Summary
Alzheimer’s disease (AD) is characterized by cerebral deposition of β-amyloid (Aβ) peptides, which are generated from amyloid precursor protein (APP) by β- and γ-secretases. APP and the secretases are membrane associated, but whether membrane trafficking controls Aβ levels is unclear. Here, we performed an RNAi screen of all human Rab-GTPases, which regulate membrane trafficking, complemented with a Rab-GTPase-activating protein screen, and present a road map of the membrane-trafficking events regulating Aβ production. We identify Rab11 and Rab3 as key players. Although retromers and retromer-associated proteins control APP recycling, we show that Rab11 controlled β-secretase endosomal recycling to the plasma membrane and thus affected Aβ production. Exome sequencing revealed a significant genetic association of Rab11A with late-onset AD, and network analysis identified Rab11A and Rab11B as components of the late-onset AD risk network, suggesting a causal link between Rab11 and AD. Our results reveal trafficking pathways that regulate Aβ levels and show how systems biology approaches can unravel the molecular complexity underlying AD.
Introduction
Alzheimer’s disease (AD) is the most common form of dementia and is characterized by the cerebral deposition of β-amyloid (Aβ) peptides in the form of amyloid plaques (De Strooper, 2010 and Frisoni et al., 2011). The amyloid cascade hypothesis postulates that Aβ peptides trigger a series of pathological events leading to neurodegeneration (Huang and Mucke, 2012 and Selkoe, 2011b). Aβ peptides are liberated from the transmembrane amyloid precursor protein (APP) by the sequential actions of β-secretase and γ-secretase (Thinakaran and Koo, 2008 and Willem et al., 2009). β-secretase activity is conferred by a transmembrane aspartyl protease, also termed BACE1 (β-site APP-cleaving enzyme 1) (Vassar et al., 1999), whereas γ-secretase is a multimeric transmembrane protein complex composed of presenilin-1 (PS1)/PS2, nicastrin, Aph-1, and PEN-2 (Annaert and De Strooper, 2002 and Selkoe and Wolfe, 2007). Familial mutations in APP, PS1, or PS2 that increase the production of the amyloidogenic Aβ42 peptide have been associated with early-onset AD (Borchelt et al., 1996 and Duff et al., 1996). However, there is only limited insight into the cause of late-onset AD (LOAD), which contributes to more than 95% of cases. Genetic modifiers of LOAD may also regulate Aβ production, raising the possibility that genes regulating APP metabolism might impact the risk for AD (Andersen et al., 2005, Rogaeva et al., 2007 and Selkoe, 2011a).
Several lines of evidence support an important role for membrane trafficking in the amyloidogenic processing of APP and hence in AD pathogenesis (Rajendran and Annaert, 2012 and Thinakaran and Koo, 2008). APP and BACE1 are transmembrane proteins that are synthesized in the endoplasmic reticulum (ER), matured in the Golgi complex, and then transported to the plasma membrane and into endosomes via endocytosis (Small and Gandy, 2006 and Thinakaran and Koo, 2008). The endolysosomal compartment has been implicated as one of the major sites for Aβ generation (Cataldo et al., 2000, Haass et al., 1992 and Koo and Squazzo, 1994). Recent work has revealed that BACE1 cleavage of APP occurs predominantly in early endosomes, and endocytosis of APP and BACE1 is essential for β cleavage of APP, and Aβ production (Kinoshita et al., 2003, Rajendran et al., 2006 and Sannerud et al., 2011). The pH of endosomes (pH 4.0–5.0) is optimal for BACE1 activity, which also explains the requirement for endocytosis (Kalvodova et al., 2005 and Vassar et al., 2009). In contrast, α-secretase cleavage of APP, which precludes Aβ production, occurs at the plasma membrane (Lichtenthaler, 2011). Components of the γ-secretase complex are also synthesized in the ER, but their assembly and maturation require the coordinated regulation of the ER-Golgi-recycling circuit (Spasic and Annaert, 2008).
We previously showed that β cleavage of APP occurs in a Rab5-EEA1-positive membrane compartment and that endocytosis is essential for Aβ generation (Rajendran et al., 2006). Targeting a transition-state BACE1 inhibitor to endosomes inhibited Aβ production in cultured cells and mice (Rajendran et al., 2008). Interestingly, proteins that belong to the retromer family, which transport cargo from early endosomes to the Golgi, have also been implicated in AD (Rogaeva et al., 2007 and Small et al., 2005). These AD risk genes regulate the residency of APP and BACE1 in the early endosomal compartment, therefore regulating Aβ generation (Siegenthaler and Rajendran, 2012). Similarly, proteins of the GGA family have been shown to traffic BACE1 from endosomes to the Golgi, and their depletion led to increased amyloidogenic processing of APP (He et al., 2005, Tesco et al., 2007 and von Arnim et al., 2006). Although APP is known to be routed from endosomes to Golgi via the retromer and retromer-associated proteins including SORL1 and VPS26 (Andersen et al., 2005, Morel et al., 2013, Small and Gandy, 2006, Rogaeva et al., 2007, Small et al., 2005 and Siegenthaler and Rajendran, 2012), nothing much is known about BACE1 recycling from endosomes. A better understanding of the specific trafficking mechanisms involved in Aβ production will thus provide further insights into disease pathogenesis and potentially provide novel therapeutic strategies for treating this currently untreatable disease.
Rab GTPases regulate many aspects of membrane protein trafficking, acting as membrane organizers on cellular compartments that mediate vesicular trafficking and aid in vesicle fusion (Seabra et al., 2002). They regulate vesicular trafficking both in the biosynthetic and endocytic routes, enabling cargo sorting within the different membrane compartments (Zerial and McBride, 2001). Rab GTPases switch between an inactive GDP-bound form and an active GTP-bound form, which enables vesicular fusion (Stenmark and Olkkonen, 2001). Although GDP-GTP exchange is mediated by Rab-specific GTP exchange factors (GEFs), GTP hydrolysis is achieved by the GTPase-activating proteins (GAPs) (Barr and Lambright, 2010). In humans, there are 60 Rab proteins and 39 RabGAPs. Overexpression of RabGAPs depletes the active form of Rab proteins and increases the inactive, GDP-bound form, thereby preventing their normal function (Yoshimura et al., 2007). Here, we systematically analyzed the role of the cell’s major membrane-trafficking processes in Aβ production by the combined use of RNAi-mediated silencing of all Rab GTPase proteins and an overexpression screen of the RabGAPs.
Results
Development of a Multiplexing Platform to Quantitatively Assay sAPPβ and Aβ Levels
We first developed a multiplexing assay to quantitatively screen for Aβ and soluble ectodomain of APP β (sAPPβ), the two products of the amyloidogenic processing of APP (Figure 1A), from a single sample. Simultaneous measurements of Aβ and sAPPβ from a single sample in the same well would indicate whether a “hit” affected the β cleavage of APP (changing sAPPβ levels) or the γ cleavage/fate of Aβ (changing Aβ levels) (Figure 1B). Thus, intermeasurement variations are avoided, which increases the sensitivity and signal-to-noise ratio allowing high-throughput measurements. We used an electrochemiluminescence (ECL) assay platform because it provides high sensitivity and robust signal detection. In each well of a 384-well plate, one spot of anti-Aβ40 and one of anti-sAPPβ were separately spotted (Figure 1C). Two other spots were coated with BSA for background and nonspecific binding controls. Upon binding to the analytes in the samples, electrochemical signals from the ruthenium-labeled respective secondary antibodies were detected through an image-based capture. Because the captured antibodies are spatially positioned at distinct spots in the well, the values corresponded to the amount of the analyte bound to the specific capture antibody. As a proof of principle, we studied the binding of synthetic Aβ40 and recombinant sAPPβ in the same well. We incubated different concentrations of synthetic Aβ peptides in the assay plate and found a linear dependency of the signal to the amount of peptide used at concentrations lower than 50 ng/ml. At concentrations above 50 ng/ml, we observed saturation (Figure 1D). Similar saturation kinetics was observed with recombinant sAPPβ protein, suggesting that this system perfectly recapitulates binding reactions. When only Aβ was added to the well, background signals were observed for sAPPβ, and vice versa. When both Aβ and sAPPβ were added in the same well, specific signals were detected (Figure 1D), demonstrating a high degree of specificity and no cross-reactivity between the two analytes. In addition, ECL detection allowed us to detect concentrations as low as 10–100 pg of Aβ and sAPPβ.
