Abstract
Type I interferons (IFNs) are multifunctional cytokines that regulate immune responses and cellular functions but also can have detrimental effects on human health. A tight regulatory network therefore controls IFN signaling, which in turn interferes with medical interventions. The JAK-STAT signaling pathway transmits the IFN extracellular signal to the nucleus for alterations of gene expression. STAT2 is a well-known essential and specific positive effector of type I IFN signaling. Here, we report that STAT2 is also a previously unrecognized crucial component of the USP18-mediated negative feedback control in both, human and murine cells. We found that STAT2 recruits USP18 to the type I IFN receptor subunit IFNAR2 via its constitutive membrane-distal STAT2 binding site. This mechanistic coupling of effector and negative feedback functions of STAT2 provides novel strategies in treatment of IFN signaling related human diseases.
Introduction
Type I interferon (IFN) signaling has emerged as a highly complex regulatory network coordinating the host’s defense against pathogens and cancer via expression of over 300 IFN-stimulated genes (ISGs) 1,2. Proteins encoded by ISGs include cytokines and chemokines that modulate innate and adaptive immune responses, enzymes that specifically block growth and survival of pathogens, and transcription factors and other regulators that affect cell proliferation and survival. Many studies from human genetic diseases and mouse models have demonstrated that IFNs are essential for immune responses against infections and cancer development 3,4. Therefore, IFNs have been successfully used to treat viral infections, auto-immune disorders, and cancer 5. Most recently, it has been revealed that autonomous IFN responses in cancer cells are required for successful anticancer therapies, including conventional chemotherapies, targeted anticancer treatment, radiotherapy, and immunotherapy 4,6. However, it is also known that high dose IFN therapies cause severe acute and chronic side effects 7,8. Furthermore, excess IFN production or dysregulated IFN signaling contributes to pathogenic process in patients with systemic lupus erythematosus, Sjogren’s syndrome, systemic sclerosis, rheumatoid arthritis and in the rare genetic disorders known as interferonopathies 9,10. Together, these findings indicate that accurate fine-tuning of the IFN system is crucial for human health and for therapeutic interventions.
The binding of type I IFNs to the receptor subunits IFNAR1 and IFNAR2 induces the activation of their associated Janus family tyrosine kinases TYK2 and JAK1, respectively 11. Activated TYK2 and JAK1 in turn phosphorylate IFNAR2-associated STAT2 and STAT1, which results in formation of the DNA binding STAT1/STAT2/IRF9 ternary complex IFN-stimulated gene factor 3 (ISGF3). ISGF3 promotes expression of genes with the IFN-stimulated response element in their promoters 12,13. Signaling patterns elicited by type I IFNs strongly depend on the cellular and physiological context 14. An intricate interplay of receptor dimerization dynamics and spatiotemporal modulation of IFN signaling by multiple positive and negative intracellular regulators 1,15 and by endocytosis 16 likely contribute to signaling plasticity 17. Among these regulators, the ubiquitin-specific protease USP18, which we identified during analysis of gene expression in a leukemia fusion protein mouse model 18,19, plays a most intriguing role. USP18 is an enzyme that removes an ubiquitin-like modifier, ISG15, from conjugated proteins 20. However, USP18 expression is strongly stimulated by IFN treatment and exerts negative regulation of type I interferon signaling, which is independent of its enzymatic activity 21. By competing with JAK1 for binding IFNAR2, USP18 may interfere with cytosolic stabilization of signaling complexes, which is likely mediated by the Janus kinases. It thereby reduces ligand binding, receptor dimerization and downstream signaling in a complex, IFN affinity-dependent manner 21–23. Interestingly, in human cells, ISG15 directly regulates USP18 stability 24. Furthermore, critical functions of USP18 in IFN mediated immune responses are demonstrated in mouse and human models 25–32 suggesting the broad impact of USP18 in immune responses and therapeutic potential of modulating USP18 inhibitory effects.
While quantifying effector interactions with IFNAR2 by live cell protein micropatterning assays, we recently observed that recruitment of STAT2 is affected by USP18 33, suggesting a functional crosstalk between these proteins. Among the seven mammalian STAT proteins, which are activated by diverse cytokines 34, STAT2 is unique in being selectively involved in type I and type III IFN signaling. Here, we investigated in detail the role of STAT2 in IFNAR desensitization by USP18 using live cell micropatterning for real time protein interaction assays and single-molecule imaging in combination with protein biochemical approaches. We found that, beyond being a key effector of IFN signaling, STAT2 is essential for USP18-mediated inhibition of JAK-STAT signaling. STAT2 directly interacts with USP18 and thus mediates its recruitment to IFNAR2. In turn, anchored USP18 interferes with receptor dimerization and JAK phosphorylation. Elucidating this previously unrecognized requirement for STAT2 in negative feedback regulation will expand the potential for local or systemic modulation of IFN signaling in treating human disease.
