Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1–E2F pathway activity.
The ribosomal protein S6 kinase (S6K) is involved in growth control, but less is known about its function in plants. The findings show here that S6 kinases in Arabidopsis affect chromosome stability and interact with the RBR1–E2F pathway to inhibit cell proliferation.
Keywords: cell proliferation, chromosome instability, E2F, retinoblastoma, S6 kinase
Abstract
The 40S ribosomal protein S6 kinase (S6K) is a conserved component of signalling pathways controlling growth in eukaryotes. To study S6K function in plants, we isolated single- and double-knockout mutations and RNA-interference (RNAi)-silencing lines in the linked Arabidopsis S6K1 and S6K2 genes. Hemizygous s6k1s6k2/++ mutant and S6K1 RNAi lines show high phenotypic instability with variation in size, increased trichome branching, produce non-viable pollen and high levels of aborted seeds. Analysis of their DNA content by flow cytometry, as well as chromosome counting using DAPI staining and fluorescence in situ hybridization, revealed an increase in ploidy and aneuploidy. In agreement with this data, we found that S6K1 associates with the Retinoblastoma-related 1 (RBR1)–E2FB complex and this is partly mediated by its N-terminal LVxCxE motif. Moreover, the S6K1–RBR1 association regulates RBR1 nuclear localization, as well as E2F-dependent expression of cell cycle genes. Arabidopsis cells grown under nutrient-limiting conditions require S6K for repression of cell proliferation. The data suggest a new function for plant S6K as a repressor of cell proliferation and required for maintenance of chromosome stability and ploidy levels.
Introduction
Cell growth and proliferation is tightly integrated with available nutrients, cellular energy levels, developmental signals and stress factors through the Target of rapamicin (TOR) kinase signalling pathway (Wullschleger et al, 2006; Diaz-Troya et al, 2008; Ma and Blenis, 2009). One downstream effector of TOR is the ribosomal protein S6 kinase (S6K), a master regulator of growth that tunes the translational capacity of cells through the phosphorylation of ribosomal protein S6 (RPS6) (Meyuhas, 2008). Knockout mutations in the S6K genes in mice and Drosophila indeed resulted in drastic reduction of cell sizes (Montagne et al, 1999; Pende et al, 2004), but surprisingly in mice this was not paralleled with a compromised protein synthesis (Pende et al, 2004). Similarly, mutations of the S6K phosphorylation sites on RPS6 affected cell size, but not protein synthesis, suggesting that S6K regulates cell size checkpoint independent of translation (Pende et al, 2004; Ruvinsky et al, 2005). The inhibition of TOR kinase through specific drugs also identified both cell cycle and cell growth regulation downstream of TOR (Feldman et al, 2009; Thoreen et al, 2009).
How TOR can regulate cell size was first identified in fission yeast, where it was shown that TOR restrains the entry into mitosis by regulating the inhibitory phosphorylation of Cdc2 by Wee1 kinase (Petersen and Nurse, 2007; Hartmuth and Petersen, 2009). The involvement of TOR and S6K in cell size checkpoint seems to be conserved. In Drosophila cells, the activation of TOR signalling can delay the entry into mitosis and thus increase cell size (Wu et al, 2007), whereas silencing of S6K1 resulted in a reduced cell size through increasing the rate cells enter into mitosis (Bettencourt-Dias et al, 2004). In budding yeast, the homologue of S6K, Sch9 was also shown to regulate cell size, as well as nutrient signalling and ageing (Jorgensen et al, 2004; Urban et al, 2007; Steffen et al, 2008). Sch9 also has important functions to reprogram gene expression between growth and stress conditions (Roosen et al, 2005; Pascual-Ahuir and Proft, 2007; Smets et al, 2008).
S6Ks are members of the AGC family (PKA, PKG, PKC) of serine/threonine kinases and are also present in plants (Bögre et al, 2003). In Arabidopsis, there are two S6K genes, S6K1 and S6K2, having highly similar sequence and arranged in tandem duplication on chromosome 3. It was shown that Arabidopsis S6K2 is able to carry out conserved signalling functions, because it could be activated by the growth hormone, insulin, in a TOR-dependent manner, when introduced into human cells (Turck et al, 1998, 2004). Correspondingly, as in other organisms, the Arabidopsis S6K functions in a complex with RAPTOR, it is activated by PDK1 and can phosphorylate RPS6 (Mahfouz et al, 2006; Otterhag et al, 2006). RPS6 phosphorylation in plants also leads to the selective recruitment of ribosomal mRNAs to polysomes and thus regulates the switch of translational capacity between growth promoting and stress conditions (Turck et al, 2004). The growth hormones, auxin and cytokinin enhance RPS6 phosphotylation in cell culture (Turck et al, 2004), whereas stress factors, such as heat and oxidative stress rapidly block it (Williams et al, 2003). In agreement with reduced RPS6 phosphorylation upon stress, osmotic stress was shown to inactivate the Arabidopsis S6K1 that was dependent on RAPTOR levels, and S6K1 over-expression resulted in an increased sensitivity to osmotic stress (Mahfouz et al, 2006).
Plant growth is the result of cell proliferation within meristems and cell enlargement outside the proliferative zone. The tor mutant in Arabidopsis has an arrested embryo development at a stage when cell elongation takes place, indicating that AtTOR might not be required for early proliferative but for cell elongation-driven growth (Menand et al, 2002). Cell proliferation in the tor mutant is also unaffected during endosperm development, but there are defects in cytokinesis, suggesting that TOR might have mitotic functions also in plants (Menand et al, 2002). S6K could also regulate elongation growth, as suggested by the over-expression of a lily S6K (LS6K1) gene in Arabidopsis that resulted in decreased cell elongation in flower organs (Tzeng et al, 2009). AtTOR expression was correlated with active cell proliferation and growth (Menand et al, 2002). S6K1 is also expressed in meristematic regions both in Arabidopsis (Zhang et al, 1994) and in lilly (Tzeng et al, 2009), as well as in cells that are actively elongating within the root (Zhang et al, 1994).
The transition from cell proliferation to cell differentiation is regulated by the Retinoblastoma-related 1 (RBR1)–E2F pathway in higher plants (Magyar, 2008), although in the algae Chlamydomonas all components of the RB pathway, including RB, E2F and DP, control cell size (Fang et al, 2006). The retinoblastoma protein is known to inhibit the transcription factor activity of E2Fs by masking their transactivation domains and by globally repressing promoters through the recruitment of chromatin remodelling enzymes (Magyar, 2008; van den Heuvel and Dyson, 2008). In Arabidopsis, RBR1 is the single homologue of Retinoblastoma, and from the six identified E2F-related genes, three are able to form complexes with RBR1, but they differently regulate the expression of genes involved in cell proliferation: E2FC is a transcriptional repressor (del Pozo et al, 2006), whereas E2FA and E2FB are activators (de Veylder et al, 2002; Magyar et al, 2005). The RBR1-bound or -free forms of E2F complexes constitute a regulatory network for the control of cell proliferation and exit to differentiation.
In this work, we studied the function of Arabidopsis S6K1 and S6K2 by analysing single mutants, a hemizygous s6k1s6k2/++ double mutant and s6k1(XVE-RNA interference (RNAi))-silenced plants. Homozygous s6k1s6k2 mutants were not recovered, probably because of the requirement of both S6K1 and S6K2 in male gametophyte development. Plants with reduced S6K1 and S6K2 levels had increased chromosome number and became aneuploid. Arabidopsis S6K1 can interact with the RBR1–E2F pathway and inhibit cell proliferation. In agreement with this notion, we found that depletion of RBR1, ectopic over-expression of E2FA together with DPA and elevation of E2FA expression under its own promoter control also lead to increased ploidy, similarly to depletion of S6K1 and S6K2 in the s6k1s6k2/++ and s6k1(XVE-RNAi) plants. Our data suggest that, in plants, S6K negatively regulates cell division as part of a signalling pathway connected to the RBR1–E2F transcriptional switch and its deregulation can influence the incident rate of polyploidization and lead to chromosome instability.
Results
s6k1s6k2/++ mutants show size variation, reduced fertility and increase in ploidy level
The S6K1 (ATPK6, At3g08730) and S6K2 (ATPK19, At3g08720) genes of Arabidopsis are organized in a tandem direct repeat arrangement in chromosome 3. On the basis of NASCARRAY data, S6K1 is highly expressed in mature pollen (five times more than in microspores) and in sperm cells, whereas S6K2 shows limited expression in pollen, but higher expression (it remains to 25% of S6K1) in sperm cells (nucleus in G1). Furthermore, S6K1 is induced by UV, oxidative and genotoxic stresses specifically in shoot, and co-expressed with genes involved in circadian rhythm (e.g. CCA1; Supplementary Table I). S6K2 is expressed in developing seeds and induced by ABA and salt treatment in the root. S6K2 is strongly co-expressed with genes involved in stress responsive regulation of plant growth, such as BONZAI1 (Yang and Hua, 2004). Although with some overlap, AtTOR, S6K1 and S6K2 have distinct domains of expression in the root. AtTOR highest expression is in the meristem, whereas the S6K1 transcript accumulates in the elongation zone, and the S6K2 in the differentiation zone, indicating that S6K1 and S6K2 might regulate the exit from proliferative growth (Supplementary Figure 1A) (Winter et al, 2007).