Figure 1. A Multiplexing Platform for the Detection of Amyloidogenic Processing of APP.
(A) Schematic of the screen.
(B) Cartoon of APP cleavage by β- and γ-secretases.
(C) Cartoon of ECL detection system.
(D) Incubation of synthetic Aβ (Aβ40, black) and recombinant sAPPβ (red) either individually or together gives specific signals.
(E) Supernatants from HEK (HEK-sweAPP) or HeLa cells overexpressing the Swedish mutant of APP (HeLa-sweAPP) and primary cortical neurons from Arc/sweAPP Tg analyzed for Aβ (black) and sAPPβ (white for HEK and HeLa; gray for the cortical neurons) levels.
(F and G) Specificity of the ECL-multiplex assay platform. Signals for Aβ40 (black) and WT sAPPβ (gray) from conditioned medium of HEK-WTAPP (F) or HEK-sweAPP (G) analyzed with capture antibodies specific for Aβ and WT sAPPβ.
(H and I) Signals for Aβ40 (black) and swe-sAPPβ (gray) from conditioned medium of HEK-WTAPP cells (H) or HEK-sweAPP cells (I).
Error bars indicate SD. See also Figures S1A and S1B.
We then proceeded to see if Aβ and sAPPβ could be measured from cultured cells and neurons from transgenic mice. Both Aβ and sAPPβ could be measured from HEK and HeLa cells stably expressing the pathogenic Swedish mutant of APP, which causes familial early-onset AD (Figure 1E). Conditioned medium collected at different times shows that both Aβ and sAPPβ gradually accumulated in a time-dependent manner (Figure 1E). Increase in the volume of the conditioned supernatants produced a linear increase in the measurements demonstrating assay robustness (Figures S1A and S1B). We could also quantify sAPPβ and Aβ in cortical neurons from mice expressing human APP with Arctic/Swedish (Arc/Swe) mutations (Figure 1E). Sample-swapping (Figure 1F) experiments showed that no significant signal was observed for the β-cleaved ectodomain of Swedish APP (sAPPβsw) when measured with the plate coated in capture antibodies, which recognized the β-cleaved ectodomain of wild-type (WT) APP (sAPPβWT). Similarly, no signal was obtained for sAPPβ when we swapped the sAPPβWT supernatants on the sAPPβsw-coated plates. However, robust signals for Aβ were detected in all conditions. Thus, the assay is highly specific for both Aβ and sAPPβ.
RNAi Screen of Human Rab Proteins Identifies Proteins Involved in Aβ and sAPPβ Production
To study the role of all human Rab proteins in the regulation of amyloidogenic APP processing, we first performed an RNAi screen (Table S1) in cells that robustly produce Aβ and sAPPβ (Rajendran et al., 2006 and Rajendran et al., 2008) and assayed these products using the multiplexing ECL assay system (Figures 2A and 2B). We included APP, BACE1, and Pen2 (a subunit of the γ-secretase) as positive controls. As expected, silencing of APP and BACE1 decreased both Aβ and sAPPβ levels, whereas silencing of Pen2 decreased Aβ levels, but not sAPPβ levels (Figures 2A and 2B), further validating the assay. Quantification was based on the ECL counts, normalized to the cell viability counts, and relative to that of the scrambled control (medium GC containing siRNA oligo [MedGC]). Apart from Rab36, silencing of the other Rab proteins did not significantly alter cell viability (Figure S1C). The screen identified Rabs that significantly decreased both Aβ and sAPPβ (Figures 2A and 2B), including Rab3A, Rab11A, Rab36, and Rab17 (Figures S2A and S2B). Rab36, a GTPase involved in late endosome positioning, was a top hit in the screen, decreasing both Aβ and sAPPβ (Figure 2C). A secondary validation screen reproduced all the selected hits (Figure S2C). Knockdown efficiency of the relevant genes was confirmed by RT-PCR (Figure S2D). Although our analysis accounts for toxicity, and quantifies Aβ and sAPPβ as a relative count of viable cells, Rab36 depletion was consistently toxic in all the screens (Figure S1C). Therefore, we removed it from further analysis.
Figure 2. RNAi Screen of Genome-wide Rabs Identifies Rabs Involved in Regulating the Levels of Aβ and sAPPβ.
(A and B) Graph showing the levels of Aβ (A) and sAPPβ (B) from the Rab siRNA screen. HeLa-sweAPP cells were transfected with Scrambled oligo (negative control), or pooled siRNA against APP, BACE1, PEN2, and the 60 Rabs and assayed for Aβ and sAPPβ. Error bars indicate SEM.
(C) List of genes that had the strongest effect on Aβ and sAPPβ levels after siRNA-mediated silencing along with the Z score and t test values.
(D) 2D plot representing the levels of Aβ and sAPPβ from the Rabs siRNA screen. Positive controls APP, BACE1, and PEN2 are indicated in green, and negative control Scrambled is indicated in blue. Rabs silencing that led to the strongest decrease in both Aβ and sAPPβ is indicated in red. All statistics were performed using the two-tailed t tests and one-way ANOVA (Table S4).
See also Figures S1C, S2, and S3.
Rab11A, the major Rab protein involved in the slow recycling of cargo proteins from early endosomes to the cell surface (Sönnichsen et al., 2000), was the second top hit in our analysis. In addition, silencing of the isoform, Rab11B, also reduced Aβ levels (Figure 2A). Interestingly, all the isoforms of Rab3, except Rab3C, decreased both Aβ and sAPPβ levels. Rab3 proteins are involved in synaptic function (Schlüter et al., 2004) and in the fast axonal transport of APP (Szodorai et al., 2009). Silencing of Rab3A and Rab3B decreased overall APP levels, suggesting that Rab3 plays a role in the trafficking and maintenance of APP levels (Figure S2E).
Silencing of Rab44, Rab6A, and Rab10 decreased Aβ levels without affecting sAPPβ (Figure 2C), implying that these GTPases affect either γ-secretase cleavage or the fate of Aβ. Interestingly, knockdowns of several Rabs also increased both sAPPβ and Aβ levels. To understand how these trafficking pathways contributed to both Aβ and sAPPβ levels, we plotted the normalized values of Aβ and sAPPβ from the screen in a 2D plot and observed a largely correlative curve for both the values, indicating that the Rab proteins responsible for regulating β cleavage are also responsible for controlling Aβ levels and that any perturbation at the level of BACE1 cleavage of APP is the rate-limiting step in Aβ production (Figure 2D).
