Abstract
The replication of rotavirus is a complex process that is orchestrated by an exquisite interplay between the rotavirus non-structural and structural proteins. Subsequent to particle entry and genome transcription, the non-structural proteins coordinate and regulate viral mRNA translation and the formation of electron-dense viroplasms that serve as exclusive compartments for genome replication, genome encapsidation and capsid assembly. In addition, non-structural proteins are involved in antagonizing the antiviral host response and in subverting important cellular processes to enable successful virus replication. Although far from complete, new structural studies, together with functional studies, provide substantial insight into how the non-structural proteins coordinate rotavirus replication. This brief review highlights our current knowledge of the structure-function relationships of the rotavirus non-structural proteins, well as fascinating questions that remain to be understood.
1. Introduction
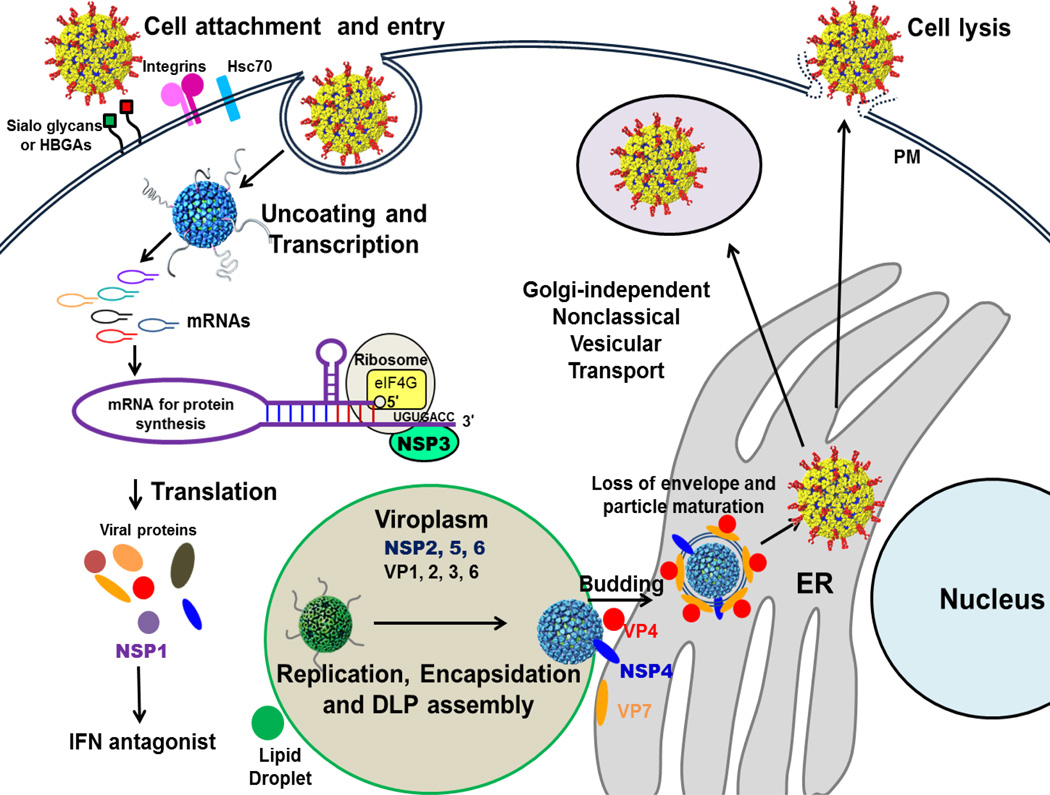
The rotavirus genome consists of 11 double-stranded (ds)RNA segments, and each segment encodes one protein with the exception of segment 11, which in some rotavirus strains codes for 2 proteins, NSP5 and NSP6 [1,2]. Of these proteins encoded by the viral genome, six are structural (VPs) and the remaining are non-structural (NSPs). The complex architecture of the mature rotavirus particle, with three concentric capsid layers, elegantly integrates the necessary elements required for cell entry and endogenous transcription of viral mRNAs [3,4]. Removal of the outer layer during the process of cell entry triggers the endogenous transcriptase activity of the resulting double-layered particles [1]. The capped transcripts are released through aqueous channels at the 5-fold axes of these intact particles [5]. Once these initial transcripts are translated, the rotavirus NSPs then coordinate various stages of genome replication and viral assembly by adapting and modifying the cellular machinery, which leads to productive release of mature particles through cell lysis. A brief overview of the rotavirus replication cycle is provided in Figure 1
Figure 1. The rotavirus replication cycle.
The rotavirus triple layered particles (TLPs) first attach to sialo-glycans [78] or histo-blood group antigens (HBGAs) [79] on the host cell surface, followed by interactions with other cellular receptors, including integrins and Hsc70 [80]. Virus is then internalized by receptor-mediated endocytosis. Removal of the outer layer, triggered by the low calcium of the endosome, results in the release of transcriptionally active double-layered particles (DLPs) into the cytoplasm. The DLPs start rounds of mRNA transcription and these mRNAs are used to translate viral proteins. Once enough viral proteins are made, the RNA genome is replicated and packaged into newly made DLPs in specialized structures called viroplasms, which require lipid droplet components for formation [44]. The newly made DLPs bind to NSP4, which serves as an endoplasmic reticulum (ER) receptor, and bud into the ER. During this process, transient enveloped particles are seen in the ER. Removal of the transient membrane and assembly of the outer capsid proteins VP4 and VP7 result in the maturation of the TLPs. The progeny virions are released through cell lysis. In polarized epithelial cells, particles are released by a non-classical vesicular transport mechanism. IFN: interferon; PM: plasma membrane.
Although the functional roles served by NSP2, NSP3, NSP4, and NSP5 during rotavirus replication are relatively well characterized and are discussed below, the roles of NSP1 and NSP6 remain less clear. Due to the lack of structural studies on NSP1 and NSP6, only a brief description of their function is included in this review. Early studies indicated that NSP1, which exhibits significant sequence variation among rotavirus strains, is not essential for rotavirus replication in cultured cells; however, more recent studies implicate NSP1 in host-range restriction, in countering innate host antiviral response, and in suppressing induction of apoptosis during early stages of infection, to promote viral growth [6–9] (See article by Angel, Franco and Greenberg). Unlike the other rotavirus NSPs, NSP6 is not encoded by all rotavirus strains. For those strains that do express NSP6, it is translated from an open reading frame out-of-phase with that of NSP5 in segment 11 [2]. This 12-kDa protein has a high turnover rate and is degraded within 2 hours of synthesis [10]. NSP6 has shown sequence independent nucleic acid binding, with similar affinities for ssRNA and dsRNA [10]. Although some studies have suggested that this protein localizes to the viroplasm [10], the precise role of NSP6 in rotavirus replication remains to be characterized.
