. The overlap of the T1 histograms of these metabolites indicates that they are probably characteristic to within very few percent making individual measurements unnecessary.
Acknowledgments
We thank Drs. Andrew A. Maudsley of the University of Miami and Brian J. Soher of Duke University for their SITools-FITT software and Dr. Joanne Morris and Ms. Shannon Luboyeski for animal veterinary care. This work was supported by NIH Grants EB01015, NS050520, NS050041, NS051129, NS059331, AI028691 and NS040237. The MGH A.A. Martinos Center for Biomedical Imaging is also supported by National Center for Research Resources grant number P41RR14075 and the Mental Illness and Neuroscience Discovery (MIND) Institute.
References
- 1.Lentz MR, Westmoreland SV, Lee V, Ratai EM, Halpern EF, Gonzalez RG. Metabolic markers of neuronal injury correlate with SIV CNS disease severity and inoculum in the macaque model of neuroAIDS. Magn Reson Med. 2008;59(3):475–484. doi: 10.1002/mrm.21556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, D’Arceuil H, He J, Duggan M, Gonzalez G, Pryor J, de Crespigny A. MRI of spontaneous fluctuations after acute cerebral ischemia in nonhuman primates. J Magn Reson Imaging. 2007;26(4):1112–1116. doi: 10.1002/jmri.21131. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Cabezas MA, Rico B, Sanchez-Gonzalez MA, Cavada C. Distribution of the dopamine innervation in the macaque and human thalamus. Neuroimage. 2007;34(3):965–984. doi: 10.1016/j.neuroimage.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 4.Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC, Snyder B, Larkin K, Liu J, Orkin J, Fang ZH, Smith Y, Bachevalier J, Zola SM, Li SH, Li XJ, Chan AW. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453(7197):921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen JF, Backes WH, Nicolay K, Kooi ME. 1H MR spectroscopy of the brain: absolute quantification of metabolites. Radiology. 2006;240(2):318–332. doi: 10.1148/radiol.2402050314. [DOI] [PubMed] [Google Scholar]
- 6.Juchem C, Merkle H, Schick F, Logothetis N, Pfeuffer J. Region and volume dependencies in spectral line width assessed by 1H 2D MR chemical shift imaging in the monkey brain at 7 T. Magn Reson Imaging. 2004;22(10):1373–1383. doi: 10.1016/j.mri.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Gruetter R, Tkac I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. 2000;43(2):319–323. doi: 10.1002/(sici)1522-2594(200002)43:2<319::aid-mrm22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 8.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40(6):822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 9.Dong Z, Dreher W, Leibfritz D. Toward quantitative short-echo-time in vivo proton MR spectroscopy without water suppression. Magn Reson Med. 2006;55(6):1441–1446. doi: 10.1002/mrm.20887. [DOI] [PubMed] [Google Scholar]
- 10.Marzola P, Osculati F, Sbarbati A. High field MRI in preclinical research. Eur J Radiol. 2003;48(2):165–170. doi: 10.1016/j.ejrad.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Goelman G, Liu S, Hess D, Gonen O. Optimizing the efficiency of high-field multivoxel spectroscopic imaging by multiplexing in space and time. Magn Reson Med. 2006;56(1):34–40. doi: 10.1002/mrm.20942. [DOI] [PubMed] [Google Scholar]
- 12.Cheverud J, Falk D, Vannier M, Konigsberg L, Helmkamp R, Hildebolt C. Heritability of brain size and surface features in rhesus macaques (Macaca mulatta) J Hered. 1990;81(1):51–57. doi: 10.1093/oxfordjournals.jhered.a110924. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan JT, Garwood M, Collins CM, Liu W, DelaBarre L, Adriany G, Andersen P, Merkle H, Goebel R, Smith MB, Ugurbil K. 7T vs. 4T: RF power, homogeneity, and signal-to-noise comparison in head images. Magn Reson Med. 2001;46(1):24–30. doi: 10.1002/mrm.1156. [DOI] [PubMed] [Google Scholar]
- 14.Van de Moortele PF, Akgun C, Adriany G, Moeller S, Ritter J, Collins CM, Smith MB, Vaughan JT, Ugurbil K. B(1) destructive interferences and spatial phase patterns at 7 T with a head transceiver array coil. Magn Reson Med. 2005;54(6):1503–1518. doi: 10.1002/mrm.20708. [DOI] [PubMed] [Google Scholar]
- 15.Fleysher R, Fleysher L, Liu S, Gonen O. TriTone: a radiofrequency field (B(1))-insensitive T(1) estimator for MRI at high magnetic fields. Magn Reson Imaging. 2008;26(6):781–789. doi: 10.1016/j.mri.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreis R, Slotboom J, Hofmann L, Boesch C. Integrated data acquisition and processing to determine metabolite contents, relaxation times, and macromolecule baseline in single examinations of individual subjects. Magn Reson Med. 2005;54(4):761–768. doi: 10.1002/mrm.20673. [DOI] [PubMed] [Google Scholar]