RabGAP-Overexpression Screen Identifies Overlapping Hits with the Rab RNAi Screen
Because RNAi can have off-target effects leading to false-positive results, we complemented our RNAi screen with an overexpression screen of all 39 RabGAPs in the human genome, which suppress Rab function by accelerating the hydrolysis of GTP. In this screen, we overexpressed each RabGAP in the same cellular system used for the RNAi screen and quantitatively measured Aβ, sAPPβ levels, and cell viability (Figures 3 and S1D). Many RabGAPs were found to affect sAPPβ and Aβ levels (Figure 3). RN-TRE, a GAP that has previously been shown to affect amyloidogenic processing of APP (Ehehalt et al., 2003), reduced both Aβ and sAPPβ. TBC1D10B, a GAP for Rab35, Rab27A, Rab22A, Rab31, and Rab3A, decreased both Aβ and sAPPβ (Frasa et al., 2012). However, only Rab3A silencing via RNAi decreased Aβ and sAPPβ (Figures 2 and 3). EVI5, a RabGAP for Rab11 (Frasa et al., 2012 and Laflamme et al., 2012), decreased both Aβ and sAPPβ. In addition, one other RabGAP for Rab11, namely TBC1D15, decreased both Aβ and sAPPβ. Intriguingly, TBC1D14, which alters Rab11 localization and subsequently delays recycling of transferrin to the plasma membrane (Longatti et al., 2012), decreased Aβ levels (Figure 3C). These results independently identified Rab11 as an important regulator of sAPPβ and Aβ production.
Figure 3. RabGAP Screen Identifies GAPs and Rabs in the Regulation of Aβ and sAPPβ Levels.
(A) Graph showing the levels of Aβ (black) and sAPPβ (gray) after RabGAP plasmid overexpression. HeLa-sweAPP cells were transfected with plasmid expressing RabGAP fused with GFP or a plasmid expressing GFP (negative control) and assayed for Aβ and sAPPβ. Error bars indicate SEM.
(B) Graphs showing Z scores of the effects on sAPPβ and Aβ levels after RabGAP overexpression.
(C) Table showing RabGAPs that had the strongest effect on Aβ and/or sAPPβ levels along with their cognate Rab(s). The asterisk (*) indicates that the TBC1D14 is not a GAP for Rab11 but has been shown to perturb the localization and function of Rab11 (Longatti et al., 2012).
See also Figure S1D.
Epistasis Mini Array Profiling Identifies Distinct Trafficking Routes
We next analyzed whether the identified Rabs regulated Aβ production through the same trafficking pathway or through distinct mechanisms. To this end, we performed an epistasis mini array profiling (EMAP) screen (Schuldiner et al., 2005) wherein we silenced all the hits against all the hits in a 14 × 14 matrix to determine whether a combined knockdown gives rise to an aggravating (noninteracting) or alleviating (interacting) phenotype (i.e., β cleavage of APP). Here, again, single knockdowns of either Rab3A or Rab11A decreased sAPPβ levels (Figure S3). However, when they were silenced together, a further decrease (an aggravating phenotype) was observed, which indicates that Rab3A and Rab11A act independently and contribute to β cleavage via distinct trafficking routes. Most of the genes that were identified as hits did not interact with each other, with the exception that a double knockdown of Rab10/Rab23 or Rab10/Rab25 showed an alleviating phenotype, suggesting that these Rabs regulate the same membrane-trafficking pathways.
Validation of Rab11A as a Hit
Because Rab11A was identified as one of the strongest hits in both the RNAi screen and the RabGAP screen, we independently validated this finding in a different cellular model. siRNA-mediated silencing of Rab11A in HEK cells stably expressing WTAPP led to a strong reduction of both Aβ and sAPPβ levels (Figure S4A), similar to the effect observed upon silencing of Rab11A in HeLa cells stably expressing sweAPP (Figure 4A). Consistent with this, western blotting analysis with APP C terminus antibodies revealed a strong reduction in β-C-terminal fragment (CTF) levels (C99) with an increase in α-CTF levels (Figure 4B). This suggests that Rab11A silencing markedly affected β cleavage of APP and also Aβ levels. Western blotting with Rab11A antibodies showed that Rab11A silencing efficiently knocked down the protein (Figure 4B) (with >90% efficiency). In addition, Rab11A silencing led to a decrease in all three species of Aβ (Aβ38, Aβ40, and Aβ42) (Figure 4C) and reduced amyloid levels in the cell lysates, suggesting that Rab11A regulates Aβ production rather than secretion (Figure S4B). Silencing of Rab11A using four different siRNAs singly also led to a decrease in Aβ without affecting cell viability, thus demonstrating the specificity of the observed effect (Figure 4D). All siRNAs against Rab11A also silenced Rab11A expression (Figure 4D, inset).
Figure 4. Rab11 Regulates BACE1 Cleavage of APP in Model Cell Lines and Primary Neurons from WT and APP Transgenic Mice.
(A) HeLa-sweAPP cells were transfected with scrambled oligo (MEDGC, negative control) or pooled siRNA against APP and Rab11A and assayed for Aβ (dark gray) and sAPPβ (light gray). Error bars indicate SD.
(B) HeLa-sweAPP cells were transfected with scrambled oligo or pooled siRNA against Rab11A, and the lysates were assayed by western blotting as indicated.
(C) HeLa-sweAPP cells were transfected with scrambled oligo or pooled siRNAs against APP and Rab11A and assayed for Aβ38 (light gray), Aβ40 (dark gray), and Aβ42 (black). Error bars indicate SEM.
(D) HeLa-sweAPP cells were transfected with scrambled oligo or pooled siRNA against APP, single siRNA (si1–si4) against Rab11A, and pooled siRNA against Rab11A and assayed for Aβ40 (dark gray) and cell viability (light gray), and the lysates were assayed by western blotting as indicated (inset). Error bars indicate SD.
(E) HeLa-sweAPP cells were transfected with PCDNA 3.1 (negative control), Rab11A DN, and Rab11B DN and assayed for Aβ40 (dark gray) and sAPPβ (light gray). Error bars indicate SEM.
(F) HeLa-sweAPP cells were transfected with scrambled oligo or pooled siRNA against APP, BACE1, and Rab11FIP1–Rab11FIP5 and assayed for sAPPβ. Error bars indicate SEM.
(G–J) Silencing of Rab11 in primary neurons from APP transgenic and WT mice reduces β cleavage of APP and Aβ levels.
(G) Rab11A and Rab11B were silenced in primary cortical and hippocampal neurons isolated from APP transgenic mice expressing the Swedish mutation in APP, and the secreted levels of Aβ and sAPPβ were measured. The scrambled oligo was used as a negative control and siRNA targeting APP as positive control. Error bars indicate SD.
(H) Western blotting with Rab11A and GAPDH antibodies from control and Rab11-silenced primary neurons.
(I) Rab11A and Rab11B were silenced in primary cortical and hippocampal neurons isolated from WT mice, and the secreted levels of endogenous Aβ40 and Aβ42 were measured. The scrambled oligo was used as a negative control and siRNA targeting APP as positive control. Error bars indicate SD.
(J) Western blotting with Rab11A and GAPDH antibodies from control and Rab11-silenced primary WT neurons. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with scrambled MEDGC or PCDNA3.1. n.s., not significant. All statistics were performed using the two-tailed t tests. Similar significance was also observed with one-way ANOVA (Dunnett’s a posteriori analysis) (Table S5). See also Figures S4A–S4E.
To further validate these results, we employed a different strategy to interfere with Rab11 function. Because Rab proteins exist in both a GTP-bound and GDP-bound form, we overexpressed the GDP-locked (dominant negative; DN) mutant forms of Rab11A (the ubiquitously expressed isoform) and Rab11B (the isoform highly expressed in neurons) in cells to phenocopy the effect of Rab11 silencing. Similar to silencing of Rab11A, overexpression of Rab11A DN reduced Aβ and sAPPβ levels significantly. Overexpression of Rab11B DN (which would also interfere with the function of WT Rab11) also significantly reduced Aβ and sAPPβ levels (Figure 4E).