Despite their distinct expression patterns, single insertion mutants for S6K1 (s6k1-1, Salk_148694) or S6K2 (s6k2-1, Salk_128183 and s6k2-2, Salk_13334) are still viable and have no readily discernable phenotypes (Supplementary Figure 2). However, the homozygous s6k2-2 mutant did show seed abortion (∼30%) (Supplementary Figure 2F), and the screening of homozygous s6k2-1 individual plants for DNA content by flow cytometry and for abnormal trichome branching allowed us to identify a line with a mixture of both diploid and aneuploid DNA content (Supplementary Figure 2G), suggesting that reduced S6K2 expression can lead to chromosome instability.
We have also identified a double s6k1s6k2 mutation in our library of Arabidopsis T-DNA insertion lines by systematic sequencing of T-DNA insert boundaries (Szabados et al, 2002). In the mutant line A199L, we found a T-DNA integration event that affected both S6K1 and S6K2 by generating a deletion of ∼2 kb. Sequencing of PCR fragments spanning the T-DNA insert borders and genomic DNA junctions showed that the deletion removed coding sequences of the C-terminal part of S6K1 (last exon), the N-terminal part of S6K2 (first two exons) and the intergenic region between the two S6K genes (Supplementary Figure 3A and C). Southern blotting with T-DNA right and left border probes indicated the presence of three tandem T-DNA copies within the S6K1 and S6K2 locus (Supplementary Figure 3B, D, E and F).
Genotyping the M3 offspring of line A199L with T-DNA and gene-specific primers revealed the absence of homozygous knockout plants. Upon self-pollination, heterozygous A199L plants yielded about 1:1 segregation of hygromycin resistant and sensitive offspring. To assess the source of distorted segregation, we performed reciprocal crosses between wild-type (Col-0) and 29 hemizygous s6k1s6k2/++ plants and determined the segregation of the T-DNA-encoded hygromycin resistance marker and the presence of T-DNA sequences in the adjacent S6K1 and S6K2 loci by PCR. Fertilization of wild type with s6k1s6k2/++ pollen produced very few seeds (274 seeds/29 crosses, 130 non-viable) and revealed a dramatic reduction of recovery of male-derived double-knockout allele in the viable progeny (3.47%; 5 HygR:139 HygS; Supplementary Table II). In the reciprocal cross, fertilization of s6k1s6k2/++ plants with wild-type pollen provided normal seed yield and an expected 1:1 segregation of wild-type and mutant S6K alleles (465 HygR:439 HygS). According to this result, we found very little viable pollen in s6k1s6k2/++ anthers (Figure 1B). Although pollen amounts might not be limiting for fertility, this seemed not to be the case, as we found 23 to 91% of aborted seeds in siliques in different individual plants (Figure 1C).
Figure 1.
Developmental abnormalities in s6k1s6k2/++ plants. (A) Flowers of WT and s6k1s6k2/++ plants. Scale bar, 1 mm. (B) Anthers from s6k1s6k2/++ and WT plants stained with Alexander dye. Arrows point to pollen. Note the different scale bars, both representing 50 μm. (C) Dissected siliques from WT and s6k1s6k2/++ plants. Arrows point to aborted seeds. Scale bar, 1 mm. (D) Chromosome number in cells of WT leaf epidermis cell, WT pollen meiocyte, s6k1s6k2/++ leaf epidermis cell, s6k1s6k2/++ pollen meiocyte and WT trichomes. Upper row: DAPI staining of chromocentres (CCs) showing a diploid cell (1), meiotic diploid cell in metaphase I (2), tetraploid somatic cell (3), meiotic tetraploid cell in anaphase I (4) and polytenic trichome cell (5). Middle row: FISH with a centromere-specific probe. Lower row: FISH with a specific probe for chromosome 1 pericentromeric regions. Small bars show pairs of chrom.1. A similar increase in chromosome numbers was also found in petal epidermal cells of both s6k1s6k2/++ and s6k1(XVE-RNAi) line 3 plants. Scale bars, 2.5 μm. (E) Flow cytometry measurements of DNA content from flower cells of wild-type (WT-2n), tetraploid wild-type (WT-4n) and s6k1s6k2/++ seedlings. (F) DNA content measurements of leaf no. 1 and 2, 15 DAG. (G) Scanning electron micrographs and corresponding drawings from epidermal leaf surface (third leaf at day 30) of WT and s6k1s6k2/++ plants. Scale bars, 50 μm. (H) Distribution of leaf epidermal cell sizes of s6k1s6k2/++ and WT plants. Variation in cell sizes within classes were analysed from multiple leaf samples and areas (n=299 cells for s6k1s6k2/++ mutants and n=265 cells for WT).
To found the cause for aborted male gametophytes, we analysed meiosis in anthers of wild-type and s6k1s6k2/++ plants by DAPI staining and determined the number of chromocentres (CCs; heterochromatin aggregates that correspond to centromeres in mitotic stages; Fransz et al, 2002) in pollen meiocytes. Surprisingly, in s6k1s6k2/++ mutants, we found an increased (mostly doubled) number of CCs in these cells (Figure 1D). Wild-type male meiocytes have five bivalent chromosome pairs in metaphase I, rather than 10 as found in the s6k1s6k2/++ anthers, which is specifically obvious during late anaphase I, when the chromosome pairs segregate (Figure 1D). This suggests that s6k1s6k2/++ plants possessed an increased chromosome number already before meiosis. Labelling of the same male meiocytes with centromere- and chromosome 1-specific probes by fluorescence in situ hybridization (FISH) further confirmed this result. We found 10 pairs of segregating chromosomes and 2 pairs of chromosome 1 during late anaphase I of s6k1s6k2/++ pollen meiocytes (Figure 1D), but were unable to detect any chromosome segregation abnormalities during meiosis I and II (Supplementary Figure 4). To clarify whether this ploidy increase also affected somatic cells, we performed similar analysis in epidermal cells of leaves and petals. DAPI staining and FISH analysis revealed a similar increase in ploidy levels in the s6k1s6k2/++ mutant when compared with wild type (Figures 1D, 2E and F). Furthermore, flow cytometry measurement of the DNA content of proliferating leaves 1 and 2 at 15 days after germination (DAG) from s6k1s6k2/++ mutant compared with wild-type diploid (WT-2n) and tetraploid (WT-4n) plants revealed an increase comparable with the DNA content of leaves 1 and 2 of a tetraploid plant (Figure 1F). Similarly, fully expanded leaves and flowers of s6k1s6k2/++ plants revealed an increase in DNA content when compared with WT-2n (Figure 1E; Supplementary Figure 5). Although, we observed morphological phenotypes typical of tetraploid Arabidopsis plants, such as large flowers, increased pollen grain size (Figure 1A and B), the extent of phenotypic instability and size variation found in the s6k1s6k2/++ mutant is higher than expected from a stable tetraploid (Koornneef et al, 2003; Yu et al, 2009) (Supplementary Figure 6A and B). Moreover, some of the phenotypes identified, such as narrow leaves, were reminiscent of aneuploid swarm from a diploid to tetraploid cross (Henry et al, 2005). To clarify which of the described phenotypes are due to ploidy changes, we analysed the offspring of the s6k1s6k2/++ mutant grown in the absence of the T-DNA-derived hygromicin marker. These plants were then analysed for the presence of T-DNA in S6K1 and S6K2 loci, their trichome branching (Supplementary Figure 6C) and ploidy level (Supplementary Figure 7). As expected from their higher ploidy levels, s6k1s6k2/++ offspring had trichomes with more than three branches. However, s6k1s6k2/++ plants showed the highest trichome branching (six branches), which was not detected in the plants with wild-type S6K1S6K2 loci. Flow cytometry analysis of flowers, compared with WT-2n and WT-4n, indicated the existence of aneuploidy (Supplementary Figure 7). Moreover, we identified an individual plant (s6k1s6k2/++ #8) with a mixture of both aneuploid (close to triploid) and tetraploid DNA content within the same flower, an indication of a chimera tissue, which confirms results showing that aneuploid plants can develop largely over-branched trichomes even if not proportional to the increase in their DNA content (Yu et al, 2009). Moreover, the aneuploidy measured by flow cytometry did not necessarily correlate with the severity of morphological changes in the s6k1s6k2/++ plants.
Figure 2.