2. Structural studies of rotavirus NSPs
Rotaviruses comprise seven distinct groups (A to G) based on the seroreactivity of the inner capsid protein VP6 (See article by Matthijnssens and van Ranst). Groups A–C are found in both humans and animals, whereas rotaviruses of groups D–G have been found only in animals to date [1]. Structural aspects of several group A rotavirus NSPs have been analyzed using X-ray crystallography and/or electron cryomicroscopy (cryo-EM) techniques. We briefly review the current status of these studies in the context of their function in the rotavirus replication cycle.
NSP3
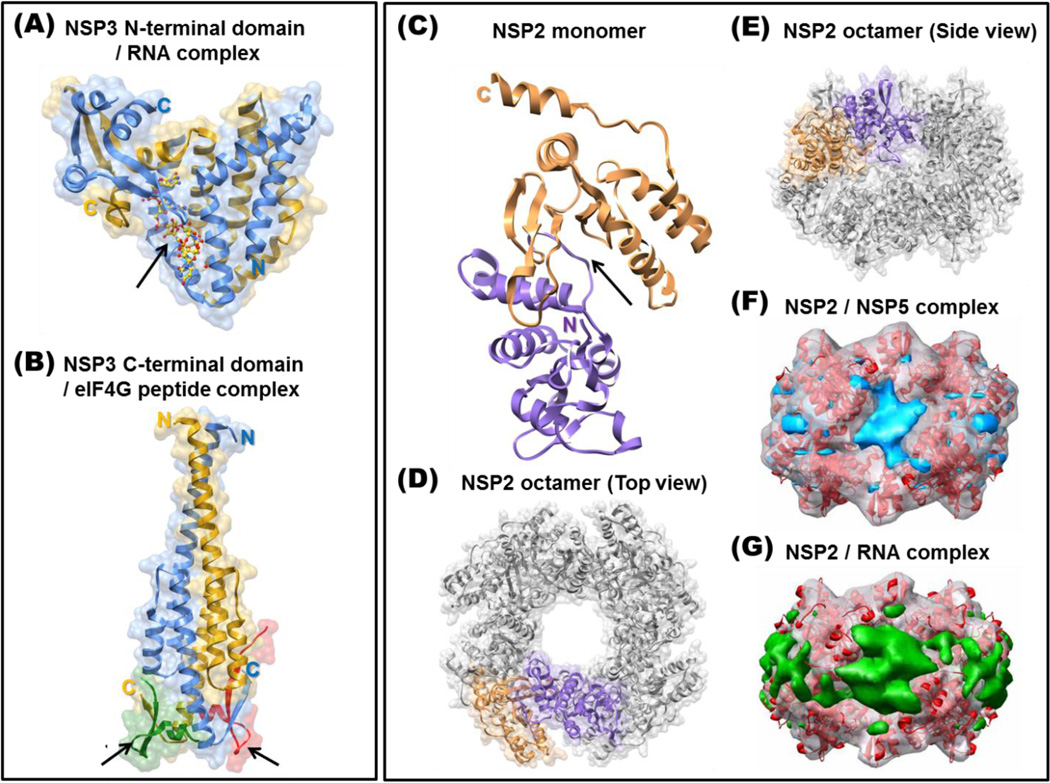
NSP3, a 36.4 kDa basic protein encoded by segment 7 of rotavirus strain SA11, is proposed to facilitate translation of the rotaviral mRNA transcripts and to suppress host protein synthesis through antagonism of the poly A binding protein (PABP) [11–15] (Also see article by Lopez and Arias). Rotaviruses rely on the host translation machinery to produce the viral proteins encoded by their genome. In host cells, only capped and polyadenylated mRNA are efficiently translated. This process involves recognition of the 5' cap (m7GpppN) by eIF4E, and of the poly-A tail by PABP. PABP binds to cellular factor eIF4G, a scaffolding protein that physically links PABP and eIF4E, to facilitate circularization of the cellular mRNA, which is important for efficient initiation of translation [16]. For rotaviruses, although the transcripts are capped during endogenous transcription by the structural protein VP3, they are not polyadenylated [17,18]. Rotaviruses overcome the lack of a poly-A tail on their mRNAs by using a consensus sequence (5'-GUGACC-3') for group A rotaviruses at the 3' ends. This is specifically bound by NSP3, which serves as a functional analogue of PABP. While the N-terminal domain of NSP3 (NSP3-N) binds to the 3' consensus sequence [**19], the C-terminal domain (NSP3-C) interacts with eIF4G, resulting in circularization of the viral mRNA through the cap binding protein eIF4E [**20]. During viral infection, NSP3 binds to eIF4G with greater affinity than does PABP, which supports the model that PABP is evicted from eIF4G thereby increasing the translation of viral transcripts [**20]. Cellular protein synthesis may be arrested as a consequence of NSP3 hijacking PABP from cellular mRNA. The structural basis for NSP3 activity is demonstrated by crystallographic studies of NSP3-N in complex with the consensus 3' mRNA sequence [**19] and of NSP3-C bound to a peptide corresponding to its binding site on eIF4G [**20] (Figure 2A and 2B). NSP3-N forms a heart-shaped asymmetric dimer with an extensive dimeric interface consisting of structurally similar N-terminal fragments, and structurally different C-terminal fragments. These intertwine to create a highly basic tunnel that is ideally suited for specific recognition of the rotaviral 3' consensus sequence. The C-terminal domain of NSP3 is predominantly α-helical and forms a symmetric dimer with two hydrophobic pockets for eIF4G binding (Figure 2B). Thermal denaturation and filter binding studies indicate that apo-NSP3 likely exists in a metastable structure that forms a highly stable complex upon binding to the consensus sequence [**19]. Such a tight interaction not only promotes translation of the rotavirus mRNA but also prevents degradation of the rotavirus mRNA by cellular nucleases [**19]. This observation raises an interesting question as to how the 3'-end of the viral RNA is freed for priming negative-strand synthesis by VP1 during replication. One possibility is that VP1 binds to an overlapping sequence at the 3'-end of the mRNA and displaces NSP3. A more likely possibility, given that replication takes place exclusively in the viroplasm, is that separate pools of transcripts are utilized for translation and replication [21]. Although the role of NSP3 in viral translation and the model involving its simultaneous interactions with viral mRNA and eIF4G (Figure 2A and 2B) are widely accepted, recent studies using selective silencing of NSP3 and eIF4G in permissive cell lines have questioned this model [**22]. These studies suggest that NSP3 interaction with eIF4G is not obligatory for viral translation but is required for shut-down of cellular protein synthesis, and that the tight interaction between NSP3 and the 3' consensus region serves to protect viral mRNA from degradation and prevent it from entering the replication pool [**22].