Results
USP18 interacts with STAT2
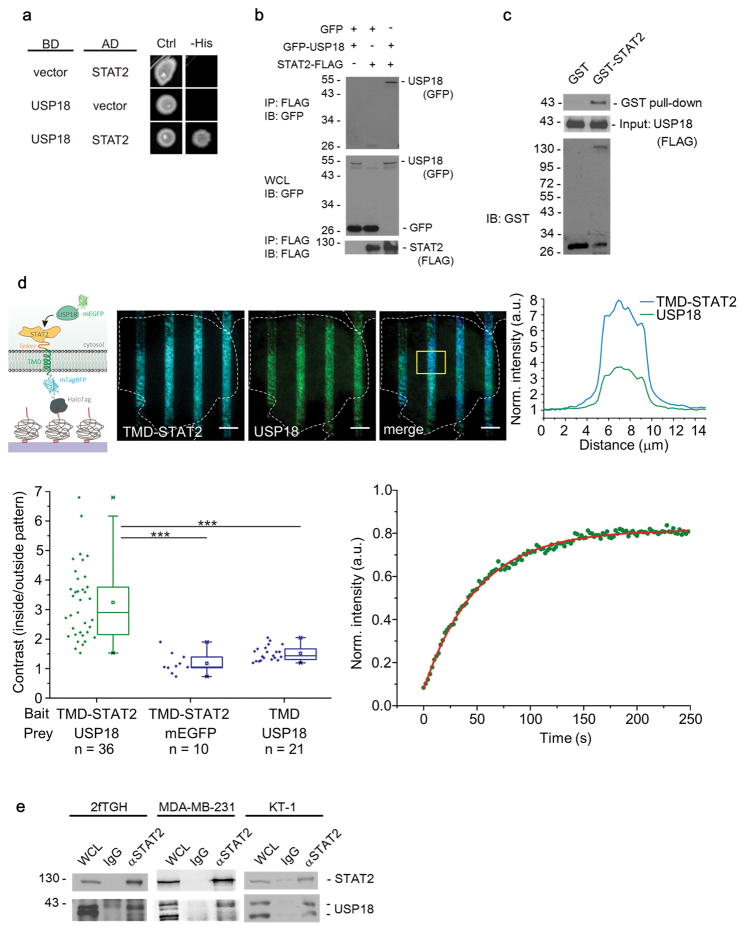
The role of USP18 in IFN signaling is independent of its ISG15 deconjugating activity but relies on its interaction with IFNAR2 21,35. To identify proteins that may regulate USP18 function in the IFN signaling pathway, we conducted a yeast two-hybrid screen using the wild type human, full-length USP18 and its active site mutant (C64A) as bait proteins. We identified 11 independent clones encoding STAT2. Likewise, direct interaction of STAT2 and USP18 was detected in a targeted yeast two-hybrid assay (Fig. 1a). Co-immunoprecipitation showed interaction between exogenously expressed USP18 and STAT2 (Fig. 1b). Furthermore, a pull-down analysis revealed the direct binding of biochemically purified STAT2 and USP18 (Fig. 1c).
Figure 1. USP18 interacts with STAT2.
(a) Yeast two-hybrid analysis of the direct interaction between USP18 and STAT2. (a),(b) Overnight cultures from strains pJ69-4a/pJ69-4alpha carrying two-hybrid plasmids with the indicated coding sequences were spotted onto selective media for plasmid maintenance (-Leu, -Tryp = Ctrl for growth control) and indicators for interactions (-His, in addition to -Leu, -Tryp).
(b) Immunoblot (IB) analysis of whole cell lysates (WCL) and anti-FLAG immunoprecipitates (IP) derived from 293T cells 24 hours after co-transfection with plasmids encoding GFP, GFP-USP18, and STAT2-FLAG.
The number for left side of column shows molecular weight (kDa).
(c) GST pull-down assay to demonstrate that the STAT2 directly associate with USP18.
(d) Immobilization of STAT2 for probing direct interaction with USP18 as depicted in the cartoon. U5A cells transfected with mEGFP-USP18 (green channel) and STAT2 intracellular fused to a transmembrane domain (TMD) and extracellular fused to mTagBFP as well as HaloTag (blue channel). Scale bars: 10 μm. Representative image of 36 cells analyzed. Intensity profiles of all channels within the yellow ROI depicted in the merged image (right panel). Quantitative analysis of the recruitment of mEGFP-USP18 to micropatterned STAT2 as determined by the contrast of the fluorescence intensities inside and outside the patterns. As negative controls, U5A cells were transfected with mEGFP-USP18 and HaloTag-mTagBFP-TMD as well as mEGFP and HaloTag-mTagBFP-TMD-STAT2. n: number of cells analyzed obtained from two independent experiments. Significance was quantified using the two-sample Kolmogorov-Smirnov test. *** P < 0.001. (lower left plot). Fluorescence recovery of USP18 recruited to micropatterned STAT2 and monoexponential fit of the recovery curve (representative of 8 cells analyzed), lower right plot).