Finally, we assessed the effect of silencing the expression of evolutionarily conserved Rab11 family-interacting proteins on β cleavage of APP in the cells (Lindsay and McCaffrey, 2002). Knockdown of four of the five FIP family proteins (Shiba et al., 2006), with the exception of Rab11FIP1 (retromer-like), decreased sAPPβ levels (Figure 4F). Thus, not only Rab11 but also Rab11-interacting proteins, which were identified as Rab11-effector proteins, reduced β cleavage of APP.
Rab11 Silencing in Primary Neurons from WT and APP Transgenic Mice Reduces Aβ Levels
We next studied if Rab11 also regulated Aβ production in the neuronal context. Silencing Rab11A and Rab11B in primary neurons isolated from APP transgenic mice significantly decreased both sAPPβ and Aβ levels (Figure 4G). Both RT-PCR and western blotting analysis show efficient silencing of Rab11A and Rab11B (Figures S4C, S4D, and 4H; Table S2). Knockdown of Rab11A and Rab11B in primary neurons isolated from WT mice again reduced Aβ (both Aβ40 and Aβ42) levels (Figures 4I and 4J). Moreover, silencing either Rab11A or Rab11B also significantly decreased Aβ levels (Figure S4E). Thus, Rab11 is crucial for β cleavage and Aβ generation.
Rab11 Controls Membrane Trafficking of BACE1
We hypothesized that Rab11 must regulate either at the level of APP or at the level of BACE1. Rab11 silencing or Rab11A DN expression did not affect the total cellular levels of APP or BACE1 (Figures 4B and S4F, respectively). To analyze protein localization, we performed live-cell imaging of cells transfected with either APP-YFP or BACE1-YFP using a wide-field microscope maintained under a temperature-controlled environment. BACE1-YFP prominently localized to the perinuclear endocytic-recycling compartment (ERC) and was also found in numerous highly motile vesicular and tubular structures dispersed throughout the cell (Figure 5A; Movie S1). Inspection of individual carriers revealed rapid transport of tubulo-vesicular structures along a curvy linear trajectory, suggesting active transport along microtubules (Movie S1). Indeed, brief treatment of cells with the microtubule-disrupting reagent nocodazole inhibited dynamic transport of BACE1 and induced the formation of enlarged immobile vesicles (data not shown). The distribution and dynamics of BACE1 trafficking, but not of APP, were markedly altered by expression of Rab11AS25N, the DN mutant of Rab11 (Rab11A DN) (Figure 5A; Movies S1 and S2). The length of the BACE1-positive tubular carriers emanating from the ERC was significantly enhanced by Rab11A DN overexpression but remained highly dynamic, suggesting that GTPase activity of Rab11 is involved in the fission of BACE1 tubular carriers and/or their tethering and fusion with the target.
Figure 5. BACE1 Colocalizes with Rab11 Compartments, and Its Trafficking Is Altered upon Rab11 Dysfunction.
(A) Live-cell images of COS cells transfected with BACE1-YFP or YFP-APP along with empty vector (control) or Rab11A DN. Expression of Rab11A DN protein is shown in the insets.
(B) COS cells were transiently cotransfected with BACE1-YFP (green) and empty vector, HA-tagged Rab11A WT or S25N DN mutant, and fixed and stained with HA (data not shown) and TfR antibodies (red) to label recycling endosomes. The colors in the first two panels are green for BACE1-YFP and red for TfR. Insets are the zoomed-in images of the corresponding images.
See also Figure S5A and Movies S1 and S2.
Rab11 GTPase is involved in recycling many cargos from the ERC, including the recycling endosome marker transferrin receptor (TfR) (Sönnichsen et al., 2000). TfR as well as internalized transferrin become associated with a distinct tubular network in cells expressing Rab11 DN (Choudhury et al., 2002 and Wilcke et al., 2000). To address whether BACE1 follows the recycling route from recycling endosomes, we first checked whether BACE1 colocalizes with TfR. BACE1 significantly colocalized with TfR (Figure 5B). Not only the steady-state levels of TfR but also internalized transferrin colocalized with BACE1 in Rab11 compartments (data not shown). In control cells and those transfected with Rab11A WT, BACE1-YFP colocalized with TfR in the ERC as well as in numerous vesicles and short tubulo-vesicular carriers (Figure 5B). However, in cells expressing Rab11A DN, BACE1 and TfR were found in elongated tubular structures that emanated from the ERC (Figure 5B). Neither the prominent perinuclear localization of APP in the TGN nor the dynamics of APP-positive structures was disturbed by Rab11A DN expression (Figure 5A). Thus, BACE1 localization and trafficking are akin to that of TfR, which requires recycling via the Rab11-mediated pathway for transferrin internalization. Because Rab11 regulates the recycling of TfR through the slow recycling route, it may also regulate the recycling of BACE1, thus affecting both the cell surface as well as the early endosomal trafficking of BACE1.
Rab11 Affects the Recycling of BACE1
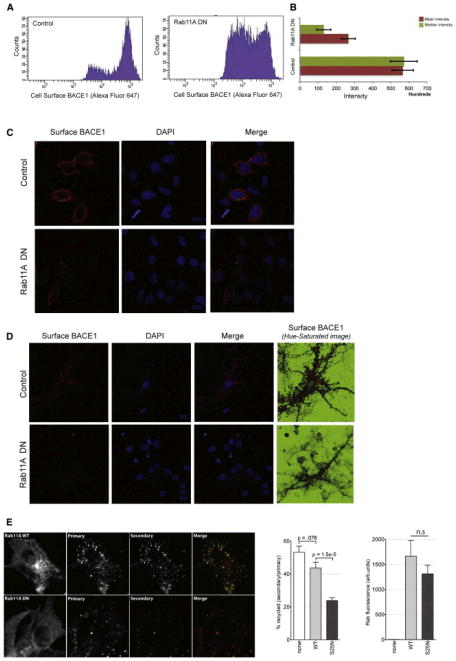
Because BACE1 accumulated in elongated tubulo-vesicular structures in cells expressing Rab11A DN, we hypothesized that under these conditions, BACE1 is unable to be trafficked to the plasma membrane and thus is unavailable for reinternalization to early endosomes, the site of Aβ production. To test this, we performed two different experiments. First, we tested if Rab11A regulated cell surface levels of BACE1 using FLAG-tagged BACE1 (Figure S5A). Here, we used both FACS (fluorescence-activated cell sorting) as well as immunofluorescence on cells transfected with either control or Rab11A DN-expressing plasmids. Cells expressing Rab11A DN indeed displayed reduced cell surface levels of BACE1 (Figures 6A and 6B). Similar results were obtained using imaging of cell surface-labeled BACE1 in HeLa (Figure 6C) and primary neurons (Figure 6D). Rab11A DN did not reduce the total levels of BACE1 (Figure S4F), in line with the observations that under Rab11A DN-expressing conditions, BACE1 accumulates in the tubulo-vesicular compartments and is not rerouted to degradative compartments.
Figure 6. Rab11 Regulates the Surface Recycling of BACE1 and Is Essential for Maintenance of BACE1 Cell Surface Levels.
(A) HeLa-swAPP cells were transfected with either control plasmids or the DN mutant of Rab11A and Flag-tagged BACE1. Cell surface levels of BACE1 were determined via FACS on nonpermeabilized cells using FLAG antibodies and Alexa 647-coupled secondary antibodies.
(B) The mean and the median intensity of the cell surface levels of BACE1 are lower in Rab11A DN-expressing cells.
(C) Microscopy of cell surface BACE1 on nonpermeabilized HeLa cells expressing BACE1-Flag. Rab11A DN-expressing cells have marked decrease in cell surface BACE1.