- 17.Gonen O, Arias-Mendoza F, Goelman G. 3D localized in vivo 1H spectroscopy of human brain by using a hybrid of 1D-Hadamard with 2D-chemical shift imaging. Magn Reson Med. 1997;37(5):644–650. doi: 10.1002/mrm.1910370503. [DOI] [PubMed] [Google Scholar]
- 18.Goelman G, Liu S, Fleysher R, Fleysher L, Grossman RI, Gonen O. Chemical-shift artifact reduction in Hadamard-encoded MR spectroscopic imaging at high (3T and 7T) magnetic fields. Magn Reson Med. 2007;58(1):167–173. doi: 10.1002/mrm.21251. [DOI] [PubMed] [Google Scholar]
- 19.Mareci T, Brooker H. Essential considerations for spectral localization using indirect gradient encoding of spatial information. J Magn Reson. 1991;92:229–246. [Google Scholar]
- 20.Goelman G, Liu S, Gonen O. Reducing voxel bleed in Hadamard-encoded MRI and MRS. Magn Reson Med. 2006;55(6):1460–1465. doi: 10.1002/mrm.20903. [DOI] [PubMed] [Google Scholar]
- 21.Weiss GH, Gupta RK, Ferretti JA, Becker ED. The choice of optimal parameters for measurement of spin-lattice relaxation times. I. mathematical formulation. J Magn Reson. 1980;37:369–379. [Google Scholar]
- 22.Posse S, Schuknecht B, Smith ME, van Zijl PC, Herschkowitz N, Moonen CT. Short echo time proton MR spectroscopic imaging. J Comput Assist Tomogr. 1993;17(1):1–14. doi: 10.1097/00004728-199301000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Marion D, Ikura M, Bax A. Improved solvent suppression in one- and two-dimensional NMR spectra by convolution of time domain data. J Magn Reson. 1989;84:425–430. [Google Scholar]
- 24.Du YP, Parker DL, Davis WL, Cao G. Reduction of partial-volume artifacts with zero-filled interpolation in three-dimensional MR angiography. J Magn Reson Imaging. 1994;4(5):733–741. doi: 10.1002/jmri.1880040517. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein MA, Fain SB, Riederer SJ. Effect of windowing and zero-filled reconstruction of MRI data on spatial resolution and acquisition strategy. J Magn Reson Imaging. 2001;14(3):270–280. doi: 10.1002/jmri.1183. [DOI] [PubMed] [Google Scholar]
- 26.Cheng HL, Wright GA. Rapid high-resolution T(1) mapping by variable flip angles: accurate and precise measurements in the presence of radiofrequency field inhomogeneity. Magn Reson Med. 2006;55(3):566–574. doi: 10.1002/mrm.20791. [DOI] [PubMed] [Google Scholar]
- 27.Tofts P. In: Quantitative MRI of the Brain measuring changes caused by Disease. Tofts P, editor. Chichester: John-Wiley & Sons limited; 2003. [Google Scholar]
- 28.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153(2):155–177. doi: 10.1006/jmre.2001.2340. [DOI] [PubMed] [Google Scholar]
- 29.Valette J, Park JY, Grohn O, Ugurbil K, Garwood M, Henry PG. Spectroscopic imaging with volume selection by unpaired adiabatic pi pulses: theory and application. J Magn Reson. 2007;189(1):1–12. doi: 10.1016/j.jmr.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 30.de Graaf RA, Nicolay K, Garwood M. Single-shot, B1-insensitive slice selection with a gradient-modulated adiabatic pulse, BISS-8. Magn Reson Med. 1996;35(5):652–657. doi: 10.1002/mrm.1910350505. [DOI] [PubMed] [Google Scholar]
- 31.Treier R, Steingoetter A, Fried M, Schwizer W, Boesiger P. Optimized and combined T1 and B1 mapping technique for fast and accurate T1 quantification in contrast-enhanced abdominal MRI. Magn Reson Med. 2007;57(3):568–576. doi: 10.1002/mrm.21177. [DOI] [PubMed] [Google Scholar]
- 32.Bottomley PA, Ouwerkerk R. The dual-angle method for fast, sensitive T1 measurement in vivo with low-angle adiabatic pulses. J Magn Reson Ser B. 1994;104:159–167. [Google Scholar]
- 33.Fleysher L, Fleysher R, Liu S, Zaaraoui W, Gonen O. Optimizing the precision-per-unit-time of quantitative MR metrics: Examples for T(1), T(2), and DTI. Magn Reson Med. 2007;57(2):380–387. doi: 10.1002/mrm.21144. [DOI] [PubMed] [Google Scholar]
- 34.Traber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging. 2004;19(5):537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- 35.Mlynarik V, Gruber S, Moser E. Proton T (1) and T (2) relaxation times of human brain metabolites at 3 Tesla. NMR Biomed. 2001;14(5):325–331. doi: 10.1002/nbm.713. [DOI] [PubMed] [Google Scholar]