(e) Interaction of endogenous USP18 and endogenous STAT2 in 2fTGH, MDA-MB-231, and KT-1 cells. Lysates from cells treated with IFNα (1000 U/ml) for 24 hours were immunoprecipitated with IgG or anti-STAT2 and immunoblotted with STAT2 or USP18 antibodies.
To quantify the interaction between STAT2 and USP18 in live cells, a cell micropatterning approach for spatially controlled immobilization of bait proteins in the plasma membrane via the HaloTag 33 was employed (Supplementary Fig. 1a). This technique quantifies equilibrium binding to a target protein within the cell in situ and thus nicely complements co-immunoprecipitation data with respect to their functional relevance. Co-localization of STAT2 and USP18 together with micropatterned IFNAR2 confirmed constitutive interaction of both STAT2 and USP18, with IFNAR2 (Supplementary Fig. 1b–d). Interaction dynamics were analyzed by fluorescence recovery after photobleaching (FRAP) revealing similar rate constants for STAT2 (τSTAT2: 79 s ± 28 s) and USP18 (τUSP18: 103 s ± 33 s, Supplementary Fig. 1e), suggesting simultaneous interaction of both proteins with IFNAR2. We next examined IFNAR2-independent interaction of STAT2 and USP18 in IFNAR2-deficient U5A cells by using cell micropatterning. A fusion protein of STAT2 with a transmembrane domain (TMD) as well as extracellular mTagBFP and HaloTag (HaloTag-mTagBFP-TMD-STAT2) was used as bait (Fig. 1d). Co-expression of this construct with mEGFP-tagged USP18 revealed strong co-localization within micropatterns as quantified by the contrast between the patterned and non-patterned regions in the cells. No significant contrast was observed when only mEGFP was co-expressed with STAT2 as bait or when USP18 was co-transfected with a transmembrane domain that was not fused to STAT2 (Fig. 1d, boxplot). Monitoring the exchange kinetics of intracellular USP18 bound to micropatterned STAT2 by FRAP (Fig. 1d) yielded a mean interaction lifetime of τ: 53 s ± 28 s (8 cells analyzed). Furthermore, using 2fTGH, MDA-MB-231, and KT-1 cells, we confirmed the interaction of endogenous USP18 and STAT2 by co-immunoprecipitation assays (Fig. 1e). Together, these different approaches clearly established that STAT2 directly binds to USP18.
STAT2 is required for USP18-mediated inhibition of type I IFN signaling
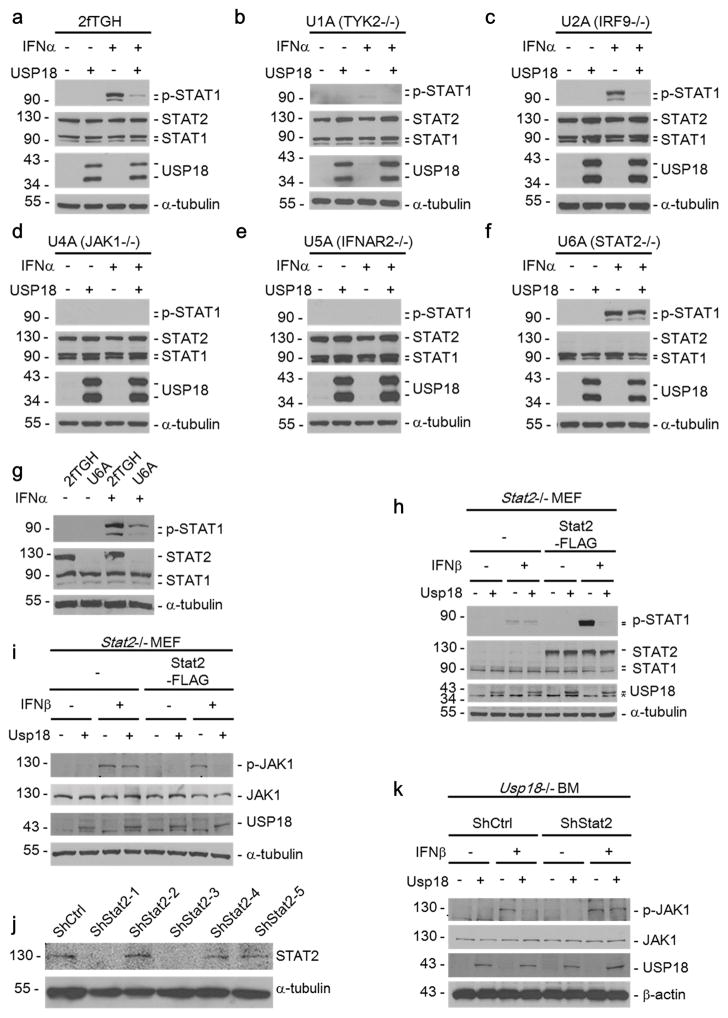
To examine the role of STAT2 in USP18-mediated desensitization of type I IFN signaling, we employed U series cell lines derived from human fibrosarcoma 2fTGH cells, U1A (TYK2−/−), U2A (IRF9−/−), U4A (JAK1−/−), U5A (IFNAR2−/−), and U6A (STAT2−/−) 36,37. These cells were stably transduced with the MIP control or MIP-USP18 (Fig. 2a–e). As expected, in cells lacking TYK2 (U1A), JAK1 (U4A), and IFNAR2 (U5A), phosphorylation of STAT1 was not observed after IFNα treatment (Fig. 2b, 2d, and 2e) since important components of the signaling pathway were missing. In 2fTGH and U2A cells, USP18 expression clearly reduced STAT1 phosphorylation upon IFNα treatment (Fig. 2a and 2c), indicating that USP18 inhibits IFN signaling upstream of IRF9, as reported previously 21. Interestingly, expression of USP18 did not affect STAT1 phosphorylation in STAT2 