Figure 2. NSP3 and NSP2 structures.
(A) The homodimer of the N-terminal RNA binding domain of NSP3 (residues 4–164) is shown in ribbon diagram in blue and golden yellow with a transparent surface. The 3' consensus sequence of rotavirus mRNA (5'-GUGACC-3') bound by NSP3 is shown in a yellow ball-and-stick representation, with oxygen atoms shown in red, and is indicated by an arrow (PDB ID: 1KNZ). (B) The structure of the NSP3 C-terminal domain (residues 206–315) homodimer in complex with eIF4G peptides is also presented in ribbon diagram with transparent surface. The NSP3 C-terminal domain homodimer is shown in blue and golden yellow, and the bound eIF4G peptides (in red and green) are indicated with arrows (PDB ID: 1LJ2). Structures of NSP2 (C–E) and its complexes with NSP5 (F) and RNA (G). The NSP2 monomer is shown in ribbon diagram, with its N- and C-terminal domains colored in purple and brown, respectively (PDB ID: 1L9V) (C). The catalytic cleft between two domains is indicated by an arrow. NSP2 self-assembles into octamers, as shown. The N-and C-terminal domains of one NSP2 molecule are colored and the remaining subunits in the functional octamer are shown in gray. The NSP2 octamer is shown as viewed along the 4-fold axis (D) and along one of the two 2-fold axes (E). Cryo-EM reconstructions of NSP2 with bound NSP5 (F, with NSP5 in blue) or RNA (G, with RNA in green) are shown as transparent surfaces with NSP2 (red ribbon) fitted into the density map.
NSP2
Critical roles for NSP2 in the formation of the viroplasm and in genome replication/encapsidation are well established. In addition to its interactions with NSP5 in the formation of viroplasms [23], NSP2 also appears to interact directly or indirectly with other structural proteins present in the viroplasm, such as VP2 [24], which forms the innermost capsid layer of the virion [25], and VP1, the viral polymerase [26]. NSP2 is a multi-functional enzyme: along with sequence-independent ssRNA binding and nucleic acid helix destabilizing activities [*27], it exhibits Mg2+22 -dependent nucleoside triphosphatase (NTPase) [*28], RNA triphosphatase (RTPase) [29], and nucleoside diphosphate (NDP) kinase activities [*30].
X-ray crystallographic and cryo-EM studies have revealed a structural basis for some of these activities [*30, **31, *32] (Figure 2C–2G). NSP2 has two distinct domains that are separated by a deep cleft for nucleotide binding and hydrolysis of γ-phosphate [**31]. Inside the cleft, a highly conserved H225 functions as a catalytic residue for the observed enzymatic activities. Both in the crystal structure and in solution, NSP2 forms a doughnut shaped octamer with a 35 Å-wide central hole and four grooves on the sides, which are predominantly lined by basic residues [**31,*32]. As shown by cryo-EM studies, both NSP5 (residues 66–188) and RNA bind to the basic grooves of the NSP2 octamer [*32]. Consistent with this structural observation, competition experiments show that NSP5 competes with ssRNA for binding to NSP2 [*32]. These observations suggest that one of the functions of NSP5 is to regulate NSP2-RNA interactions during genome replication. More recently, it has been shown in vitro that tubulin directly interacts with NSP2, which may account for the observations that viroplasm formation during rotavirus infection disrupts the microtubule network and that tubulin granules co-localize with the viroplasm [*33,34]. Cryo-EM studies indicate that tubulin binds to the same basic grooves on the octamer as do NSP5 and RNA, and binding studies show that tubulin competes with the binding of NSP5 and RNA [*33]. Although the role of microtubule depolymerization during rotavirus infection remains unclear, it is possible that this is a mechanism to disrupt cellular functions to the advantage of the virus.
The ssRNA-binding and helix-destabilizing activities of NSP2, neither of which depends upon the NTPase activity of NSP2, are likely required for relaxing mRNA templates in preparation for dsRNA synthesis [*27]. However, the roles for the observed NTPase, RTPase, and NDP kinase activities of NSP2 during rotavirus replication remain unclear. These enzymatic activities all rely on a single highly conserved H225 residue. Complementation assays have shown that an H225A mutation abrogates genome replication/encapsidation but not viroplasm formation, indicating an essential role of these enzymatic activities for replication [*35]. Initially, it was proposed that NSP2 functions as a molecular motor, akin to a helicase, by making use of the energy derived from NTP hydrolysis to assist in genome replication/encapsidation. However, several characteristics of NSP2, such as its octameric self-assembly, absence of any recognizable helicase motifs, lack of significant conformational change upon nucleotide binding and hydrolysis, use of histidine as a catalytic residue, and formation of a relatively stable phosphohistidine intermediate during NTP hydrolysis, clearly distinguishing NSP2 from typical helicases and suggests the role for its NTPase activity is not in energy transduction.
An unusual aspect of the rotavirus genome is the absence of a γ-phosphate at the 5′-end of the minus-strand RNA [17,18]. It is possible that removal of the terminal phosphate is necessary as a defense against the antiviral retinoic acid inducible gene I (RIG-I) pathways, which are triggered by RNA-triphosphates [36], and the RTPase activity of NSP2 may be used in this context. The role for the NDP kinase activity of NSP2, which inherently relies on NTPase activity, may be to regulate nucleotide pools in the viroplasm such that they are sufficient to support genome transcription and replication by the viral polymerase [*30]. Altogether, these studies reveal that NSP2 is a multi-functional enzyme. It is possible that NSP2 uses a different combination of catalytic activities or switches between activities depending on its interactions with ligands.