(D) Immunofluorescence of cell surface BACE1 on nonpermeabilized primary neurons transfected with BACE1-Flag. Also in neurons, Rab11A DN-expressing cells have a marked decrease in cell surface BACE1.
(E) Recycling of BACE1 is significantly decreased in Rab11A DN-expressing cells. Cells were transfected with BACE-FLAG/Rab11A constructs (WT and DN) and incubated with Alexa 647-conjugated M1 FLAG primary antibodies for 30 min at 37°C, followed by washing and incubation with Alexa 488 secondary antibodies at 37°C for 30 min. This allows the primary antibody to bind the surface/cycling BACE1 and label the endosomal BACE population, and the secondary antibody to only bind the BACE1 that recycled to the surface. A ratio of the secondary to the primary gives the amount recycled in this time.
See also Figures S4F and S5B.
Next, we specifically looked at the effect of Rab11A in the recycling pool of BACE1 using a quantitative ratiometric assay to measure recycling by differential labeling. To this end, cells were labeled with Alexa 647-conjugated M1 anti-FLAG for 30 min at 37°C, to allow antibodies to bind surface BACE1 and be internalized. After internalization, cells were incubated with Alexa 488-conjugated secondary antibodies for 30 min at 37°C, to detect internalized anti-FLAG-bound receptors that recycled to the surface. In the Rab11A DN-expressing cells, BACE1 recycling was significantly decreased, confirming that Rab11 functions to recycle BACE1 from the endosomes to the plasma membrane (Figure 6E). Quantitation of the fluorescence signal clearly revealed that the recycling pool of BACE1 is markedly decreased in cells expressing Rab11A DN. Two parallel controls were used: a surface control, where both incubations were performed at 4°C; and an endocytosis control, where the first incubation was at 37°C and the second at 4°C (Figure S5B). Cells without BACE1 did not show any labeling under these conditions. Together, these results suggest that vesicular trafficking regulated by Rab11 GTPase is critically important for the homeostatic regulation of BACE1 trafficking and Aβ production in endocytic organelles.
Rab11 Genetic Variability in LOAD
We used exome-sequencing data from 170 neuropathologically assessed controls and 185 patients with LOAD to study if variants in Rab11A were associated with AD. Our results revealed a significant association of a variant in Rab11A (rs117150201; uncorrected p value = 0.01), thus establishing a potential association between Rab11 and AD (Table S3).
Network Analysis of GWAS Genes Linked to LOAD Identifies Rab11A and Rab11B as Interacting Proteins
To determine whether other genes linked with LOAD risk also acted via Rab11, we used protein-protein interaction (PPI) network analysis to study the interaction partners of proteins linked to LOAD risk. We used the protein products of top ten genes linked to LOAD (AlzGene; http://www.alzgene.org) and created a two-level depth interactome of these proteins. In the cluster with Bin1, a risk gene that is strongly associated with LOAD risk, we identified both Rab11A and Rab11B and their interacting proteins, Rab11FIP1–Rab11FI5 (Figure S6). In our RNAi-based screen, Rab11A, Rab11B, and all the FIPs (except for FIP1) regulated Aβ levels, further supporting the findings that the LOAD risk gene, Bin1, could contribute to amyloid production via Rab11. Interestingly, the Bin1 cluster also included Arf6, a known player in BACE1 endocytosis to early endosomal compartments (Sannerud et al., 2011) and GGA3, which regulates early endosomal residency of BACE1 (Tesco et al., 2007), further supporting the link between Bin1, Rab11 and BACE1 endocytosis and recycling. These results from this unbiased network analysis strongly link Rab11 to LOAD risk.
Rabs and Membrane-Trafficking Pathways in Amyloid Secretion
Using a systems approach, our screen also identified other regulatory active Rabs, and it provides insights into membrane-trafficking pathways that play crucial roles in amyloid production and secretion. This allowed us to present a comprehensive road map for APP processing (Figure 7). Most of the hits were Rabs involved in the regulation of the trafficking either to or from early endosomes (Galvez et al., 2012 and Pfeffer and Aivazian, 2004), confirming the critical role of early endosomes in Aβ generation (Kinoshita et al., 2003, Koo and Squazzo, 1994, Rajendran et al., 2006 and Small and Gandy, 2006). In addition to Rab11, several other Rabs were identified to also regulate Aβ levels. Silencing of Rab2A, for instance, increased Aβ and sAPPβ levels (Figure 2), and the cognate GAP, TBC1D11, showed a similar effect (Figure 3). Rab2A is involved in the maintenance of Golgi structure and function. It interacts with the atypical protein kinase C (aPKC) (Tisdale, 2003) that positively regulates α cleavage of APP via α-secretase (Nitsch et al., 1993).
Figure 7. A Road Map of APP Processing and Aβ Production.
Integrated map of the Rabs identified in the screen as regulators of APP processing and Aβ production/secretion. Rabs indicated in green positively regulate Aβ levels, and those indicated in red are negative regulators. Note that the majority of the Rabs regulate trafficking to and from early endosomes.
On the other side, Rab5C silencing significantly increased sAPPβ and Aβ, whereas the other Rab5 isoforms did not. Rab5C is involved in the endocytosis and recycling of cell surface molecules (Onodera et al., 2012). Silencing of Rab5C dramatically increased the cellular levels of APP (Figure S2E), which explains the increase in sAPPβ and Aβ.
The Rab proteins that have been implicated in exosome release, namely Rab27 and Rab35 (Hsu et al., 2010 and Ostrowski et al., 2010), negatively regulated Aβ and sAPPβ levels (Figure 2). This is consistent with our previous work showing that very small amounts of Aβ can be released via exosomal vesicles, and suggests that the exosomal pathway is connected to an intracellular degradation mechanism and that inhibition of the exosomal pathway reroutes the cargo for effective secretion.
Discussion
To rule out potential off-target effects of the RNAi screen, we complemented it with five independent experimental conditions: (1). The first was an unbiased Rab-GAP overexpression screen; a paired RNAi screen (of all human Rab GTPases) and an overexpression screen (of RabGAPs) were performed; (2) Rab11 DN phenocopied the effect of RNAi of Rab11. (3) All four different siRNAs against Rab11 decreased Aβ levels. (4) Rab11 RNAi had the same effect in many different cells, including primary neurons. Finally, (5) silencing Rab11-associated proteins such as EHD or FIPs produced similar effect.
The results from our RNAi, RabGAP, and EMAP screens provide a valuable list of all the human Rab proteins that affect Aβ and sAPPβ levels and their nature of interaction. We also silenced different isoforms of a particular Rab by means of RNAi, and indeed, in many cases, we found isoform-specific effects on Aβ levels.
Silencing of Rab4A, Rab6A, and Rab10 decreased only Aβ. Conversely, silencing of Rab8A, Rab25, Rab14, and Rab34 significantly increased only Aβ. Such Aβ-specific effects are likely the result of either an altered γ-secretase cleavage of APP or a change in secretion and/or degradation of the produced Aβ. Of particular interest is Rab8, which increased Aβ levels when depleted in cells. Rab8 is involved in various physiological functions such as exocytosis and cilia formation. Interestingly, expression of a mutant PS1 (A260V) in PC12 cells dramatically decreased Rab8 levels, thus affecting trafficking via Golgi (Kametani et al., 2004). We also identified some hits that specifically decreased sAPPβ levels without affecting Aβ. Although this at first seems counterintuitive, it implies the possibility that specific Rabs play a role in the release/secretion of sAPPβ without affecting the processing of APP. However, we see a clear overall correlation of sAPPβ and Aβ, which suggests two things: (1) β cleavage is rate limiting and thus determines cellular and secreted Aβ levels, or (2) compartments that define β cleavage are also involved in γ cleavage of APP to generate Aβ (Kaether et al., 2006 and Rajendran et al., 2006).