deficient U6A cells (Fig. 2f), suggesting that STAT2 is required for USP18-mediated inhibition. It should be noted that IFN-induced STAT1 phosphorylation was much weaker in U6A cells than in 2fTGH cells, since STAT2 supports STAT1 phosphorylation (Fig. 2g) 38. To further verify the critical role of STAT2, we used murine embryonic fibroblasts derived from Stat2 knockout mice (Stat2−/− MEF). In these cells, ectopic Usp18 did not exert an inhibitory effect on IFNβ-induced STAT1 phosphorylation (Fig. 2h, lanes 1–4). STAT2 transduction strongly increased IFNβ-stimulated STAT1 phosphorylation, but in this context ectopic USP18 exerted a strong inhibitory effect (Fig. 2h, lanes 5–8). These results indicate that, in addition to physically interacting with USP18, STAT2 plays a critical role in mediating the negative effect of USP18 on Type I IFN signaling.
Figure 2. STAT2 is required for USP18-mediated inhibition of Type I IFN signaling.
(a–f) MIP or MIP-USP18 expressing 2fTGH, U1A, U2A, U4A, U5A, and U6A cells were treated with IFNα (1000 U/ml) for 15 minutes. The cell lysates were immunoblotted with the indicated antibodies.
(g) IB analysis of STAT1 phosphorylation in 2fTGH and U6A cells in the presence or absence of (1000 U/ml) IFNα.
(h–i) Stat2−/− MEFs were infected with MIP control (−) or MIP-Usp18 (+) retroviruses, either in the presence or absence of rescue with C-terminally FLAG-tagged Stat2 cDNA. Where indicated cells were treated with either mouse IFNβ (500 U/ml) for 15 minutes (h) or 30 minutes (i), before cell lysates were collected and analyzed by Western blotting with indicated antibodies.
(j) Validation of Stat2 knockdown. Ba/F3 cells were infected with control or Stat2-targeting shRNA lentivirus. After 5 days puromycin selection, Stat2 expression was examined by Western blotting.
(k) Usp18−/− bone marrow cells were infected with pCX4-bsr control (−) or pCX4-bsr-Usp18 (+), either in the presence or absence of control (shCtrl) or Stat2-knockdown (shStat2–3) shRNA expression. Two days following double drug selection (puromycin and blasticidin), cells were either left untreated (−) or treated (+) with mouse IFNβ (500 U/ml) for 30 minutes. Cell lysates were collected and analyzed by Western blotting with indicated antibodies.
To investigate whether the inhibitory effect of STAT2 plus USP18 can be detected upstream of STAT1 phosphorylation, we examined JAK1 phosphorylation. Exogenously expressed USP18 did not affect JAK1 phosphorylation in Stat2−/− MEFs, but clearly diminished JAK1 phosphorylation in Stat2-transduced Stat2−/− MEFs (Fig. 2i), indicating that STAT2 is critical for USP18-mediated inhibition of JAK1 phosphorylation. To confirm this finding, we also selected an effective Stat2 shRNA to knockdown Stat2 expression (Fig. 2j). Neither control shRNA nor Stat2-specific shRNA affected IFNβ-induced JAK1 phosphorylation in Usp18−/− primary murine bone marrow cells (Fig. 2k). In contrast, expressing a Stat2 shRNA, but not a control shRNA, abolished the negative effect of exogenously expressed USP18 on JAK1 phosphorylation. These results further support that STAT2 is required for USP18-mediated inhibition, which is upstream of STAT phosphorylation.