NSP5
Despite various properties that are attributed to NSP5 from in vivo and in vitro studies, the only role firmly established for this protein is as a binding partner of NSP2 in the formation of viroplasms (Figure 2F). NSP5 is a dimeric phosphoprotein, rich Ser and Thr residues, that undergoes O-linked glycosylation [37–40]. It exists in multiple isoforms, from a 28-kDa hypophosphorylated form to 32-kDa hyperphosphorylated form, during the course of rotavirus replication [38,39]. However, the mechanism(s) by which NSP5 is phosphorylated during infection is unclear; self-phosphorylation through autokinase activity and the possible involvement of cellular kinases such as casein kinases I and II and a role of phosphorylated NSP5 in viroplasm formation have been suggested [41–43]. Viroplasm formation requires components of lipid droplets, however, the mechanism and proteins of such droplets involved in viroplasm formation have not been elucidated [44]. The formation of viroplasm-like structures by expressing NSP2 and NSP5 alone is calcium regulated and directed by a C-terminal helical domain within NSP5 [45]. In addition to NSP2, NSP5 has been shown to interact with other rotavirus proteins such as VP1 [46], VP2 [47] and NSP6 [48], and also with single and double-stranded RNA in a sequence-independent manner [49]. These studies reveal NSP5 is involved in many processes such as the dynamics and regulation of viroplasms and an adapter to integrate the various functional properties of NSP2 with other rotavirus proteins during viral genome replication/encapsidation [50].
Unlike for NSP3 and NSP2, detailed structural characterization of NSP5 using x-ray crystallography has so far been unsuccessful. The only three-dimensional structural analysis reported to date is a cryo-EM of a deletion construct of NSP5 (residues 68–188) in complex with NSP2, which indicates that the N-terminal 68 residues are dispensable for NSP2 binding [32]. The deletion was necessary to avoid precipitation upon mixing of full-length NSP5 with NSP2. More recent studies using a variety of biophysical techniques on various NSP5 recombinant full-length and deletion constructs provide further insight into its structural characteristics. These studies indicate that full-length NSP5 forms decamers, comprised of five dimers in which the C-terminal residues interact with each other at the center with projecting dimeric arms [**51]. Based on these studies, it is suggested that such a quaternary organization of NSP5 would allow simultaneous interactions with various binding partners. While only some of the dimeric arms in the decameric organization interact with the NSP2 octamer due to structural constraints, the other protomers are free to interact with other binding partners, VP1, VP2 and RNA, to perform functions complementary to viroplasm formation. However, this idea needs to be validated by further in vitro and in vivo studies.
NSP4
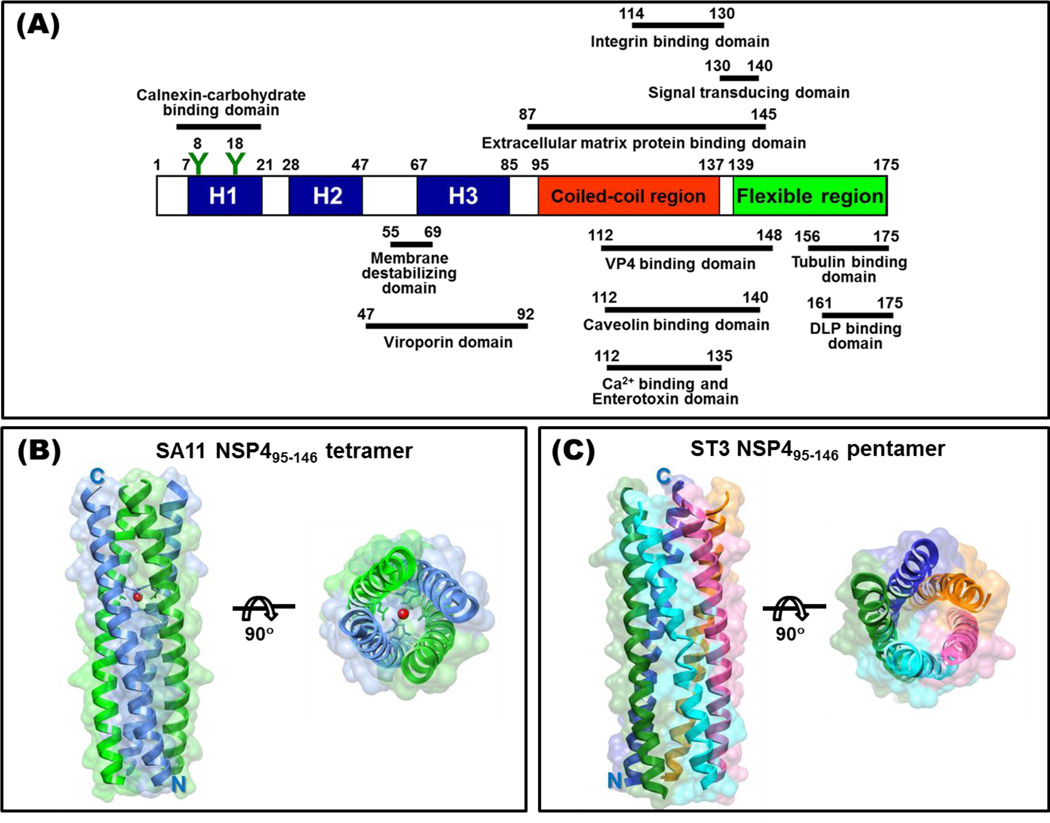
NSP4, encoded by gene segment 10, is a 175 amino acid, multifunctional protein and many of the NSP4 functions have been mapped to distinct domains within the protein (Figure 3A). NSP4 is synthesized as an ER transmembrane glycoprotein and consists of three hydrophobic domains (H1–H3) with two N-linked high mannose glycosylation sites oriented to the luminal side of the ER in the H1 domain [52,53], the H2 transmembrane domain traverses the ER bilayers and serves as an uncleaved signal sequence, a viroporin domain formed by a cluster of positively charged residues and an amphipathic α-helix (H3) [**54] followed by an extended cytoplasmic domain containing a coiled-coil region and a flexible C-terminus [1]. Relatively little is known about the N-terminus, likely due to the hydrophobic and membrane associated properties of this region. The H3 domain is highly amphipathic and was recently shown to disrupt cellular calcium homeostasis by viroporin-mediated release of ER calcium stores [**54]. A number of functional and protein binding sites have been mapped to the NSP4 cytoplasmic coiled-coil domain, including integrin I domains, caveolin, and VP4 [**55, 56, 57]. A secreted form of NSP4, which contains the integrin I domain binding site, is involved in diarrhea induction through interaction with cellular plasma membrane integrin I domains and possibly other receptors, signaling a phospholipase C-dependent increase in intracellular calcium and subsequent release of chloride through the calcium activated chloride channel transmembrane protein 16A (TMEM16A) [**55,*58,**59]. Finally, the NSP4 C-terminal flexible region binds microtubules [60] and acts as an intracellular receptor for DLPs to facilitate infectious particle assembly [61]. Other NSP4 activities for which the functional sites have not been fully mapped include disruption of plasma membrane integrity [62], inhibition of sodium absorption by epithelial sodium channels (ENaC) and sodium glucose transporter 1 (SGLT1) [*58], and remodeling of the cellular microtubule and actin networks [63,64].