Reducing S6K transcripts in s6k1(XVE-RNAi) plants also leads to ploidy changes. S6K1 (A) and S6K2 (B) transcript levels in s6k1s6k2/++ mutants and the corresponding WT control determined by quantitative RT–PCR. S6K1 (C) and S6K2 (D) transcript levels in s6k1(XVE-RNAi) seedlings (line #3 and #6) and the corresponding WT control in control (−) and 5 μM β-estradiol-treated conditions (+), determined by quantitative RT–PCR. All samples were collected at day 30 after sowing. (E) Percentage of nuclei from leaf epidermal cells having <10 (%<10), 10, and >10 CCs (%>10) from WT (n=59), 5 μM β-estradiol-treated (+) s6k1(XVE-RNAi) line 3 (n=50) plants and s6k1s6k2/++ (n=106) mutants. (F) Percentage of CCs in nuclei from petal epidermal cells in WT (n=166), in 5 μM β-estradiol-treated (+) s6k1(XVE-RNAi) line 3 plants (n=67) and in s6k1s6k2/++ mutants (n=415). (G) Scanning electron micrographs of trichomes from WT and s6k1s6k2/++ leaves. Scale bar, 200 μm. (H) Percentage of trichomes with 3,4,5 and 6 branches in XVE-RNAi empty vector control without β-estradiol (control, −) (n=227), and treated with 5 μM β-estradiol (treated, +) (n=108), s6k1(XVE-RNAi) line 3 control (−) (n=227) and β-estradiol treated (+) (n=292), WT plants (n=279) and s6k1s6k2/++ mutants (n=326). (I) Summary of flow cytometry measurements of DNA content from cells in the first and second leaves at 15 DAG of XVE-RNAi empty vector control and s6k1(XVE-RNAi) constructs.
It is known that increased ploidy is accompanied with increased cell size (Galbraith et al, 1991). Therefore, we determined the cell size distribution of epidermal pavement cells from fully developed leaf 3 of wild-type and growth-arrested s6k1s6k2/++ plants, and found a higher proportion of small cells in the leaf epidermis of s6k1s6k2/++ mutants (Figure 1G and H). However, the total cell number in the leaf has not increased in the s6k1s6k2/++ leaves compared with wild type (Supplementary Figure 6D), indicating that cell elongation rather than cell proliferation was impaired. Cell elongation is accompanied by endoreduplication in Arabidopsis leaves, and our flow cytometry analysis of s6k1s6k2/++ expanding leaves 15 DAG (Figure 1F) showed that the 2C peak was missing, as expected from the higher chromosome number; however, the reduction in 16C cells, when compared with WT-4n leaves, suggests a decrease in endoreduplication.
Silencing of S6K1 and S6K2 also results in increased ploidy
We could not determine the exact moment, during the mutant isolation procedure, when the s6k1s6k2/++ line A199L became polyploid. To independently evaluate whether S6K1 and S6K2 genes are linked to the observed change in ploidy, we have generated RNAi lines, in which both S6K genes are silenced through the expression of a β-estradiol-inducible S6K1 RNAi construct, s6k1(XVE-RNAi) (Zuo et al, 2000).
By quantitative RT–PCR, we found that in hemizygous s6k1s6k2/++ mutant, the transcript levels were 53–84% for S6K1 and 40–70% for S6K2 compared with wild type (Figure 2A and B). This result was confirmed by northern analysis of S6K transcript levels in s6k1s6k2/++ mutants (Supplementary Figure 3G and H). Interestingly, we detected, in one individual, a truncated S6K1 mRNA, smaller than the expected S6K1 transcript before the T-DNA insert site, which probably corresponds to the N-terminal region of S6K1. We find unlikely, but cannot rule out, that this low level of short RNA could generate an S6K protein fragment that exerts a dominant negative effect over the wild-type function.
In s6k1(XVE-RNAi), lines 3 and 6 grown in the absence of β-estradiol inducer, the S6K1 and S6K2 transcript levels were already lower than in wild-type plants because of leaky basal expression of the RNAi construct. However, upon β-estradiol treatment, the S6K1 and S6K2 transcript levels were further reduced in line 3 to 25% of S6K1 and to 37% of S6K2 compared with wild type. The degree of silencing was slightly less in line 6 with reduction of S6K1 to 50% and S6K2 to 57% of wild-type levels (Figure 2C and D). We have not recovered viable silenced lines with a complete loss of S6K levels. Similarly to s6k1s6k2/++ mutants, the s6k1(XVE-RNAi) lines also showed large flowers and high level of aborted seeds (Supplementary Figure 6E and I).
To determine how S6K silencing affects the DNA content, s6k1(XVE-RNAi) line 3 was compared with s6k1s6k2/++ mutant and wild type by performing flow cytometry analysis of developed leaves and flowers (Figure 2I; Supplementary Figure 5). The 2C and 4C peaks of wild-type and XVE-RNAi control plants were replaced by 4C and 8C peaks both in the s6k1s6k2/++ and s6k1(XVE-RNAi) flower samples, an organ that normally remains diploid and does not show endoreduplication cycles. The DNA content also increased in leaves of s6k1s6k2/++ and s6k1(XVE-RNAi) plants compared with wild-type and XVE-RNAi control plants. Therefore, we also determined the number of chromosomes by counting CCs in the s6k1(XVE-RNAi) line 3 compared with the line transformed with empty vector, and found a similar doubling of chromosome number as in the s6k1s6k2/++ mutants both in epidermal cells of leaves and petals (Figure 2E and F). The doubled chromosome number is indicative for the occurrence of endomitosis (chromosome duplication without cell division), rather than a change in the endoreduplication cycle, where the repeated rounds of DNA replication result in unseparated sister chromatids, such as seen in wild-type trichomes, a cell type with endoreduplication (Figure 1D).
Leaves from s6k1s6k2/++ and s6k1(XVE-RNAi) plants revealed an increase in trichome branching, a phenotype known to correlate with ploidy changes (Hulskamp, 2004). Correspondingly, trichomes developed on leaves of s6k1s6k2/++ plants had increased number of branches compared with the wild type (Figure 2G). The majority of trichomes on wild-type plants had three branches (n=279) and only around 10% had four, whereas about half of the trichomes on s6k1s6k2/++ (n=326) leaves developed four branches (Figure 2H). The previously screened s6k1(XVE-RNAi) lines (# 3, 6 and also line #2) had up to 70% of trichomes with four branches after β-estradiol induction of s6k1(XVE-RNAi) when compared with the empty vector control lines. Later, we found that increased trichome branching was already present without inducer, shown for the s6k1(XVE-RNAi) line 3 (n=292), but upon β-estradiol treatment, the proportion of trichomes with five branches has substantially increased (Figure 2H). This increase in DNA content found in the s6k1s6k2/++ mutant, three independent s6k1(XVE-RNAi) lines and in one offspring of s6k2-1, suggests that S6K could be required for maintenance of stable chromosome numbers.
As chromosome instability accumulates in each generation, we analysed T1 primary transformants expressing the s6k1(XVE-RNAi) construct in a wild-type (Col-0; n=20) or s6k2-2 mutant (n=27) background (Supplementary Figure 8A). We determined trichome branching (a sensitive visible measure for increased ploidy and aneuploidy) and DNA content by flow cytometry of leaves before and after transfer to β-estradiol-containing medium. We have confirmed the silencing of S6K1 in selected lines using Q-RT–PCR (Supplementary Figure 8B and C). As in our previous analysis, several independent lines developed over-branched trichomes already in the absence of β-estradiol, although after 3 days of induction, the percentage of four-branched trichomes increased. Importantly, some newly developed leaves have reverted to normal and had the majority of trichomes with three branches, indicating that ploidy change can occur in somatic sectors (Supplementary Figure 8A). We found that one T1 s6k1(XVE-RNAi)-silenced line in the s6k2-2 mutant background showed increase in its DNA content, which was confirmed by mixing isolated nuclei from this line and WT-2n (Supplementary Figure 8D). We cannot clarify whether this increase is due to (1) the expression, shortly after flower transformation, of the s6k1(XVE-RNAi) construct in undeveloped flower buds still undergoing meiosis, leading to unreduced gametes that could be fertilized by normal haploid gametes; (2) the s6k2-2 background mutant and (3) a conjunction of the two possibilities.
A change in ploidy is not reversible, but nevertheless we attempted to do genetic complementation of the hemizygous s6k1s6k2/++ mutant by expressing both S6K1 and S6K2 under the control of their own promoter. Analysis of T1 lines revealed increased trichome branching, large flowers and growth variability similar to the original s6k1s6k2/++ mutant (data not shown).
In summary, the reduction of both S6K1 and S6K2 levels in the s6k1s6k2/++ mutant and s6k1(XVE-RNAi) lines resulted in higher incidence of ploidy changes when compared with single mutants, indicating that S6K1 and S6K2 are not fully equivalent. However, we cannot exclude that mutations in one S6K gene could lead to some compensation from the other available S6K.