To further examine the mechanistic requirement for STAT2 in USP18-dependent negative regulation of IFN signaling we examined expression of 23 IFN-inducible genes in Stat2−/− MEFs transduced with control- or Usp18- expression vectors, and with or without exogenous reintroduction of Stat2. As expected, the majority of genes tested (21 of 23) were not induced by IFN treatment in the absence of STAT2 (representative genes shown in Supplementary Figure 2a). We did, however, identify two genes (Irf9 and Cxcl9) that showed weak induction of expression in the absence of STAT2 (Supplementary Figure 2b). Importantly, USP18-mediated inhibition of gene expression for these ISGs was only observed upon reintroduction of STAT2 (Supplementary Figure 2a and 2b). These observations fully support the essential biological role of STAT2 for IFN-responsive signal transduction 39. Furthermore, both of our biochemical and gene expression analyses demonstrate the mechanistic requirement for STAT2 in USP18-mediated inhibition of interferon signaling.
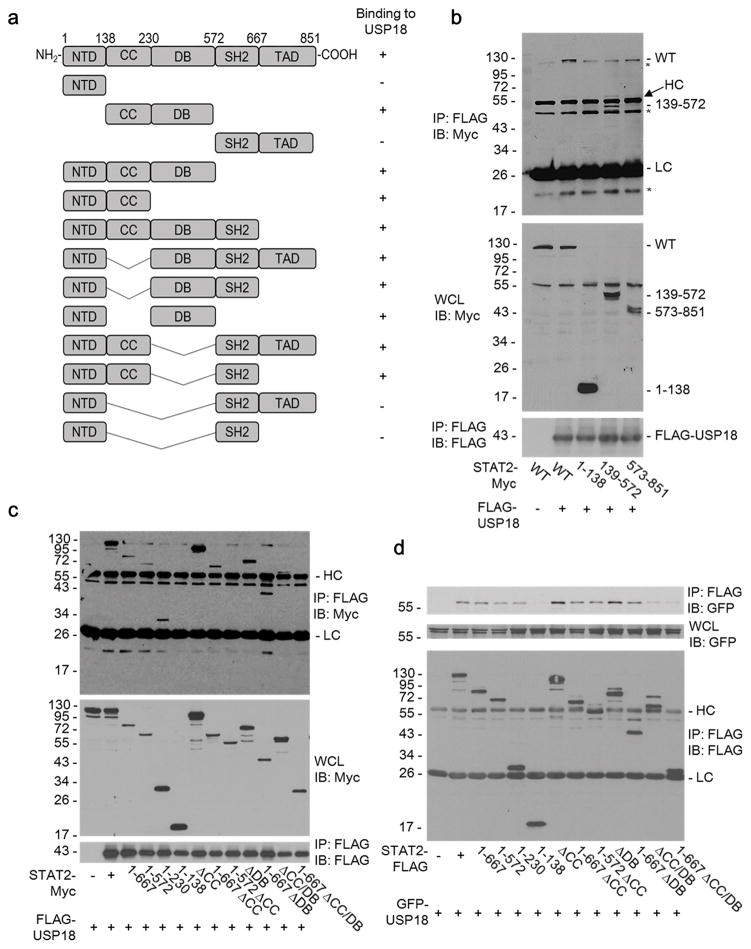
Both coiled-coil (CC) and DNA binding (DB) domains of STAT2 are involved in STAT2 interaction with USP18
Structurally, STAT2 can be divided into N-terminal (NTD), coiled-coil (CC), DNA binding (DB), linker domain (LD), Src homology 2 (SH2) dimerization, and C terminal transactivation domains (Fig. 3a). In this study, we included the linker domain in DB, since it has been reported that the linker domain is required for the appropriate DB structure 40. To understand the mechanism of STAT2 in USP18-mediated inhibition of IFN signaling for potential therapeutic applications, we performed co-immunoprecipitation assays using a set of STAT2 deletion mutants (Fig. 3a). FLAG-tagged USP18 (FLAG-USP18) was coexpressed with Myc-tagged STAT2 (STAT2-Myc) or either of three fragments of STAT2 (aa 1-136 of NTD, aa 139-572 with CC and DB domains, and aa 573-861 containing SH2 and transactivation domains). NTD and the C-terminal region of STAT2 did not co-precipitate with USP18. However, the protein with only CC and DB domains of STAT2 significantly interacted with USP18 (Fig. 3b). Furthermore, using USP18 and additional mutants of STAT2, immunoprecipitation of either USP18 (Fig. 3c) or STAT2 (Fig. 3d) revealed that the CC-only or DB-only-deleted STAT2 were able to associate with USP18. However, deletion of both CC and DB domains of STAT2 led to a loss of co-precipitation with USP18. Thus, results from this series of interaction analyses suggest that both CC and DB domains of STAT2 contribute to its interaction with USP18.