Figure 3. NSP4.
(A) Schematic of NSP4 functional domains and cellular and viral protein interaction domains. Oligomeric domains are mainly in the alpha amphipathic helix (H3) and the coiled-coil region. (B) Ribbon representation of the tetramer of SA11 NSP495–146 with a transparent surface (PDB ID: 2O1K). Two NSP4 molecules in an asymmetric unit are shown in blue and green, respectively. The red dot indicates the location of a divalent metal binding site, possibly binding Ca2+. (C) The pentameric structure of ST3 NSP495–146 is shown as a ribbon diagram with a transparent surface (PDB ID: 3MIW). Each NSP4 chain is colored differently [**77].
RNAi-mediated knockdown of NSP4 demonstrates NSP4 is essential for rotavirus replication, transcription, and morphogenesis [65–67] yet how NSP4 contributes to these processes remains poorly understood. The ability of NSP4 to differentially interact with multiple viral and cellular proteins, including calnexin [68], laminin-3, fibronectin [69], caveolin [56], integrin I domains [**55], and tubulin [60] (Figure 3A) may be attributed to localization of NSP4 to different cellular compartments (ER [70], the ER-Golgi intermediate compartment (ERGIC), LC3-positive autophagosomes surrounding viroplasms [71], microtubules [60,63], membrane rafts [72,73], the exofacial surface of the plasma membrane [74], and the extracellular matrix [**55,69]) during different stages of infection and possibly to distinct forms or conformations of this multifunctional protein.
Although NSP4 has been found in dimeric, tetrameric, pentameric and higher ordered multimeric structures, including heterooligomeric complexes with VP4 and VP7, structural data are limited to the highly conserved region from amino acids 95–135 [75, *76,**77]. This region, which encompasses the enterotoxigenic peptide 114–135, which elicits diarrhea in neonatal mice, forms a parallel, tetrameric coiled-coil (Figure 3B), with the hydrophobic core interrupted by three polar amino acids (Gln109, Glu120, Gln123) and two of the four Glu120 residues and four Gln123 residues coordinating a central calcium ion [*76,**77]. When this NSP4 fragment is purified in acidic conditions, it forms a concentration-dependent pentameric coiled-coil structure (Figure 3C) without the bound calcium ion at Glu120/Gln123 [**77]. These results suggest that NSP4 is pleomorphic and that pH, calcium, and protein concentration may influence the NSP4 conformation and oligomeric state. Structural data are needed on the different intracellular and extracellular forms of NSP4 to augment biochemical and functional studies addressing the many functions that NSP4 performs during rotavirus infection.
3. Conclusions and future perspectives
Together with functional studies, structural studies on the NSPs have provided significant insight into mechanistic aspects of rotavirus replication and have revealed novel molecular mechanisms of regulation of protein function. However, many questions remain about how these proteins interact, coordinate and modulate both viral and cellular function. 1) What is the structure of NSP1? How does NSP1 antagonize the host antiviral response and target IRFs for proteasome degradation? What is the structural basis for the suggested E3-like ligase activity of NSP1? 2) Although individual domain structures of NSP3 are known, what is the structure of the full-length NSP3? Is NSP3 interaction with the elongation factor complex required only for translation of viral mRNA or does this interaction play a role in shut-off of host protein synthesis? 3) The structure of NSP2 and the structural basis for its enzymatic activities are relatively well understood, but what is the precise role for its enzymatic activities in viroplasm dynamics and genome replication/encapsidation? The atomic details of NSP2 interactions with its binding partners during viral replication should clarify new mechanisms of the multiple activities of this protein during replication. 4) What is the structure of full-length NSP5, and will this support the proposed decameric model and reveal the mechanistic basis for its role in the viroplasm and in replication? 5) The current structures of small fragments of NSP4 that indicate alternate tetramer and pentamer oligomeric conformations stimulate interest in having the structure of the full-length protein to answer: what is the structural basis of NSP4’s intriguing properties as a viral enterotoxin, viroporin, intracellular-receptor for DLP binding and subsequent budding into the ERwhere the outer capsid proteins are acquired and infectious particles lose their transient envelope and mature? 6) What is the structure and function of NSP6? Finally, do the complex combinations of interactions of NSPs with each other and with specific cellular proteins explain the emerging concept that human rotavirus transmission and spread may require particular constellations of non-outer capsid genes? (See article by Matthijnssens and Van Ranst). Continued structural studies using X-ray crystallography and cryo-EM, including single particle reconstructions and possibly electron tomography of infected cells, may provide answers to these questions. Further molecular insight should help explain how this model enteric virus with a segmented genome orchestrates the complex process of replication, including encapsidation of a correct set of 11 dsRNA segments into each particle. It will be fascinating to explore the unique processes of enveloped to non-enveloped particle maturation, including interactions with intracellular membranes and production of infectious rotavirus particles.
Highlights.
NSP3 structures provide mechanistic insight into its function during translation.
NSP2 octamers integrate enzymatic and ligand binding activities during replication.
NSP2 shows a novel NDP kinase activity through a phospho-histidine intermediate.
Biophysical studies suggest a novel decameric model for multifunctional NSP5.
Observed tetrameric and pentameric states of NSP4 may be relevant for its function.