S6K negatively regulates cell proliferation
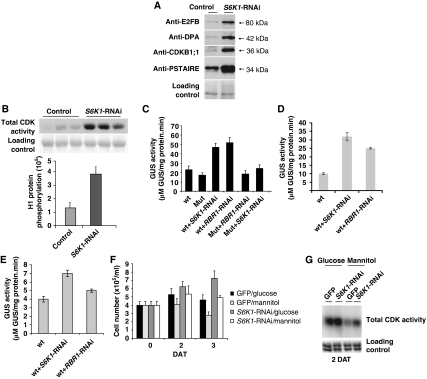
To investigate whether deregulated cell proliferation could be the reason for the change in ploidy levels in the s6k1s6k2/++ mutant and s6k1(XVE-RNAi) lines, we used a protoplast transformation system to transiently silence S6K1 and S6K2 levels and evaluate how this short-term silencing might affect cell division, the expression of cell cycle regulators and cyclin-dependent kinase activity. Protoplasts prepared from cultured Arabidopsis cells were transformed with high efficiency (typically 40–60% as determined by transformation with a GFP expression construct), and were growing and dividing 1–3 days after transformation (DAT) (Supplementary Figure 9D). To determine the efficiency of RNAi constructs in reducing the levels of expression of specific genes, the RNAi constructs were co-transformed with epitope-tagged S6K1 cDNA expression constructs followed by detection of the target proteins with western blotting. As both S6K1 RNAi and S6K2 RNAi constructs could effectively silence S6K1, in these assays, we refer to total S6K levels (Supplementary Figure 9A and C). When S6K expression was silenced in protoplasts for 3 DAT, we found a remarkable increase in the levels of cell cycle regulatory proteins, including E2FB, DPA, CDKB1;1 and CDKA (Figure 3A). Therefore, we analysed in detail how S6K would specifically regulate the cell cycle.
Figure 3.
Cell cycle proteins and CDK activities are upregulated in cells with silenced S6K levels. (A) Detection of E2FB, DPA, CDKB1;1, PSTAIRE-containing CDKA protein levels in mock-transformed (control) Arabidopsis cells, and in cells transformed with the S6K1-RNAi construct, 3 days after transformation (DAT). (B) Total CDK activities of purified CDKs by binding to p13suc1 Sepharose beads from mock (control) and S6K1-RNAi-transformed Arabidopsis cells 3 DAT (triplicates). Upper panel: autoradiogram showing histone H1 phosphorylation by CDKs; lower panel: quantification of the same phosphorylation signal. (C) The CDKB1;1 WT promoter (wt) and CDKB1;1 mutant (Mut) promoter, where the consensus E2F-binding site was mutated (Boudolf et al, 2004) were fused to the GUS-reporter gene and GUS activity measured in mock (control) cells and in cells transformed with S6K1-RNAi and RBR1-RNAi constructs in early stationary stage at 3 DAT. (D) Determination of activity of the RNAR2 WT promoter (wt) fused to the GUS-reporter gene in mock-transformed (control) cells and in cells transformed with S6K1-RNAi and RBR1-RNAi constructs in early stationary stage at 3 DAT. (E) Activity of the CycD3;1 WT promoter (wt) fused to the GUS-reporter gene was determined in similar cells as described in (D). (F) Cell numbers counted in cultures GFP-transformed and cultured in the presence of glucose (GFP/glucose) or mannitol (GFP/mannitol); or transformed with S6K1-RNAi and cultured in presence of glucose (S6K1-RNAi/glucose) or mannitol (S6K1-RNAi/mannitol). Cell numbers were counted at day of transformation (0), and at 2 and 3 DAT. (G) Detection of total CDK activities from the transformed cultures described in (F) at 2 DAT.
It is known that high levels of CDK activity characterize actively proliferating cells. To determine whether CDK activity was increased in S6K-silenced cells, we purified total CDK through binding to p13suc1 protein beads and monitored CDK activity by phosphorylation of histone H1. We found a three-fold increase in CDK activity in the S6K-silenced cells compared with cells mock transformed (control) (Figure 3B).
CDKB1;1 is a mitosis-specific gene, expressed only in dividing cells (Porceddu et al, 2001; Sorrell et al, 2001) and its promoter contains an E2F-binding element that is regulated by E2FA (Boudolf et al, 2004). We transformed Arabidopsis protoplasts with a CDKB1;1 promoter-fused GUS-reporter gene (wt) and with a similar GUS construct that carried a mutated version of CDKB1;1 promoter (Mut) from which the E2F-binding site was removed. Three DAT, the activity of the CDKB1;1 promoter was largely elevated in cells with silenced S6K levels. As expected, a similar activation of the CDKB1;1 promoter in response to silencing of the transcriptional repressor protein RBR1 was observed with an RBR1-RNAi construct that was previously confirmed to almost eliminate the co-transformed RBR1-GFP construct, and deplete the endogenous RBR1 protein, proportionally to the transformation efficiency (Supplementary Figure 9B and C). Silencing RBR1 and S6K did not significantly alter the activity of the mutant CDKB1;1 promoter, indicating that both RBR1 and S6K repress the CDKB1;1 promoter activity through the E2F promoter element (Figure 3C).
To determine whether other known E2F-target genes are deregulated when S6K is silenced, we also tested promoter-GUS constructs for the G1-specific CycD3;1 and the S phase-specific RNR2 genes (Figure 3D and E) (Horvath et al, 2006). S6K silencing significantly elevated the activity of both promoters 3 DAT, an effect that was more pronounced than RBR1 silencing.
As the function of S6K in nutrient signalling pathways has been well characterized in yeast and animal systems, we decided to investigate whether Arabidopsis S6K regulates cell proliferation in response to nutrient availability. Cell suspension protoplasts regenerate cell wall and begin to divide between 1 and 2 days in culture in the presence of the hormone, auxin, and sufficient fermentable glucose, but halt proliferation when glucose is replaced by mannitol, a non-fermentable carbon source (Figure 3F). We transformed protoplasts with GFP (control) or with s6k1(XVE-RNAi) constructs and determined cell number 2 and 3 DAT, both in glucose- and mannitol-containing cultures. We found an increase in cell number in glucose-containing media after 2 days, and this was higher when S6K was silenced, especially at 3 DAT. In mannitol-containing media, the increase in cell number occurred only when S6K levels were reduced (Figure 3F). We determined total CDK activity at 2 DAT, and found it to be elevated in the glucose-containing media when S6K was silenced, although this increase was somewhat lower than described in Figure 3B. In mannitol-containing media, total CDK activity decreased in the control samples (GFP), but was still elevated in S6K-silenced cells (Figure 3G). Our results show that under nutrient-limiting conditions, such as glucose starvation, S6K negatively affects cell proliferation.
S6K1 associates with RBR1 and E2FB in vivo
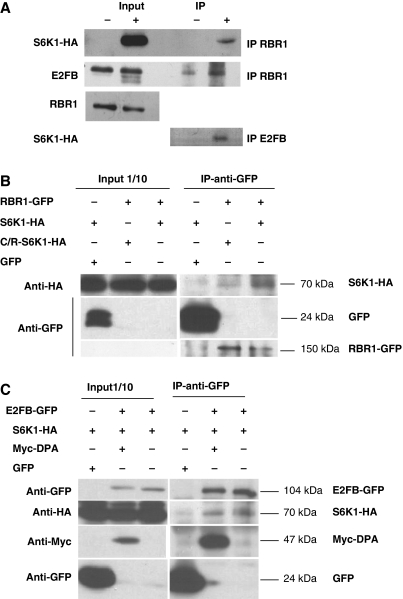
The results obtained so far indicated that Arabidopsis S6Ks are negative regulators of cell proliferation, and affect the activity or expression of several cell cycle regulators that constitute the RBR1–E2F pathway. To gain more insight into S6K regulation of this pathway, we investigated whether S6K1 associates with RBR1. Owing to the lack of a specific antibody against the Arabidopsis S6K1 protein, we generated a stably transformed Arabidopsis cell suspension line expressing a haemagglutinin (HA) epitope-tagged S6K1 (S6K1-HA) under the control of β-estradiol-inducible promoter. Over-expression of S6K1-HA had no detectable effect on the proliferation of these cells cultured in the presence of auxin (data not shown). We were able to induce S6K1-HA expression by β-estradiol, and detected S6K1-HA in the induced sample after immunoprecipitation with an RBR1-specific antibody (Figure 4A). As a positive control, we also tested for the presence of endogenous E2FB in the RBR1 immunocomplex. Surprisingly, we found more E2FB associated with RBR1 when S6K1 was over-expressed (Figure 4A). As E2FB is known to directly interact with RBR1 (Magyar et al, 2005), we used an E2FB-specific antibody to immunoprecipitate E2FB from the same input samples as for RBR1 and could readily detect S6K1-HA in the immunocomplex when its expression was induced (Figure 4A).
Figure 4.