Figure 3. Both the coiled-coil (CC) and DNA binding (DB) domains of STAT2 are involved in the interaction with USP18.
(a) A schematic drawing of STAT2 domain structure and the associated deletion mutants used in this study. The ability of a given deletion mutant to interact with USP18 (+ or − binding) is indicated to the right.
(b–c) IB analysis of WCL and anti-FLAG IP derived from 293T cells 24 hours after co-transfection with plasmids encoding FLAG-USP18 and either the full-length STAT2-Myc (WT) or the indicated deletion mutants.
(d) IB analysis of WCL and anti-FLAG IP derived from 293T cells 24 hours after co-transfection with plasmids encoding GFP-USP18 and either STAT2-FLAG or the indicated deletion mutants. Asterisks are used to indicate non-specific bands. HC = Heavy Chain, LC = Light Chain.
Coiled-coil and DNA binding domains of STAT2 are important for USP18-mediated inhibition of type I IFN signaling
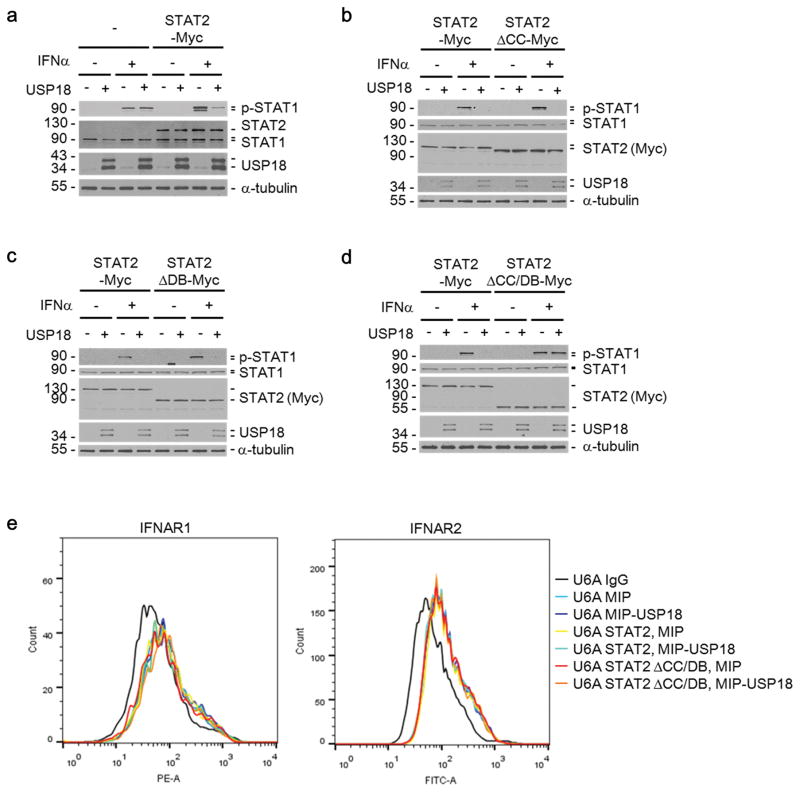
To examine the role of the STAT2-USP18 interaction in negative feedback regulation of IFN signaling, we expressed in U6A cells an empty vector control, full length STAT2, or deletion mutants of STAT2. USP18 expression reduced STAT1 phosphorylation in STAT2 expressing U6A cells (Fig. 4a). Furthermore, the negative effect of USP18 was detected in the presence of STAT2 lacking the CC domain (Fig. 4b) or the DB domain (Fig. 4c). In contrast, although STAT2 lacking both CC and DB domains still promoted IFNα-induced STAT1 phosphorylation, it did not support the inhibitory function of USP18 (Fig. 4d), suggesting that the interaction of these two proteins via CC and DB domains of STAT2 is crucial for the effect of USP18 on IFN signaling. Importantly, expression of USP18 and different STAT2 constructs did not affect cell surface levels of the IFN receptor subunits IFNAR1 and IFNAR2 in these cell lines (Fig. 4e).
Figure 4. The coiled-coil (CC) and DNA binding (DB) domains of STAT2 are important for USP18-mediated inhibition of Type I IFN signaling.