Acknowledgments
We acknowledge the support from NIH grants AI36040 (to BVV), AI 080656 and P30 DK56338 (to MKE), and the Robert Welch foundation (Q1279) to BVV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Estes MK, Greenberg HB. Rotaviruses. In: Knipe PMH DM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. edn 6th. Lippincott Williams & Wilkins; 2012. In press. [Google Scholar]
- 2.Mattion NM, Mitchell DB, Both GW, Estes MK. Expression of rotavirus proteins encoded by alternative open reading frames of genome segment 11. Virology. 1991;181:295–304. doi: 10.1016/0042-6822(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawton JA, Estes MK, Prasad BV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 6.Arnold MM, Patton JT. Diversity of interferon antagonist activities mediated by NSP1 proteins of different rotavirus strains. J Virol. 2011;85:1970–1979. doi: 10.1128/JVI.01801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bagchi P, Dutta D, Chattopadhyay S, Mukherjee A, Halder UC, Sarkar S, Kobayashi N, Komoto S, Taniguchi K, Chawla-Sarkar M. Rotavirus nonstructural protein 1 suppresses virus-induced cellular apoptosis to facilitate viral growth by activating the cell survival pathways during early stages of infection. J Virol. 2010;84:6834–6845. doi: 10.1128/JVI.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graff JW, Ettayebi K, Hardy ME. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000280. e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graff JW, Ewen J, Ettayebi K, Hardy ME. Zinc-binding domain of rotavirus NSP1 is required for proteasome-dependent degradation of IRF3 and autoregulatory NSP1 stability. J Gen Virol. 2007;88:613–620. doi: 10.1099/vir.0.82255-0. [DOI] [PubMed] [Google Scholar]
- 10.Rainsford EW, McCrae MA. Characterization of the NSP6 protein product of rotavirus gene 11. Virus Res. 2007;130:193–201. doi: 10.1016/j.virusres.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Poncet D, Aponte C, Cohen J. Rotavirus protein NSP3 (NS34) is bound to the 3' end consensus sequence of viral mRNAs in infected cells. J Virol. 1993;67:3159–3165. doi: 10.1128/jvi.67.6.3159-3165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vende P, Piron M, Castagne N, Poncet D. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3' end. J Virol. 2000;74:7064–7071. doi: 10.1128/jvi.74.15.7064-7071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keryer-Bibens C, Legagneux V, Namanda-Vanderbeken A, Cosson B, Paillard L, Poncet D, Osborne HB. The rotaviral NSP3 protein stimulates translation of polyadenylated target mRNAs independently of its RNA-binding domain. Biochem Biophys Res Commun. 2009;390:302–306. doi: 10.1016/j.bbrc.2009.09.115. [DOI] [PubMed] [Google Scholar]
- 15.Chung KT, McCrae MA. Regulation of gene expression by the NSP1 and NSP3 non-structural proteins of rotavirus. Arch Virol. 2011;156:2197–2203. doi: 10.1007/s00705-011-1117-6. [DOI] [PubMed] [Google Scholar]
- 16.Sachs A. Physical and Functional Interactions between the mRNA Cap structure and Poly(A) Tail. In: Sonenberg JWBH N, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; 2000. pp. 447–467. [Google Scholar]
- 17.Imai M, Akatani K, Ikegami N, Furuichi Y. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J Virol. 1983;47:125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCrae MA, McCorquodale JG. Molecular biology of rotaviruses. V. Terminal structure of viral RNA species. Virology. 1983;126:204–212. doi: 10.1016/0042-6822(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 19. Deo RC, Groft CM, Rajashankar KR, Burley SK. Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer. Cell. 2002;108:71–81. doi: 10.1016/s0092-8674(01)00632-8. This paper shows that NSP3 is a functional homologue of PABP for rotavirus and binds viral mRNA 3' consensus sequences.
- 20. Groft CM, Burley SK. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. These studies provide a structural basis for how NSP3 circularizes mRNA for translation by interacting with viral mRNA 3' consensus sequences and eIF4G.
- 21.Silvestri LS, Taraporewala ZF, Patton JT. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol. 2004;78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Montero H, Arias CF, Lopez S. Rotavirus Nonstructural Protein NSP3 is not required for viral protein synthesis. J Virol. 2006;80:9031–9038. doi: 10.1128/JVI.00437-06. This paper questions the the proposed obligatory interaction of NSP3 with intitation transcription complex during viral mRNA translation.
- 23.Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J Gen Virol. 1999;80(Pt 2):333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- 24.Patton JT, Silvestri LS, Tortorici MA, Vasquez-Del Carpio R, Taraporewala ZF. Rotavirus genome replication and morphogenesis: role of the viroplasm. Curr Top Microbiol Immunol. 2006;309:169–187. doi: 10.1007/3-540-30773-7_6. [DOI] [PubMed] [Google Scholar]
- 25.Prasad BV, Rothnagel R, Zeng CQ, Jakana J, Lawton JA, Chiu W, Estes MK. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 26.Valenzuela S, Pizarro J, Sandino AM, Vasquez M, Fernandez J, Hernandez O, Patton J, Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J Virol. 1991;65:3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taraporewala ZF, Patton JT. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J Virol. 2001;75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001. These studies provided the first biochemical analysis characterizing the RNA binding properties associated with NSP2 which may play a crucial role during genome replication and encapsidation.
- 28. Taraporewala Z, Chen D, Patton JT. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. This paper presents the first characterization of an enzymatic activity associated with NSP2.
- 29.Vasquez-Del Carpio R, Gonzalez-Nilo FD, Riadi G, Taraporewala ZF, Patton JT. Histidine triad-like motif of the rotavirus NSP2 octamer mediates both RTPase and NTPase activities. J Mol Biol. 2006;362:539–554. doi: 10.1016/j.jmb.2006.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar M, Jayaram H, Carpio RV, Jiang X, Taraporwala ZF, Jacobson RH, Patton JT, Prasad BV. Crystallographic and biochemical analysis of rotavirus NSP2 with nucleotides reveals an NDP kinase like activity. J Virol. 2007;81:12272–12284. doi: 10.1128/JVI.00984-07. These structural and biochemical analyses demonstrate that NTP hydrolysis involves a phospho-histidine intermediate and that NSP2 has phosphoryl-transfer activity.
- 31. Jayaram H, Taraporewala Z, Patton JT, Prasad BV. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature. 2002;417:311–315. doi: 10.1038/417311a. Presents the first crystallographic analysis of NSP2 revealing a novel HIT-like fold, and describing the domain organization, and oligomeric state of NSP2.