S6K1 associates with RBR1 and E2FB. (A) Co-immunoprecipitation of S6K1-HA, E2FB and RBR1. Detection of S6K1-HA in both 1/10th of the input used for immunoprecipitation (IP) of RBR1 and in the RBR1-immunoprecipitate in control cells (−) or in cells where S6K1-HA expression was induced (+) with 5μM of β-estradiol. Detection of E2FB in the same samples of IP-RBR1 (second row). Detection of RBR1 in the input samples used for IP-RBR1 (third row). Detection of S6K1-HA (fourth row) in the E2FB immunoprecipitate (IP-E2FB) from the same input sample as for RBR1-IP. (B) Determination of co-immunoprecipitation of RBR1-GFP with S6K1-HA or C/R-S6K1-HA. Left panel: detection of 1/10th of the input used for IP. Right panel: detection of S6K1-HA, C/R-S6K1-HA, GFP and RBR1-GFP in the RBR1-GFP immunoprecipitate. (C) Determination of co-immunoprecipitation of E2FB-GFP with S6K1-HA or Myc-DPA. Left panel: detection of 1/10th of the input used for IP. Right panel: detection of E2FB-GFP, S6K1-HA, Myc-DPA and GFP in the E2FB-GFP immunoprecipitate.
Viral oncoproteins can bind to the pocket domain of RBR1 through their LxCxE motifs. Detailed analysis of this motif on RBR1 cellular targets showed that it is required for interaction with some of those proteins (Singh et al, 2005). Interestingly, both S6K1 and S6K2 contain an LxCxE-like motif (91/101LVxCxE96/106) amino-terminal to the kinase domain. To test whether this motif on S6K1 can mediate the interaction with RBR1, we mutated the cysteine 94 (C94) to arginine (R) within the motif (C/R-S6K1-HA) and tested the interaction of wild-type and mutated forms after co-expression with a GFP-tagged RBR1 in Arabidopsis protoplasts. RBR1-GFP levels were low and, therefore, could only be detected in the immunoprecipitate, but not in the input. Nevertheless, using the anti-GFP antibody, we could readily immunoprecipitate S6K1-HA (Figure 4B), confirming our previous results with anti-RBR1 antibody (Figure 4A). Although, the mutated C/R-S6K1-HA form was expressed to a similar level as the wild-type S6K1-HA form in the input sample, a significantly lower amount was found to associate with RBR1-GFP in the GFP immunoprecipitate, indicating that the LVxCxE motif is partially required for the RBR1–S6K1 interaction (Figure 4B). Though free GFP was expressed to a very high level and was effectively immunoprecipitated with the GFP antibody, no association of S6K1-HA was found with free GFP, showing the high specificity of this immunoclomplex assay (Figure 4B). In a similar experiment, we tested the S6K1 association with E2FB and confirmed our previous results indicating their association, possibly in an indirect way, mediated by RBR1, though we cannot rule out that S6K might also directly interact with E2FB (Figure 4C). As control, we also tested the association of E2FB with its dimerization partner DPA. As expected, we found that E2FB interacts with DPA, and this interaction was stronger than binding of S6K1 to E2FB, suggesting that the association of S6K1 to the RBR1, E2FB, DPA complex is non-stoichiometric and might be transient (Figure 4C). These findings further suggest that S6K1 modulates cell proliferation by associating with RBR1–E2F complex.
S6K1 regulates nuclear localization of RBR1
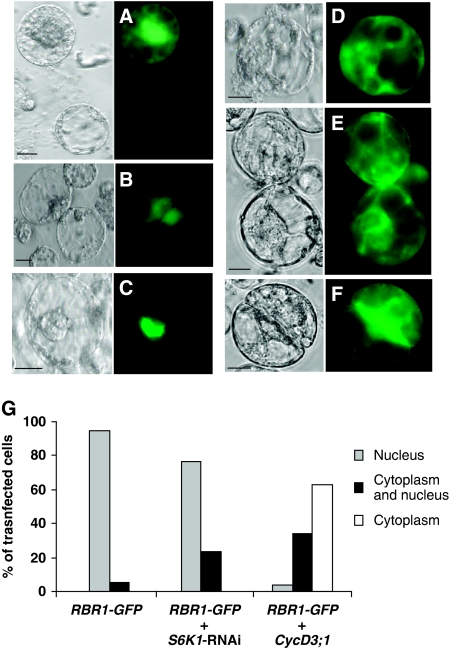
To investigate the function of the S6K1–RBR1 interaction, we analysed the localization of these regulators when expressed in Arabidopsis cells (Figure 5). Confirming the previous results (Mahfouz et al, 2006), the S6K1-GFP fusion protein was localized both in cytoplasm and nucleus (Figure 5B), whereas the RBR1-GFP fusion protein accumulated in the nucleus (95% nuclear, 5% cytoplasmic, n=705; Figure 5C). However, 36 h after co-transformation of RBR1-GFP with S6K1-RNAi, we found that RBR1-GFP was relocated from the nucleus to the cytoplasm in a significant proportion of cells (23% cytoplasmic, n=766; Figure 5D)—an effect that could not be simply attributed to the dispersion of signal during mitosis, as the mitotic index in an asynchronous protoplast culture is ∼3%. RBR1 is phosphorylated and inactivated by the Cyclin D3;1-CDKA complex (Nakagami et al, 2002). Therefore, we tested the effect of CycD3;1 over-expression on RBR1-GFP localization and found that it also resulted in re-localization of RBR1 to the cytoplasm (59% only cytoplasmic, n=448; Figure 5E and F). The quantification of percentage of cells showing the RBR1-GFP signal in different cellular compartments is summarized in Figure 5G. As expected, protoplasts maintained in culture for 2 days in the presence of auxin were still able to divide and their division frequency was increased by over-expression of CycD3;1 (Figure 5F). These results showed that CycD3;1 and S6K1 oppositely regulate the nuclear localization of RBR1. S6K1 could restrain cell proliferation by associating with and promoting the nuclear localization of RBR1.
Figure 5.
S6K regulates RBR1 cellular localization. (A) Cells transformed with GFP; (B) S6K1-GFP; (C) RBR1-GFP; (D) RBR1-GFP construct was co-transformed with S6K1-RNAi construct and cultured for 36 h. (E) RBR1-GFP construct was co-transformed with CycD3;1 and cultured for 36 h. (F) Same as in (E) cultured for 72 h. Scale bars, 10 μm. (G) Quantification of the GFP localization signal shown in (C–E). RBR1-GFP (n=705 cells); RBR1-GFP+ S6K1-RNAi (n=766 cells); RBR1-GFP+CycD3;1 (n=448 cells) (s.d. values were below 0.04 and are not visible in the graph).
Depletion of RBR1 and over-expression of E2FA under its own promoter or by the 35S viral promoter together with DPA lead to increased chromosome number
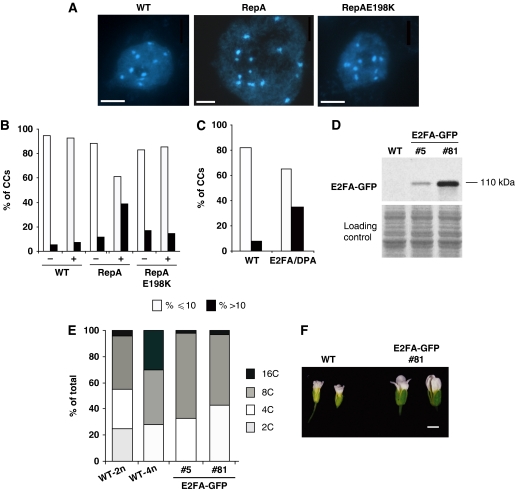
The above-described results suggested that, in Arabidopsis, S6K negatively regulates cell proliferation. We found that S6K1 can associate with RBR1 and contribute to the maintenance of RBR1 in the nucleus. Therefore, we hypothesized that the increased chromosome number observed both in the s6k1s6k2/++ mutant and s6k1(XVE-RNAi) plants could be due to compromised RBR1 function and/or increased levels of active E2Fs. Desvoyes et al (2006) had shown that inactivation of RBR1, through over-expression of RBR1-binding viral protein RepA, would release E2Fs and lead to excessive proliferation in the leaf epidermal layer, over-branched trichomes and increased DNA content. To evaluate whether the increase in DNA content found by Desvoyes et al (2006) was due to endomitosis rather than endoreduplication as suggested, we counted CC numbers in RepA over-expressors, as well as in RepA mutant (RepAE198K) plants, where the RBR1/E2F interaction is not disrupted. We found that induction of RepA expression by dexamethasone, but not of its mutant form (RepAE198K), dramatically increased the nuclear size and the number of CCs (Figure 6A and B). This phenotype could be a consequence of elevated levels of free activator type E2Fs, such as E2FA and E2FB. Therefore, we analysed available E2FA/DPA transgenic lines that showed increased cell proliferation in differentiated tissues, as well as extra rounds of endoreduplication (de Veylder et al, 2002). Similarly to RBR1 depletion, increased levels of E2FA/DPA resulted in higher number of CCs in leaf epidermal cells when compared with wild-type plants of the same developmental stage (Figure 6C), suggesting that the increased ploidy of E2FA/DPA over-expressing plants could be due, at least in part, to elevated chromosome number rather than endoreduplication. To investigate this further, we generated E2FA-GFP lines, where E2FA expression is under the control of its own promoter and selected lines with different levels of E2FA-GFP expression (Figure 6D). We found that, although in these lines E2FA expression is lower than in 35S-promoter-driven E2FA over-expressors, two lines out of 15 independent transformants showed increased flower size, similar to the s6k1s6k2/++ mutant and s6k1(XVE-RNAi) plants (Figure 6F). Correspondingly, flow cytometry analysis of the first two leaves of 15 DAG seedlings revealed the absence of a 2C peak, an indication of ploidy increase in these lines. However, the 16C endoreduplication peak was reduced when compared with true WT-4n (Figure 6E). Similarly to s6k1s6k2/++ mutant and s6k1(XVE-RNAi) plants, we observed high phenotypic variability in the offspring of E2FA-GFP line 8, and the existence of aborted seeds, although not as pronounced as in s6k1s6k2/++ (Supplementary Figure 6J). Taken together, these results pheno-copied our observations with the s6k1s6k2/++ mutant and s6k1(XVE-RNAi) plants and further confirmed our hypothesis that S6K1 might inhibit cell proliferation and its downregulation can increase the incidence of poly- and aneuploidy through its interaction and regulation of the RBR1–E2F pathway.