We identified Rab11 as a main regulator of APP processing. Both by ECL assays and through western blotting to study APP processing, we show that BACE1-mediated processing of APP is affected in Rab11-silenced cells. Interestingly, whereas we see a significant reduction in C99 levels, C83 levels are increased. This could be due to an increased α cleavage of APP or C99 or could also represent a defect in γ-secretase cleavage, demonstrating that Rab11-recycling endosomes play a crucial role in APP processing and Aβ production. Indeed, Rab11 has been shown to interact with γ-secretase component, PS (Dumanchin et al., 1999 and Wakabayashi et al., 2009). However, we did not observe any influence of Rab11A on the activity or localization of γ-secretase (data not shown).
APP endocytosis is mediated by the YENPTY cytosolic motif and is regulated by both cholesterol-dependent and clathrin-mediated endocytic pathways (Perez et al., 1999 and Schneider et al., 2008). Similarly, endocytosis of BACE1 has been shown to be both clathrin dependent and clathrin independent via the ADP-ribosylation factor 6 pathway (Prabhu et al., 2012 and Sannerud et al., 2011). So far, early endosomes, TGN, and the plasma membrane have been suggested to contribute to Aβ production (Choy et al., 2012, Chyung and Selkoe, 2003, Rajendran et al., 2006 and Sannerud et al., 2011). Although APP is known to be routed from endosomes to Golgi via the retromer and associated proteins (Andersen et al., 2005, Morel et al., 2013, Small and Gandy, 2006, Rogaeva et al., 2007, Small et al., 2005 and Siegenthaler and Rajendran, 2012), nothing much is known about BACE1 recycling from endosomes.
Our results now identify the Rab11-dependent slow recycling pathway as a trafficking route involved in Aβ generation. A recent study showed that synaptic activity mediates the convergence of APP and BACE1 in acidic microdomains that colocalize with TfR (Das et al., 2013). Because this study did not look at colocalization of APP and BACE1 in Rab11 compartments and that a significant proportion of TfR is also found in early endosomes (Sönnichsen et al., 2000), the interaction most likely occurs in early endosomes because TfR is internalized from the plasma membrane to early endosomes and then is recycled to the plasma membrane via Rab11-positive vesicles. Several studies suggest that early endosomes are the compartments where BACE1 cleaves APP, also in the neuronal context (Choy et al., 2012, Chyung and Selkoe, 2003, Rajendran et al., 2006 and Sannerud et al., 2011). The fact that retromer-associated proteins or GGA3 regulates Aβ levels is due to its involvement in sorting of APP or BACE1, respectively, from early endosomes and not recycling endosomes (Andersen et al., 2005, Morel et al., 2013, Siegenthaler and Rajendran, 2012 and Tesco et al., 2007). Furthermore, early endosomes are acidic (Maxfield and Yamashiro, 1987), and recycling endosomes are not because they lack functional vacuolar ATPase (Gagescu et al., 2000 and Schmidt and Haucke, 2007). In addition, we do not observe the β-cleaved ectodomain of APP in Rab11-positive compartments (Rajendran et al., 2006), suggesting that the compartments that Das et al. (2013) observed as stations where APP and BACE1 meet are most likely to be early endosomes and not recycling endosomes. So, how do recycling endosomes contribute to Aβ generation? Our results now clearly show that mechanistically, Rab11-containing recycling endosomes mediate the recycling of BACE1 to replenish the pool of BACE1 in early endosomes. In support of this latter notion, our time-lapse analysis shows dramatic alteration of BACE1 dynamics in cells expressing Rab11A DN mutant. In addition, we could clearly demonstrate that recycling of BACE1 from endosomes to the plasma membrane (for subsequent reinternalization) is severely impaired in the absence of functional Rab11 using both FACS and recycling assays. In addition, Rab17, another protein involved in the recycling of internalized cargo, also regulated sAPPβ and Aβ levels, thus underscoring the importance of recycling endosomes in APP processing.
Our screen and validation results uncover a trafficking route involved in Aβ production. Because BACE1 cleavage of APP is critically involved in the pathogenesis of early-onset AD (Yang et al., 2003) and in certain forms of sporadic AD (Jonsson et al., 2012), we propose that the Rab11-mediated recycling route represents a drug target for AD, which is supported by our exome-sequencing results and the unbiased PPI network analysis. Interestingly, Eps15 homology domain-containing (EHD) proteins, which form complexes with Rab11 via Rab11FIP2, were independently identified as critical regulators of dynamic BACE1 trafficking and axonal sorting in hippocampal neurons. Moreover, knockdown of EHD1 or EHD3 significantly reduced Aβ levels in primary neurons (Buggia-Prévot et al., 2013). The network model suggests that the interaction of Bin1 with Rab11 is via Arf6, a known player in BACE1 endocytosis to early endosomal compartments (Sannerud et al., 2011), and GGA3, a protein that regulates early endosomal residency of BACE1, (Tesco et al., 2007) is also involved in the network, further supporting the link between AD risk and Rab11-mediated BACE1 endocytosis and recycling.
Taken together, our results clearly establish a link for recycling endosomes in BACE1 recycling, Aβ generation, and AD and underscore the importance of systems level analysis in understanding the complexity of AD. An extended version of the Experimental Procedures can be found in the Supplemental Experimental Procedures.
Experimental Procedures
cDNA Constructs
Plasmids encoding dsRed-tagged human WT Rab11A and DN mutant (S25N) (Choudhury et al., 2002) were purchased from Addgene. C-terminally EYFP-tagged mouse BACE1 was generated by subcloning BACE1 cDNA (provided by Nabil G. Seidah), in-frame into the pEYFP-N1 vector. HA-tagged Rab11A constructs have been described by Ren et al. (1998). The sequence corresponding to the Flag epitope in the BACE1-FLAG construct was introduced after the propeptide cleavage site within the luminal domain of mouse BACE1 using the following primers: 5′-CCGGGAGACCGACTACAAGGACGATGATGACAAGGGGGGAGGATC-3′ and 5′-CCGGGATCCTCCCCCCTTGTCATCATCGTCCTTGTAGTCGGTCTC-3′.
siRNA
siRNAs were purchased from Invitrogen (stealth siRNA). The sequences and the accession codes are supplied in Table S1.
RNAi Screen
RNAi screen was performed using HeLa cells expressing the Swedish APP mutation (HeLa-sweAPP). siRNAs were transfected with a final concentration of 5 nM using Oligofectamine (Invitrogen) as transfection reagent at a concentration of 0.3 μl in a total volume of 100 μl following manufacturer’s instructions. Each siRNA transfection was performed in quadruplicate. After 24 hr, the transfection mix on the cells was replaced with fresh culture medium. At 69 hr after transfection, medium was again replaced with 100 μl fresh medium containing 10% Alamar blue (AbD Serotec). At 72 hr after transfection, Alamar blue measurements were taken using SpectraMAX GeminiXS (Molecular Devices) at an excitation wavelength of 544 nm and emission at 590 nm. Supernatant was collected and assayed for Aβ and sAPPβ. The cells in the transfected plate were lysed with 50 μl of lysis buffer for 20 min on ice.
RabGAP Screen
The RabGAP screen was performed in HeLa-sweAPP cells. Cells were seeded in 96-well plates at a density of 6,000 cells/well 1 day before transfection. Effectene (QIAGEN) was used as transfection reagent following manufacturer’s instruction using 2.5 μl of Effectene and 0.8 μl of Enhancer in a total volume of 100 μl. Transfection mix was replaced with fresh medium after 3.5 hr. At 24 hr after transfection, medium was again replaced with fresh medium containing 10% Alamar blue. At 27 hr after transfection, Alamar blue measurements were taken using SpectraMAX GeminiXS at an excitation wavelength of 544 nm and emission at 590 nm. Supernatant was collected and assayed for Aβ and sAPPβ using the ECL assay (see Supplemental Experimental Procedures). The cells in the transfected plate were lysed with 30 μl of lysis buffer, incubated for 20 min on ice, and stored at −20°C.