- 32. Jiang X, Jayaram H, Kumar M, Ludtke SJ, Estes MK, Prasad BV. Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: implications for genome replication. J Virol. 2006;80:10829–10835. doi: 10.1128/JVI.01347-06. These studies indicate overlapping binding sites for RNA and NSP5 in the NSP2 octamer and suggest a regulatory role for NSP5 in the RNA binding of NSP2.
- 33. Martin D, Duarte M, Lepault J, Poncet D. Sequestration of free tubulin molecules by the viral protein NSP2 induces microtubule depolymerization during rotavirus infection. J Virol. 2010;84:2522–2532. doi: 10.1128/JVI.01883-09. These studies suggests that NSP2 binds tubulin near the same site as RNA and NSP5.
- 34.Cabral-Romero C, Padilla-Noriega L. Association of rotavirus viroplasms with microtubules through NSP2 and NSP5. Mem Inst Oswaldo Cruz. 2006;101:603–611. doi: 10.1590/s0074-02762006000600006. [DOI] [PubMed] [Google Scholar]
- 35. Taraporewala ZF, Jiang X, Vasquez-Del Carpio R, Jayaram H, Prasad BV, Patton JT. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J Virol. 2006;80:7984–7994. doi: 10.1128/JVI.00172-06. This study explored the contribution of structural elements to the function of NSP2 in the context of infection using a novel NSP2-dependent complementation system that supports genome replication and particle assembly in mutant SA11 rotavirus with a temperature-sensitive (ts) lesion mapping to the genome segment (gene 8) encoding NSP2-infected cells containing a gene 8-specific siRNA, but only when NSP2 is expressed in trans from siRNA-resistant transcripts.
- 36.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, Endres S, Rothenfusser S. 5'-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poncet D, Lindenbaum P, L'Haridon R, Cohen J. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J Virol. 1997;71:34–41. doi: 10.1128/jvi.71.1.34-41.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afrikanova I, Miozzo MC, Giambiagi S, Burrone O. Phosphorylation generates different forms of rotavirus NSP5. J Gen Virol. 1996;77(Pt 9):2059–2065. doi: 10.1099/0022-1317-77-9-2059. [DOI] [PubMed] [Google Scholar]
- 39.Blackhall J, Munoz M, Fuentes A, Magnusson G. Analysis of rotavirus nonstructural protein NSP5 phosphorylation. J Virol. 1998;72:6398–6405. doi: 10.1128/jvi.72.8.6398-6405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welch SK, Crawford SE, Estes MK. Rotavirus SA11 genome segment 11 protein is a nonstructural phosphoprotein. J Virol. 1989;63:3974–3982. doi: 10.1128/jvi.63.9.3974-3982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackhall J, Fuentes A, Hansen K, Magnusson G. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J Virol. 1997;71:138–144. doi: 10.1128/jvi.71.1.138-144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campagna M, Budini M, Arnoldi F, Desselberger U, Allende JE, Burrone OR. Impaired hyperphosphorylation of rotavirus NSP5 in cells depleted of casein kinase 1alpha is associated with the formation of viroplasms with altered morphology and a moderate decrease in virus replication. J Gen Virol. 2007;88:2800–2810. doi: 10.1099/vir.0.82922-0. [DOI] [PubMed] [Google Scholar]
- 43.Eichwald C, Jacob G, Muszynski B, Allende JE, Burrone OR. Uncoupling substrate and activation functions of rotavirus NSP5: phosphorylation of Ser-67 by casein kinase 1 is essential for hyperphosphorylation. Proc Natl Acad Sci U S A. 2004;101:16304–16309. doi: 10.1073/pnas.0406691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung W, Gill M, Esposito A, Kaminski C, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen A, Sen N, Mackow ER. The Formation of Viroplasm-Like Structures by the Rotavirus NSP5 Protein Is Calcium Regulated and Directed by a C-Terminal Helical Domain. J Virol. 2007;81:11758–11767. doi: 10.1128/JVI.01124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arnoldi F, Campagna M, Eichwald C, Desselberger U, Burrone OR. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J Virol. 2007;81:2128–2137. doi: 10.1128/JVI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berois M, Sapin C, Erk I, Poncet D, Cohen J. Rotavirus nonstructural protein NSP5 interacts with major core protein VP2. J Virol. 2003;77:1757–1763. doi: 10.1128/JVI.77.3.1757-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres-Vega MA, Gonzalez RA, Duarte M, Poncet D, Lopez S, Arias CF. The C-terminal domain of rotavirus NSP5 is essential for its multimerization, hyperphosphorylation and interaction with NSP6. J Gen Virol. 2000;81:821–830. doi: 10.1099/0022-1317-81-3-821. [DOI] [PubMed] [Google Scholar]
- 49.Vende P, Taraporewala ZF, Patton JT. RNA-binding activity of the rotavirus phosphoprotein NSP5 includes affinity for double-stranded RNA. J Virol. 2002;76:5291–5299. doi: 10.1128/JVI.76.10.5291-5299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Contin R, Arnoldi F, Campagna M, Burrone OR. Rotavirus NSP5 orchestrates recruitment of viroplasmic proteins. J Gen Virol. 2010;91:1782–1793. doi: 10.1099/vir.0.019133-0. [DOI] [PubMed] [Google Scholar]
- 51. Martin D, Ouldali M, Menetrey J, Poncet D. Structural organisation of the rotavirus nonstructural protein NSP5. J Mol Biol. 2011;413:209–221. doi: 10.1016/j.jmb.2011.08.008. This paper describes a intriguing decameric model for NSP5 based on various biophysical analyses.
- 52.Bergmann CC, Maass D, Poruchynsky MS, Atkinson PH, Bellamy AR. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 1989;8:1695–1703. doi: 10.1002/j.1460-2075.1989.tb03561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan WK, Au KS, Estes MK. Topography of the simian rotavirus nonstructural glycoprotein (NS28) in the endoplasmic reticulum membrane. Virology. 1988;164:435–442. doi: 10.1016/0042-6822(88)90557-0. [DOI] [PubMed] [Google Scholar]
- 54. Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. MBio. 2010;1(5) doi: 10.1128/mBio.00265-10. pii: e00265-10. First demsonstration of NSP4 as a novel viroporin.
- 55. Seo NS, Zeng CQ, Hyser JM, Utama B, Crawford SE, Kim KJ, Hook M, Estes MK. Integrins alpha1beta1 and alpha2beta1 are receptors for the rotavirus enterotoxin. Proc Natl Acad Sci U S A. 2008;105:8811–8818. doi: 10.1073/pnas.0803934105. These studies implicate NSP4 enterotoxin domain in diarrhea induction through integrin binding and signalling.