Figure 6.
Depletion of RBR1 and over-expression of E2FA/DPA leads to increase in chromocentre (CC) number and polyploidy. (A) DAPI staining of CCs from leaf epidermal cells, showing a diploid cell wild type (WT) (Col-0), a polyploid cell in RepA plants and a diploid cell in mutated RepA198K plants. Scale bars, 1.6 μm. (B) Percentage of nuclei from leaf epidermal cells having <10 or 10 (%⩽10) and >10 CCs (%>10) in WT control (−) (n=372) and in 10 μM dexamethasone-treated (+) (n=379) WT plants; in RepA control (−) (n=305) and in 10 μM dexamethasone-treated (+) (n=407) plants and in RepAE198K control (−) (n=293) and in 10 μM dexamethasone-treated (+) (n=404) plants. (C) Percentage of nuclei from leaf epidermal cells having <10 or 10 (%⩽10) and >10 CCs (%>10) in WT (n=473) and in E2FA/DPA over-expressing lines (n=922). (D) Detection of E2FA-GFP protein level in flowers from the T1 generation of two independent transgenic lines expressing E2FA-GFP under the control of its native promoter. (E) Flow cytometry measurement of the first leaf pair from 15 DAG WT-2n, WT-4n and two T2-independent homozygous E2FA-GFP seedlings from lines described in (D). (F) E2FA-GFP highly expressing line #81 develops larger flowers than WT plants. Scale bar, 1 mm.
Discussion
The TOR – S6K signalling module is a conserved integrator of signals that promote growth, such as nutrient and growth factors, versus signals that restrict growth, such as starvation and stresses (Reiling and Sabatini, 2006; Diaz-Troya et al, 2008). In this work, we have studied the function of the Arabidopsis S6K1 and S6K2 genes and provide multiple lines of evidence that S6Ks in Arabidopsis negatively regulate cell proliferation and that the S6K1 and S6K2 gene dose can influence the incidence of change in ploidy level in plants.
S6K restrains cell division during cell size control and in response to stress
In this work, we have found that, similarly to other eukaryotes, including budding yeast (Jorgensen et al, 2004), fission yeast (Petersen and Nurse, 2007), Drosophila (Montagne et al, 1999; Bettencourt-Dias et al, 2004; Guertin et al, 2006; Wu et al, 2007) and human cells (Fingar et al, 2002, 2004), the Arabidopsis S6K negatively regulates mitosis. CDKB1, a plant-specific regulator for the onset of mitosis, is expressed only at late G2 and M phases and is known to be regulated by the E2F family of transcription factors (Boudolf et al, 2004). Using promoter-reporter gene constructs, we showed that CDKB1 expression is repressed by S6K through the E2F-binding element within the CDKB1 promoter. Furthermore, our analysis of growth-impaired s6k1s6k2/++ mutant plants revealed a reduction in the size of leaf epidermal cells when compared with wild type, indicating an inhibition of cell expansion. Although these plants have an increased, close to 4n, chromosome number, the cells are still smaller than the 2n WT Col-0. Moreover, we found that endoreduplication, a process that is developmentally initiated as cells exit meristematic cell proliferation, but continue to grow through elongation, was reduced. Interestingly, downregulation of S6K did not result in increased cell number, in opposition to over-expression of CycD3;1 (Dewitte et al, 2003) or E2FB (Magyar et al, 2005), where reduced cell elongation correlated with elevated cell number. Thus, S6K might specifically be required for elongation-driven growth.
The TOR-PDK-AGC kinase module in budding (Sobko, 2006) and fission yeasts also regulates stress resistance and is involved in arresting the cell cycle under stress conditions (Matsuo et al, 2003). In Arabidopsis, the expression of S6Ks is induced by a variety of stresses (Mizoguchi et al, 1995). However, over-expression of S6K1 leads to hypersensitivity to osmotic stress, but the mechanism is not known (Mahfouz et al, 2006). In cultured Arabidopsis cells, S6K activity increased when cells entered the stationary phase and became maximal upon stimulation with fresh culture medium, an indication that nutrient availability could affect S6K function (Turck et al, 2004). We have found that the inhibitory function of Arabidopsis S6Ks in cell proliferation becomes more apparent in protoplasts cultured for 2–3 days and close to entering the stationary phase, as well as when cells are starved for fermentable glucose.
Plants with reduced S6K level undergo polyploidization
The s6k1s6k2/++ mutant is gametophytic lethal on the male side, but we could not fully clarify the requirement for S6K in this developmental mechanism. S6K1 is highly expressed in mature pollen and both S6K1 and S6K2 are expressed in sperm cells. We have shown that S6K regulates CDK and E2F activities. Interestingly, CDKA, as well as an E2FA target, FBL17, are required for male sperm division (Nowack et al, 2007; Gusti et al, 2009). We found that the Arabidopsis s6k1s6k2/++ hemizygous mutants and the s6k1(XVE-RNAi) plants, that have reduced S6K levels, display an increased ploidy level. This was revealed by the presence of multi-branched trichomes, a cellular phenotype that strictly correlate with increased DNA content. Cells can accumulate higher DNA amount because of two different processes: endoreduplication and endomitosis. Although endoreduplication results in polytenic chromosomes, endomitosis leads to an increase in chromosome number (Edgar and Orr-Weaver, 2001). DAPI staining of CCs and FISH analysis of centromers in epidermal cells of leaves and petals of s6k1s6k2/++ and s6k1(XVE-RNAi) lines showed that the elevated ploidy level was due to an increase in chromosome number rather than endoreduplication. Tetraploid Arabidopsis lines are relatively stable (Yu et al, 2009), but show variable degree of multivalent chromosome pairing during meiosis (Santos et al, 2003). Although the existence of triploid and aneuploid chromosome setup is well tolerated in plants, these individuals display variation in leaf sizes, flower morphology, have increased flower size and reduced fertility (Comai, 2005; Henry et al, 2005). The s6k1s6k2/++ and s6k1(XVE-RNAi) lines also have these characteristic abnormalities typical of aneuploids (Supplementary Figure 6). Indeed, our comparative analysis of s6k1s6k2/++ segregants to diploid and WT-4n plants (Col-0) revealed that much of their DNA content deviated from 4n, an indication of aneuploidy. In some plants, we have found a mixture of ploidy states possibly originating from chimera sectors, suggesting that aneuploidy can arise in somatic tissues. We also isolated a T1 s6k2-2/s6k1(XVE-RNAi) line that became triploid, indicating the existence of diploid and haploid gamete fusion. Compared with other mutants (Ravi et al, 2008; d'Erfurth et al, 2008, 2009; Kohler et al, 2010) the frequency of this meiotic abnormality must be low, as we were unable to find meiotic defects in the s6k1s6k2/++ plants.