Live-Cell Image Acquisition and Processing
Images were acquired on a motorized NikonTe2000 microscope equipped with a Cascade II:512 CCD Camera (Photometrics) using a 100× objective (NA 1.4). During live imaging, cells were maintained at 37°C in imaging medium (140 mM NaCl, 5 mM KCl, 3 mM CaCl2, 2 mM MgCl2, 1.5 mM D-glucose, and 10 mM HEPES [pH 7.4]) in a custom-designed environment chamber. Time-lapse images were acquired at the rate of two frames/s. Images of fixed cells were acquired as 200 nm z stacks, deconvolved using Huygens software (Scientific Volume Imaging). Extended Depth of Field plugin of ImageJ software (National Institutes of Health) was used to generate single-plane projections from processed z stacks (Rasband, 1997–2012).
Recycling Assay
Cells were labeled with Alexa 647-conjugated M1 anti-FLAG for 30 min at 37°C, to allow antibodies to bind surface BACE1 and be internalized. Cells were washed twice in media, and incubated with Alexa 488-conjugated secondary antibodies for 30 min at 37°C, to detect labeled internalized receptors that recycled to the surface. Cells were fixed in 4% paraformaldehyde for 15 min, washed, mounted, and imaged. Cells were imaged using an Andor Revolution XD spinning disk system with a Nikon Eclipse Ti automated inverted microscope, using a 60× TIRF 1.49 NA objective. Images were acquired using an Andor iXon+ EM-CCD camera using Andor IQ with 488, 561, and 647 nm solid-state lasers as light sources, using identical parameters for all images. Confocal images were collected as tiff stacks and analyzed in ImageJ. All fluorescence quantitations were performed on images directly acquired from the camera with no manipulation or adjustments. To measure fluorescence, a region of interest was drawn around each cell and the mean fluorescence in each channel recorded. In each field, a region of the field without cells was chosen to estimate background. Percent recycling was calculated as the ratio of 488/647 fluorescence, after background correction. Statistical analyses were done using Microsoft Excel or GraphPad Prism (GraphPad Software).
Supplementary Material
Acknowledgments
We thank G. Yu for the HeLa-swAPP cells, T.C. Südhof for the generous gift of Rab11 constructs, and F. Barr for the generous gift of the RabGAP library. We thank M. Schwab, W. Annaert, H. Mohler, R. Nitsch, C. Hock, and the members of the L.R. Lab for critical input into the study. We thank R. Paolicelli for the help with statistics and E. Schwarz for help with the editing of the manuscript. L.R. acknowledges financial support from the Swiss National Science Foundation grant, the Novartis Foundation grant, the Velux Foundation, Bangerter Stiftung, Baugarten Stiftung, and the Synapsis Foundation. L.R. and V.U. acknowledge funding support from the European Neuroscience Campus of the Erasmus Mundus Program. G.S. was supported by an EMBO long-term fellowship and is a recipient of a Postdoc-Forschungskredit of the University of Zurich. G.T. acknowledges grant support from the National Institutes of Health (AG019070 and AG021495) and Cure Alzheimer’s Fund. V.B.-P. was partially supported by a fellowship from Alzheimer’s Disease Research Fund of Illinois Department of Public Health. This exome-sequencing work was supported in part by the Alzheimer’s Research UK (ARUK), by an anonymous donor, by the Wellcome Trust/MRC Joint Call in Neurodegeneration award (WT089698) to the UK Parkinson’s Disease Consortium whose members are from the UCL/Institute of Neurology, the University of Sheffield and the MRC Protein Phosphorylation Unit at the University of Dundee, by the Big Lottery (to K.M.), and by a fellowship from ARUK to R.L.G.. The exome-sequencing component was also supported in part by the Intramural Research Programs of the National Institute on Aging and the National Institute of Neurological Disease and Stroke, National Institutes of Health, Department Of Health and Human Services Project number ZO1 AG000950-10. Some samples and pathological diagnoses were provided by the MRC London Neurodegenerative Diseases Brain Bank and the Manchester Brain Bank from Brains for Dementia Research, jointly funded from ARUK and AS via ABBUK Ltd.
Footnotes
Consortia
The members of the AESG (Alzheimer’s Exome Sequencing Group) are Rita Guerreiro, José Brás, Celeste Sassi, J. Raphael Gibbs, Dena Hernandez, Michelle K. Lupton, Kristelle Brown, Kevin Morgan, John Powell, Andrew Singleton, and John Hardy.
Author Contributions
L.R designed the research; V.U. performed all the RNAi, RabGAP screens, and validation experiments. B.S. and J.B. performed the ECL optimization assays. G.S. performed the optimization of siRNA transfection in mouse primary neurons. V.B.-P. and G.T. performed plasmid cloning and live-cell imaging experiments. M.S. provided reagents. N.R. performed the PPI network analysis of GWAS AD gene products. M.P. integrated all the data to curate the road map for APP processing. A.L.S. and M.A.P. performed and analyzed the recycling experiments. R.L.G., J.H., and the AESG group provided the exome-sequencing data on Rab11. L.R., V.U., V.B.-P., and G.T. analyzed the data. L.R. wrote the paper, and all the authors participated in the editing of the manuscript.