(a) U6A cells, stably transduced with control (−) or C-terminally Myc-tagged STAT2 (STAT2-Myc), were infected with MIP control (−) or MIP-USP18 (+) retrovirus. With or without IFNα (1000 U/ml) treatment for 15 minutes cell lysates were collected and analyzed by Western blotting with the indicated antibodies.
(b–d) U6A cells, stably transduced to express either full-length STAT2 (STAT2-Myc), or the indicated STAT2 deletion mutant (b, STAT2ΔCC-Myc; c, STAT2ΔDB-Myc; d, STAT2ΔCC/DB-Myc) with or without IFNα (1000 U/ml) treatment for 15 minutes cell lysates were collected and analyzed by Western blotting with the indicated antibodies.
(e) Histograms showing the surface expression of IFNAR1 and IFNAR2 following infection with indicated constructs in U6A cell lines.
The N- and C-terminal regions of USP18 bind to STAT2 and IFNAR2 and are important for inhibiting the IFN response
Based on our previous report 21 and the results presented above, USP18 interacts with both IFNAR2 and STAT2. We therefore assessed which regions of USP18 were required for its interaction with STAT2 and IFNAR2. We generated six constructs for expression of selected regions of USP18 (Supplementary Fig. 3a and 4a). When co-immunoprecipitation assays were conducted with cell lysates containing FLAG-tagged USP18 and Myc-tagged STAT2, aa 1-112, 51-242, 1-242, and 243-312, but not aa 113-242 and 313-372 of USP18 interacted with STAT2, suggesting that aa 51-112 and 243-312 of USP18 are two important regions for the STAT2-USP18 interaction (Supplementary Fig. 3b). Further analysis revealed that aa 303-312 of USP18 interacted with STAT2 (Supplementary Fig. 3c). Consequently, an aa 303-312 deletion mutant of USP18 (USP18Δ303-312) was unable to suppress IFNα-induced STAT1 phosphorylation (Supplementary Fig. 3d).
We also established human KT-1 cell lines stably expressing wild-type USP18 or the non-interacting mutant USP18Δ303-312. Upon addition of IFNα, cells expressing wild-type USP18-showed strong inhibition of phosphorylation of JAK1 and STAT1. USP18Δ303-312 did not show such a negative effect (Supplementary Fig. 3e), in agreement with its loss of STAT2 interaction.
Regarding USP18 and IFNAR2 interaction, peptides comprising aa 1-112, 1-242, and 313-372, but not aa 51-242, 113-242, and 243-312 of USP18 co-immunoprecipitated with the intracellular domain (ICD) of IFNAR2 (Supplementary Fig. 4b). These results suggest that aa 1-51 and aa 313-372 of USP18 are important for the IFNAR2-USP18 interaction. Further analysis narrowed the first interaction domain to aa 36-51 of USP18 (Supplementary Fig. 4c and 4d). In line with this result, expression of aa 36-372, but not aa 51-372, of USP18 inhibited STAT1 phosphorylation upon IFNα treatment (Supplementary Fig. 4e). Taken together, these findings indicate that aa 36-51 and 313-371 of USP18 are critical for the USP18-IFNAR2 interaction and that aa 51-112 and 303-312 of USP18 are important for the USP18-STAT2 interaction. We therefore conclude that the N- and C-terminal regions (aa 36-51 and 317-371) of USP18 play important roles in the interaction with IFNAR2, and the adjacent regions (aa 51-112 and 303-312) are critical for USP18 binding to STAT2.