It is known that a number of ribosomal proteins and specifically S6 are required, in a dose-dependent manner, for suppression of cell proliferation in yeast (Bernstein et al, 2007), and suppression of tumour growth in Drosophila (Watson et al, 1992; Stewart and Denell, 1993; Volarevic et al, 2000), zebrafish (MacInnes et al, 2008), mouse (Panic et al, 2006) and human cells (Sulic et al, 2005). In fish and mammals, haplo-insufficiency, conferred by the heterozygous state of mutations affecting ribosomal protein genes, appears to depend on the activation of a p53-dependent checkpoint mechanism that regulates the translational machinery to prevent aberrant division. The inhibition of translation during mitosis (Kurabe et al, 2009) is required to prevent defective cytokinesis leading to binucleate cells (Wilker et al, 2007). Interestingly, inhibition of TOR kinase by rapamycin rescues these cytokinetic abnormalities. However, in Arabidopsis, reduced S6K levels seem to lead to chromosome instability and increased ploidy levels, although we do not know whether these effects relate to S6K function in ribosomal protein phosphorylation or S6K function to regulate the cell cycle. A mitotic function for S6K was also suggested in mammalian cells, where S6K2 activity is highest in G2 and M phases (Boyer et al, 2008; Xu et al, 2009), S6K2 is centrosome located (Rossi et al, 2007) and inhibition of S6K activity can lead to chromosome mal-segregation (Bonatti et al, 1998) and polyploidization (Ma et al, 2009).
S6K negatively regulates cell proliferation through the RBR1–E2F pathway
The mechanism by which TOR and S6K connect to cell cycle regulatory functions appears to be distinct in different organisms. In fission yeast, TOR regulates the inhibitory tyrosine phopshorylation of Cdc2, and thereby the onset of mitosis (Petersen and Nurse, 2007). In budding yeast, ribosome biogenesis directly regulates the passage through Start through Whi5, a yeast functional equivalent of the human tumour suppressor, RB (Bernstein et al, 2007). Our results show that in Arabidopsis, S6K regulates cell proliferation through the plant RB homologue, RBR1. We have multiple evidence to support this statement: (i) RBR1 interacts with S6K1 and this association partly depends on an N-terminal LxCxE-like motif (LVxCxE), which is required for interaction between RB and several of its partners (Singh et al, 2005); (ii) S6K1 promotes the nuclear localization of RBR1 and repression of CDKB1;1 gene expression through the E2F element within its promoter, as well as the repression of G1-specific CycD3;1 and S phase-specific RNR2 promoter activity and (iii) S6K silencing leads to elevated protein levels of a number of cell cycle regulators, including CDKA, CDKB, E2FB, DPA, as well as higher CDK activity.
In studies involving plants, the loss of Rb function, or increased E2F activity, was connected to an increase in endoreduplication, a developmentally regulated process in which proliferative mitotic cycles are replaced by repeated S phases without mitosis, whereas in animal cells deregulation of RB function affected ploidy levels through chromosome instability (de Veylder et al, 2002; Hernando et al, 2004; Park et al, 2005; Desvoyes et al, 2006; Sozzani et al, 2006; Lageix et al, 2007; Srinivasan et al, 2007). To clarify whether changes in RBR1 and E2F levels could lead to increased ploidy by elevated chromosome number (endomitosis), we determined the number of CCs in leaf epidermal cells of RBR1-depleted (RepA; Desvoyes, 2006) and E2FA/DPA over-expressing plants (de Veylder et al, 2002). We found that these cells had higher number of CCs and were, therefore, polyploid. Interestingly, Lageix et al (2007) reported a similar increase of CCs in leaf cells of plants expressing a nanovirus-encoded protein (Clink) that is able to bind RBR1 and repress its function. Furthermore, under nutrient depletion such as sucrose starvation, RBR1 was shown to promote G1 phase cell cycle arrest (Hirano et al, 2008), and our results confirmed that silencing of S6K, and thus release of the S6K–RBR1 block, can allow glucose-starved cells to proliferate. Taken together, these results indicate that the ploidy changes found in s6k1s6k2/++ and s6k1(XVE-RNAi) plants are, at least partly, due to loss of RBR1 function and/or increased E2F activity, and further confirm the importance of negative regulation of S6K in the maintenance of cell cycle control.
Plants produce unreduced gametes at an average rate of ∼0.5%, which can lead to polyploidization (Otto, 2007). The main route of polyploidy formation is through unreduced gametes and unstable triploid progeny that can somehow overcome the triploid block (Kohler et al, 2010), possibly because poly- and aneuploidy are well tolerated in plants (Comai, 2005; Doyle et al, 2008). Our findings suggest that the S6K1, S6K2, RBR1 and E2FA gene dose, expression levels and functions can affect the rate of polyploidization, whereas S6Ks are also part of signalling pathways that respond to nutrients and stresses, and regulate cell proliferation. These results suggest the possibility that S6K, RBR1 and E2FA may contribute to the evolutionary adaptation of plants through influencing changes in ploidy.
Materials and methods
Plant work
In the supplementary data section, we provide a detailed explanation of the plasmid constructs made, the transgenic lines generated and the approaches used for mutant isolation and analysis. Supplementary Table III lists the oligonucleotides used in this work. The description of the methods developed for analysis of siliques, pollen, leaves and petals from wild-type, s6k1s6k2/++ mutants and s6k1(XVE-RNAi) plants, as well as the evaluation of plant size, is also given in the Supplementary data section. Unless otherwise stated, all analysis of plant phenotypes has been carried out by comparison with diploid Col-0, referred as wild type or WT-2n.
RNA extraction and quantitative RT–PCR
RNA was prepared from 30 days old plants grown under sterile conditions using the RNAeasy plant RNA extraction kit from Qiagen (Germany). In total, 1 μg of RNA was used for cDNA synthesis using the Superscript III Reverse transcription kit accordingly to the manufacturer's protocol (Invitrogen). Quantitative PCR reactions were performed using an Applied Biosystems 7900HT real-time PCR system. Further details are given in the Supplementary data.
Cytological analysis, FISH and flow cytometry analysis
Flower buds and developed leaves were used for cytological analysis and FISH. For flow cytometry analysis, leaves 1 and 2 from 15-DAG seedlings and flowers from similar development stages were used. Details for these experiments are available in the Supplementary data.
Co-immunoprecipitation of S6K1, E2FB and RBR1
Co-immunoprecipitation (co-IP) was performed accordingly to Magyar et al (2005). Site-directed mutagenesis of the S6K1 coding sequence is given in the Supplementary data. Antibodies used in these experiments were as follows: anti-HA (Santa Cruz Biotechnology); anti-RBR1 (Horvath et al, 2006), anti-E2FB (Magyar et al, 2005) and anti-GFP (Roche). In each co-IP from transformed protoplasts, four samples were pooled together and the following western analysis has been carried out as described (Magyar et al, 2005).
Transformation of Arabidopsis protoplasts and analysis of cell cycle proteins
Protoplast transformation was performed as described (Magyar et al, 2005), a detailed description of constructs and antibodies tested is given in the Supplementary data.
Protein kinase assay
To determine total CDK kinase activity, total protein extract from transformed protoplasts (see above) was affinity purified by binding to p13suc1–Sepharose beads and used for protein kinase assays with 1 μg of Histone H1 as substrate (Magyar et al, 2005). The phosphorylated products were resolved in a 10% SDS–PAGE gel and the phosphorylation signal was detected using a Typhoon 9410 phosphorimager and quantified by the ImageQuant software (GE Healthcare, Sweden).