References
- 1.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annaert De Strooper B. A cell biological perspective on Alzheimer’s disease. Annu Rev Cell Dev Biol. 2002;18:25–51. doi: 10.1146/annurev.cellbio.18.020402.142302. [DOI] [PubMed] [Google Scholar]
- 3.Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchelt DR, Thinakaran G, Eckman CB, Lee MK, Davenport F, Ratovitsky T, Prada CM, Kim G, Seekins S, Yager D, et al. Familial Alzheimer’s disease-linked presenilin 1 variants elevate Abeta1–42/1–40 ratio in vitro and in vivo. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 5.Buggia-Prévot V, Fernandez CG, Udayar V, Vetrivel KS, Elie A, Roseman J, Sasse VA, Lefkow M, Meckler X, Bhattacharyya S, et al. A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer’s disease Aβ production. Cell Rep. 2013;5:1552–1563. doi: 10.1016/j.celrep.2013.12.006. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury A, Dominguez M, Puri V, Sharma DK, Narita K, Wheatley CL, Marks DL, Pagano RE. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest. 2002;109:1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choy RW, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid β (Aβ) production in the trans-Golgi network. Proc Natl Acad Sci USA. 2012;109:E2077–E2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chyung JH, Selkoe DJ. Inhibition of receptor-mediated endocytosis demonstrates generation of amyloid beta-protein at the cell surface. J Biol Chem. 2003;278:51035–51043. doi: 10.1074/jbc.M304989200. [DOI] [PubMed] [Google Scholar]
- 10.Das U, Scott DA, Ganguly A, Koo EH, Tang Y, Roy S. Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron. 2013;79:447–460. doi: 10.1016/j.neuron.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Strooper B. Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol Rev. 2010;90:465–494. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- 12.Duff K, Eckman C, Zehr C, Yu X, Prada CM, Perez-tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin 1. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 13.Dumanchin C, Czech C, Campion D, Cuif MH, Poyot T, Martin C, Charbonnier F, Goud B, Pradier L, Frebourg T. Presenilins interact with Rab11, a small GTPase involved in the regulation of vesicular transport. Hum Mol Genet. 1999;8:1263–1269. doi: 10.1093/hmg/8.7.1263. [DOI] [PubMed] [Google Scholar]
- 14.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 16.Frisoni GB, Hampel H, O’Brien JT, Ritchie K, Winblad B. Revised criteria for Alzheimer’s disease: what are the lessons for clinicians? Lancet Neurol. 2011;10:598–601. doi: 10.1016/S1474-4422(11)70126-0. [DOI] [PubMed] [Google Scholar]
- 17.Gagescu R, Demaurex N, Parton RG, Hunziker W, Huber LA, Gruenberg J. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell. 2000;11:2775–2791. doi: 10.1091/mbc.11.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvez T, Gilleron J, Zerial M, O’Sullivan GA. SnapShot: mammalian Rab proteins in endocytic trafficking. Cell. 2012;151:234–234.e232. doi: 10.1016/j.cell.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 20.He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- 21.Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Grønborg M, Möbius W, Rhee J, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 24.Kaether C, Schmitt S, Willem M, Haass C. Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic. 2006;7:408–415. doi: 10.1111/j.1600-0854.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- 25.Kalvodova L, Kahya N, Schwille P, Ehehalt R, Verkade P, Drechsel D, Simons K. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J Biol Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 26.Kametani F, Usami M, Tanaka K, Kume H, Mori H. Mutant presenilin (A260V) affects Rab8 in PC12D cell. Neurochem Int. 2004;44:313–320. doi: 10.1016/s0197-0186(03)00176-1. [DOI] [PubMed] [Google Scholar]
- 27.Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116:3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 28.Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 29.Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, Emery G. Evi5 promotes collective cell migration through its Rab-GAP activity. J Cell Biol. 2012;198:57–67. doi: 10.1083/jcb.201112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lichtenthaler SF. α-secretase in Alzheimer’s disease: molecular identity, regulation and therapeutic potential. J Neurochem. 2011;116:10–21. doi: 10.1111/j.1471-4159.2010.07081.x. [DOI] [PubMed] [Google Scholar]
- 31.Lindsay AJ, McCaffrey MW. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J Biol Chem. 2002;277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- 32.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxfield FR, Yamashiro DJ. Endosome acidification and the pathways of receptor-mediated endocytosis. Adv Exp Med Biol. 1987;225:189–198. doi: 10.1007/978-1-4684-5442-0_16. [DOI] [PubMed] [Google Scholar]
- 34.Morel E, Chamoun Z, Lasiecka ZM, Chan RB, Williamson RL, Vetanovetz C, Dall’Armi C, Simoes S, Point Du Jour KS, McCabe BD, et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nitsch RM, Slack BE, Farber SA, Borghesani PR, Schulz JG, Kim C, Felder CC, Growdon JH, Wurtman RJ. Receptor-coupled amyloid precursor protein processing. Ann N Y Acad Sci. 1993;695:122–127. doi: 10.1111/j.1749-6632.1993.tb23039.x. [DOI] [PubMed] [Google Scholar]
- 36.Onodera Y, Nam JM, Hashimoto A, Norman JC, Shirato H, Hashimoto S, Sabe H. Rab5c promotes AMAP1-PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J Cell Biol. 2012;197:983–996. doi: 10.1083/jcb.201201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 38.Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- 39.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 40.Prabhu Y, Burgos PV, Schindler C, Farías GG, Magadán JG, Bonifacino JS. Adaptor protein 2-mediated endocytosis of the β-secretase BACE1 is dispensable for amyloid precursor protein processing. Mol Biol Cell. 2012;23:2339–2351. doi: 10.1091/mbc.E11-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajendran L, Annaert W. Membrane trafficking pathways in Alzheimer’s disease. Traffic. 2012;13:759–770. doi: 10.1111/j.1600-0854.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 42.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajendran L, Schneider A, Schlechtingen G, Weidlich S, Ries J, Braxmeier T, Schwille P, Schulz JB, Schroeder C, Simons M, et al. Efficient inhibition of the Alzheimer’s disease beta-secretase by membrane targeting. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 44.Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci USA. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sannerud R, Declerck I, Peric A, Raemaekers T, Menendez G, Zhou L, Veerle B, Coen K, Munck S, De Strooper B, et al. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc Natl Acad Sci USA. 2011;108:E559–E568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlüter OM, Schmitz F, Jahn R, Rosenmund C, Südhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt MR, Haucke V. Recycling endosomes in neuronal membrane traffic. Biol Cell. 2007;99:333–342. doi: 10.1042/BC20070007. [DOI] [PubMed] [Google Scholar]
- 49.Schneider A, Rajendran L, Honsho M, Gralle M, Donnert G, Wouters F, Hell SW, Simons M. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 51.Seabra MC, Mules EH, Hume AN. Rab GTPases, intracellular traffic and disease. Trends Mol Med. 2002;8:23–30. doi: 10.1016/s1471-4914(01)02227-4. [DOI] [PubMed] [Google Scholar]
- 52.Selkoe DJ. Alzheimer’s disease. Cold Spring Harb Perspect Biol. 2011;3:a004457. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selkoe DJ. Resolving controversies on the path to Alzheimer’s therapeutics. Nat Med. 2011;17:1060–1065. doi: 10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- 54.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Shiba T, Koga H, Shin HW, Kawasaki M, Kato R, Nakayama K, Wakatsuki S. Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc Natl Acad Sci USA. 2006;103:15416–15421. doi: 10.1073/pnas.0605357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegenthaler BM, Rajendran L. Retromers in Alzheimer’s disease. Neurodegener Dis. 2012;10:116–121. doi: 10.1159/000335910. [DOI] [PubMed] [Google Scholar]
- 57.Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 59.Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spasic D, Annaert W. Building gamma-secretase: the bits and pieces. J Cell Sci. 2008;121:413–420. doi: 10.1242/jcs.015255. [DOI] [PubMed] [Google Scholar]
- 61.Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szodorai A, Kuan YH, Hunzelmann S, Engel U, Sakane A, Sasaki T, Takai Y, Kirsch J, Müller U, Beyreuther K, et al. APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. J Neurosci. 2009;29:14534–14544. doi: 10.1523/JNEUROSCI.1546-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tisdale EJ. Rab2 interacts directly with atypical protein kinase C (aPKC) iota/lambda and inhibits aPKCiota/lambda-dependent glyceraldehyde-3-phosphate dehydrogenase phosphorylation. J Biol Chem. 2003;278:52524–52530. doi: 10.1074/jbc.M309343200. [DOI] [PubMed] [Google Scholar]
- 66.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 67.Vassar R, Kovacs DM, Yan R, Wong PC. The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Arnim CA, Spoelgen R, Peltan ID, Deng M, Courchesne S, Koker M, Matsui T, Kowa H, Lichtenthaler SF, Irizarry MC, Hyman BT. GGA1 acts as a spatial switch altering amyloid precursor protein trafficking and processing. J Neurosci. 2006;26:9913–9922. doi: 10.1523/JNEUROSCI.2290-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakabayashi T, Craessaerts K, Bammens L, Bentahir M, Borgions F, Herdewijn P, Staes A, Timmerman E, Vandekerckhove J, Rubinstein E, et al. Analysis of the gamma-secretase interactome and validation of its association with tetraspanin-enriched microdomains. Nat Cell Biol. 2009;11:1340–1346. doi: 10.1038/ncb1978. [DOI] [PubMed] [Google Scholar]
- 70.Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willem M, Lammich S, Haass C. Function, regulation and therapeutic properties of beta-secretase (BACE1) Semin Cell Dev Biol. 2009;20:175–182. doi: 10.1016/j.semcdb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 72.Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL, Beach T, Sue L, Wong P, Price D, et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- 73.Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.