- 56.Parr RD, Storey SM, Mitchell DM, McIntosh AL, Zhou M, Mir KD, Ball JM. The rotavirus enterotoxin NSP4 directly interacts with the caveolar structural protein caveolin-1. J Virol. 2006;80:2842–2854. doi: 10.1128/JVI.80.6.2842-2854.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyser JM, Zeng CQ, Beharry Z, Palzkill T, Estes MK. Epitope mapping and use of epitope-specific antisera to characterize the VP5* binding site in rotavirus SA11 NSP4. Virology. 2008;373:211–228. doi: 10.1016/j.virol.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ousingsawat J, Mirza M, Tian Y, Roussa E, Schreiber R, Cook DI, Kunzelmann K. Rotavirus toxin NSP4 induces diarrhea by activation of TMEM16A and inhibition of Na(+) absorption. Pflugers Arch. 2011;461:579–589. doi: 10.1007/s00424-011-0947-0. Demonstrates that enterotoxin domain induces diarrhea by activating the recently identified Ca2+-activated Cl− channel TMEM16A, and by inhibiting Na+ absorption by the epithelial Na+ channel ENaC and the Na+/glucose cotransporter SGLT1.
- 59. Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. First paper to demonstrate that NSP4 is a viral enterotxin.
- 60.Xu A, Bellamy AR, Taylor JA. Immobilization of the early secretory pathway by a virus glycoprotein that binds to microtubules. EMBO J. 2000;19:6465–6474. doi: 10.1093/emboj/19.23.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Au KS, Chan WK, Burns JW, Estes MK. Receptor activity of rotavirus nonstructural glycoprotein NS28. J Virol. 1989;63:4553–4562. doi: 10.1128/jvi.63.11.4553-4562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Newton K, Meyer JC, Bellamy AR, Taylor JA. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J Virol. 1997;71:9458–9465. doi: 10.1128/jvi.71.12.9458-9465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang W, McCrae MA. The rotavirus enterotoxin (NSP4) promotes re-modeling of the intracellular microtubule network. Virus Res. 2012;163:269–274. doi: 10.1016/j.virusres.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 64.Berkova Z, Crawford SE, Blutt SE, Morris AP, Estes MK. Expression of rotavirus NSP4 alters the actin network organization through the actin remodeling protein cofilin. J Virol. 2007;81:3545–3553. doi: 10.1128/JVI.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silvestri LS, Tortorici MA, Vasquez-Del Carpio R, Patton JT. Rotavirus glycoprotein NSP4 is a modulator of viral transcription in the infected cell. J Virol. 2005;79:15165–15174. doi: 10.1128/JVI.79.24.15165-15174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lopez T, Camacho M, Zayas M, Najera R, Sanchez R, Arias CF, Lopez S. Silencing the morphogenesis of rotavirus. J Virol. 2005;79:184–192. doi: 10.1128/JVI.79.1.184-192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zambrano JL, Diaz Y, Pena F, Vizzi E, Ruiz MC, Michelangeli F, Liprandi F, Ludert JE. Silencing of rotavirus NSP4 or VP7 expression reduces alterations in Ca2+ homeostasis induced by infection of cultured cells. J Virol. 2008;82:5815–5824. doi: 10.1128/JVI.02719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mirazimi A, Nilsson M, Svensson L. The molecular chaperone calnexin interacts with the NSP4 enterotoxin of rotavirus in vivo and in vitro. J Virol. 1998;72:8705–8709. doi: 10.1128/jvi.72.11.8705-8709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boshuizen JA, Rossen JW, Sitaram CK, Kimenai FF, Simons-Oosterhuis Y, Laffeber C, Buller HA, Einerhand AW. Rotavirus enterotoxin NSP4 binds to the extracellular matrix proteins laminin-beta3 and fibronectin. J Virol. 2004;78:10045–10053. doi: 10.1128/JVI.78.18.10045-10053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petrie BL, Greenberg HB, Graham DY, Estes MK. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1984;1:133–152. doi: 10.1016/0168-1702(84)90069-8. [DOI] [PubMed] [Google Scholar]
- 71.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol. 2006;80:6061–6071. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sapin C, Colard O, Delmas O, Tessier C, Breton M, Enouf V, Chwetzoff S, Ouanich J, Cohen J, Wolf C, Trugnan G. Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells. J Virol. 2002;76:4591–4602. doi: 10.1128/JVI.76.9.4591-4602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Storey SM, Gibbons TF, Williams CV, Parr RD, Schroeder F, Ball JM. Full-length, glycosylated NSP4 is localized to plasma membrane caveolae by a novel raft isolation technique. J Virol. 2007;81:5472–5483. doi: 10.1128/JVI.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gibbons TF, Storey SM, Williams CV, McIntosh A, Mitchel DM, Parr RD, Schroeder ME, Schroeder F, Ball JM. Rotavirus NSP4: Cell type-dependent transport kinetics to the exofacial plasma membrane and release from intact infected cells. Virol J. 2011;8:278. doi: 10.1186/1743-422X-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maass DR, Atkinson PH. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J Virol. 1990;64:2632–2641. doi: 10.1128/jvi.64.6.2632-2641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bowman GD, Nodelman IM, Levy O, Lin SL, Tian P, Zamb TJ, Udem SA, Venkataraghavan B, Schutt CE. Crystal structure of the oligomerization domain of NSP4 from rotavirus reveals a core metal-binding site. J Mol Biol. 2000;304:861–871. doi: 10.1006/jmbi.2000.4250. The first crystal structure of the rotavirus enterotoxin domain.
- 77. Chacko AR, Arifullah M, Sastri NP, Jeyakanthan J, Ueno G, Sekar K, Read RJ, Dodson EJ, Rao DC, Suguna K. Novel pentameric structure of the diarrhea inducing region of the rotavirus enterotoxigenic protein NSP4. J Virol. 2011;85:12721–12732. doi: 10.1128/JVI.00349-11. This paper describes an alternative structure for the enterotoxin domain and suggests that NSP4 can form different conformational and oligomeric structures.
- 78.Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, von Itzstein M. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol. 2009;5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 79.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez S, Arias CF. Early steps in rotavirus cell entry. Curr Top Microbiol Immunol. 2006;309:39–66. doi: 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]