Supplementary Material
Acknowledgments
The β-estradiol-inducible expression system was made available for us by N-H Chua and its GatewayR modification by B Ulker, who are gratefully acknowledged. The three-way GatewayR vectors were kindly provided by Ben Scheres. The RepA lines were provided by Crisanto Gutierrez, the E2FA/DPA line by Lieven de Veylder. The tetraploid wild-type Arabidopsis (Col-0, referred as WT-4n) was kindly provided by Luca Comai. This work was supported by the Framework 5 EU project GVE and by BBSRC at RH, as well as by the SFB635 and AFGN grants from the Deutsche Forschungsgemeinschaft for CK at the MPIZ, Cologne. RH was funded by the Fundação para a Ciência e Tecnologia (SFRH/BPD/7164/2001).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ (2007) Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol Biol Cell 18: 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, Cooper M, Jones D, Frenz L, Glover DM (2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432: 980–987 [DOI] [PubMed] [Google Scholar]
- Bögre L, Ökrész L, Henriques R, Anthony RG (2003) Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci 8: 424–431 [DOI] [PubMed] [Google Scholar]
- Bonatti S, Simili M, Galli A, Bagnato P, Pigullo S, Schiestl RH, Abbondandolo A (1998) Inhibition of the Mr 70 000 S6 kinase pathway by rapamycin results in chromosome malsegregation in yeast and mammalian cells. Chromosoma 107: 498–506 [DOI] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Acosta JAT, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer D, Quintanilla R, Lee-Fruman KK (2008) Regulation of catalytic activity of S6 kinase 2 during cell cycle. Mol Cell Biochem 307: 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846 [DOI] [PubMed] [Google Scholar]
- de Veylder L, Beeckman T, Beemster G, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inze D (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the Ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R (2009) Turning meiosis into mitosis. PLoS Biol 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Simon M, Jenczewski E, Mercier R (2008) Mutations in AtPS1 (Arabidopsis thaliana parallel spindle 1) lead to the production of diploid pollen grains. PLoS Genet 4: e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua N, Gutierrez C (2006) Cell type-specific role of the Retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL (2008) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4: 851–865 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL (2001) Endoreplication cell cycles: more for less. Cell 105: 297–306 [DOI] [PubMed] [Google Scholar]
- Fang SC, de los Reyes C, Umen JG (2006) Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet 2: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM (2009) Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J (2004) mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol 24: 200–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16: 1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, de Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D, Harkins K, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Guntur KVP, Bell GW, Thoreen CC, Sabatini DM (2006) Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol 16: 958–970 [DOI] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P (2009) The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4: e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth S, Petersen J (2009) Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J Cell Sci 122: 1737–1746 [DOI] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Young K, Watson B, Wu H, Comai L (2005) Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C (2004) Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802 [DOI] [PubMed] [Google Scholar]
- Hirano H, Harashima H, Shinmyo A, Sekine M (2008) Arabidopsis Retinoblastoma-related protein 1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Mol Biol 66: 259–275 [DOI] [PubMed] [Google Scholar]
- Horvath B, Magyar Z, Zhang Y, Hamburger A, Bako L, Visser R, Bachem C, Bogre L (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp M (2004) Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol 5: 471–480 [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Mittelsten Scheid O, Erilova A (2010) The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet 26: 142–148 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Fransz P, de Jong H (2003) Cytogenetic tools for Arabidopsis thaliana. Chromosome Res 11: 183–194 [DOI] [PubMed] [Google Scholar]
- Kurabe N, Arai S, Nishijima A, Kubota N, Suizu F, Mori M, Kurokawa J, Kondo-Miyazaki M, Ide T, Murakami K, Miyake K, Ueki K, Koga H, Yatomi Y, Tashiro F, Noguchi M, Kadowaki T, Miyazaki T (2009) The death effector domain-containing DEDD supports S6K1 activity via preventing Cdk1-dependent inhibitory phosphorylation. J Biol Chem 284: 5050–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S, Catrice O, Deragon J-M, Gronenborn B, Pelissier T, Ramirez BC (2007) The nanovirus-encoded clink protein affects plant cell cycle regulation through interaction with the Retinoblastoma-related protein. J Virol 81: 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Yu H, Lin D, Sun Y, Liu L, Liu Y, Dai B, Chen W, Cao J (2009) S6K1 is involved in polyploidization through its phosphorylation at Thr421/Ser424. J Cell Physiol 219: 31–44 [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA (2008) Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci USA 105: 10408–10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z (2008) Keeping the balance between proliferation and differentiation by the E2F transcriptional regulatory network is central to plant growth and development. In Plant Growth Signaling, Bogre L and Beemster GT (eds). Berlin Heidelberg: Springer Vol. 10 [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bako L, Inze D, Bogre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DPS (2006) Arabidopsis Target of rapamycin interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Kubo Y, Watanabe Y, Yamamoto M (2003) Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J 22: 3073–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O (2008) Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol 268: 1–37 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K (1995) Two genes that encode ribosomal-protein S6 kinase homologs are induced by cold or salinity stress in Arabidopsis thaliana. FEBS Lett 358: 199–204 [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G (1999) Drosophila S6 kinase: a regulator of cell size. Science 285: 2126–2129 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of Retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Shirzadi R, Dissmeyer N, Dolf A, Endl E, Grini PE, Schnittger A (2007) Bypassing genomic imprinting allows seed development. Nature 447: 312–315 [DOI] [PubMed] [Google Scholar]
- Otterhag L, Gustavsson N, Alsterfjord M, Pical C, Lehrach H, Gobom J, Sommarin M (2006) Arabidopsis PDK1: identification of sites important for activity and downstream phosphorylation of S6 kinase. Biochimie 88: 11–21 [DOI] [PubMed] [Google Scholar]
- Otto SP (2007) The evolutionary consequences of polyploidy. Cell 131: 452–462 [DOI] [PubMed] [Google Scholar]
- Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, Grabusic K, Volarevic S (2006) Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol 26: 8880–8891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Ahn J, Kim Y, Kim S, Kim J, Kim W, Pai H (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Proft M (2007) The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. EMBO J 26: 3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G (2004) S6K1-/-/S6K2-/- mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol 24: 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Nurse P (2007) TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol 9: 1263–1272 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Stals H, Reichheld J-P, Segers G, De Veylder L, de Pinho Barroco R, Casteels P, Van Montagu M, Inze D, Mironov V (2001) A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J Biol Chem 276: 36354–36360 [DOI] [PubMed] [Google Scholar]
- Ravi M, Marimuthu MP, Siddiqi I (2008) Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Reiling JH, Sabatini DM (2006) Stress and mTORture signaling. Oncogene 25: 6373–6383 [DOI] [PubMed] [Google Scholar]
- Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, Cameroni E, Thevelein JM, De Virgilio C, De Moor B, Winderickx J (2005) PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol 55: 862–880 [DOI] [PubMed] [Google Scholar]
- Rossi R, Pester JM, McDowell M, Soza S, Montecucco A, Lee-Fruman KK (2007) Identification of S6K2 as a centrosome-located kinase. FEBS Lett 581: 4058–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O (2005) Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19: 2199–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JL, Alfaro D, Sanchez-Moran E, Armstrong SJ, Franklin FC, Jones GH (2003) Partial diploidization of meiosis in autotetraploid Arabidopsis thaliana. Genetics 165: 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Krajewski M, Mikolajka A, Holak TA (2005) Molecular determinants for the complex formation between the Retinoblastoma protein and LXCXE sequences. J Biol Chem 280: 37868–37876 [DOI] [PubMed] [Google Scholar]
- Smets B, De Snijder P, Engelen K, Joossens E, Ghillebert R, Thevissen K, Marchal K, Winderickx J (2008) Genome-wide expression analysis reveals TORC1-dependent and -independent functions of Sch9. FEMS Yeast Res 8: 1276–1288 [DOI] [PubMed] [Google Scholar]
- Sobko A (2006) Systems biology of AGC kinases in Fungi. Sci STKE 2006: re9. [DOI] [PubMed] [Google Scholar]
- Sorrell DA, Menges M, Healy JM, Deveaux Y, Amano C, Su Y, Nakagami H, Shinmyo A, Doonan JH, Sekine M, Murray JA (2001) Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar bright yellow-2 cells. Plant Physiol 126: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W, Knudsen ES (2007) RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J Biol Chem 282: 23867–23877 [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M (2008) Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell 133: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MJ, Denell R (1993) Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol Cell Biol 13: 2524–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S (2005) Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev 19: 3070–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Kovács I, Oberschall A Ábrahám E, Kerekes I, Zsigmond L, Nagy R, Alvarado M, Krasovskaja I, Gál M, Berente A, Rédei GP, Haim AB, Koncz C (2002) Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J 32: 233–242 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Kozma SC, Thomas G, Nagy F (1998) A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70s6k function in vivo. Mol Cell Biol 18: 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng TY, Kong LR, Chen CH, Shaw CC, Yang CH (2009) Overexpression of the lily p70(s6k) gene in Arabidopsis affects elongation of flower organs and indicates TOR-dependent regulation of AP3, PI and SUP translation. Plant Cell Physiol 50: 1695–1709 [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G (2000) Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288: 2045–2047 [DOI] [PubMed] [Google Scholar]
- Watson KL, Konrad KD, Woods DF, Bryant PJ (1992) Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA 89: 11302–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, Yaffe MB (2007) 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature 446: 329–332 [DOI] [PubMed] [Google Scholar]
- Williams AJ, Werner-Fraczek J, Chang IF, Bailey-Serres J (2003) Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol 132: 2086–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An ‘Electronic Fluorescent Pictograph' browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Cully M, Andersen D, Leevers SJ (2007) Insulin delays the progression of Drosophila cells through G2/M by activating the dTOR/dRaptor complex. EMBO J 26: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xu XY, Zhang Z, Su WH, Zhang Y, Yu YQ, Li YX, Zong ZH, Yu BZ (2009) Characterization of p70 S6 kinase 1 in early development of mouse embryos. Dev Dyn 238: 3025–3034 [DOI] [PubMed] [Google Scholar]
- Yang S, Hua J (2004) A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Haage K, Streit VE, Gierl A, Ruiz RA (2009) A large number of tetraploid Arabidopsis thaliana lines, generated by a rapid strategy, reveal high stability of neo-tetraploids during consecutive generations. Theor Appl Genet 118: 1107–1119 [DOI] [PubMed] [Google Scholar]
- Zhang SH, Lawton MA, Hunter T, Lamb CJ (1994) atpk1, a novel ribosomal protein kinase gene from Arabidopsis. I. Isolation, characterization, and expression. J Biol Chem 269: 17586–17592 [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.