STAT2 recruits USP18 to IFNAR2
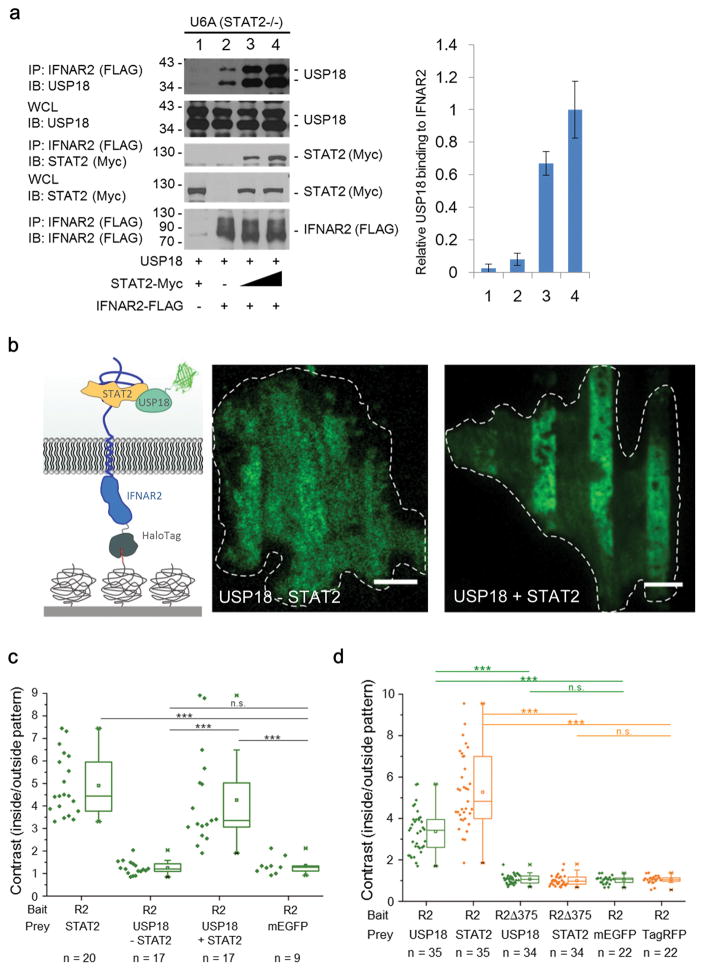
These results established that USP18 independently interacts with IFNAR2 and STAT2. Since STAT2 itself constitutively interacts with IFNAR2 33,41,42, we aimed to further examine whether interaction between USP18 and IFNAR2 was affected by STAT2. In STAT2-deficient U6A cells, the USP18-IFNAR2 interaction was up to 10-fold enhanced upon expression of STAT2 (Fig. 5a). The importance of STAT2 for USP18 recruitment could also be verified by live cell micropatterning: while no binding of USP18 to micropatterned IFNAR2 was detectable in U6A cells, a strong increase in contrast after complementation with STAT2 was observed (Fig. 5b and 5c). In HeLa cells, the endogenous expression level of STAT2 was sufficient to yield significant binding of USP18 bound to micropatterned IFNAR2 (Supplementary Fig. 5c). However, substantially increased USP18 binding was observed upon ectopic co-expression of STAT2. These results highlight the critical role of STAT2 concentration in effective recruitment of USP18 to IFNAR2. In absence of STAT2 binding of USP18 was weakened such as no significant recruitment of USP18 to micropatterned IFNAR2 was detectable in this experimental system. A comparable loss in USP18 binding to IFNAR2 was obtained in presence of STAT2 upon deletion of the STAT2 binding site on IFNAR2, which was suggested to include at least amino acids 418-444 41. The recruitment of STAT2 and USP18 to IFNAR2, C-terminally truncated at position 375, was strongly reduced (Fig. 5d and Supplementary Fig. 5c). Mapping the STAT2 and USP18 binding to IFNAR2 by cell micropatterning of further deletions and mutations (Supplementary Fig. 5d–f) confirmed aa 418-444 of IFNAR2 as a minimal interaction site for STAT2 and USP18. These results established that the interaction of USP18 and STAT2 is responsible for recruitment of USP18 to IFNAR2 and is critical for the negative effect of USP18 on type I IFN-induced JAK1 phosphorylation (Fig. 2i–k), which is upstream of type I IFN-induced STAT1 activation.
Figure 5. STAT2 recruits USP18 to IFNAR2.
(a) IB analysis of WCL and anti-FLAG IP derived from U6A cells 24 hours after co-transfection with plasmids encoding USP18, FLAG-IFNAR2, and increasing amounts of STAT2-Myc expression construct. The relative USP18 binding to IFNAR2 from three independent experiments was quantified and plotted as the ratio of IFNAR2-bound USP18 to total USP18 (right panel). Data are normalized to the maximum binding (lane 4).
(b) Recruitment of USP18 and STAT2 to micropatterned IFNAR2 in STAT2-deficient U6A cells as illustrated in the cartoon. U6A cells transfected with HaloTag-mTagBFP-IFNAR2 and mEGFP-USP18 (USP18 –STAT2) (green channel, left image) and U6A cells transfected with HaloTag-mTagBFP-IFNAR2, mEGFP-USP18 and STAT2-TagRFP-T (USP18 +STAT2) (green channel, right image). Representative images of 17 cells analyzed in two independent experiments.
(c) Recruitment of USP18 to immobilized IFNAR2 (R2) into micropatterns was quantified in U6A cells by determining the contrast of the fluorescence intensities inside and outside the patterns. For comparison, constitutive binding of STAT2 (positive control) and cytosolic mEGFP (negative control) to micropatterned full length IFNAR2 expressed in U6A cells was quantified. n: number of cells analyzed in two independent experiments. Significance was quantified using the two-sample Kolmogorov-Smirnov test. *** P < 0.001, n.s., not significant.
(d) The C-terminal STAT2 interacting region of IFNAR2 is necessary for recruiting USP18. Quantification of recruitment of mEGFP-USP18 co-expressed with STAT2-TagRFP-T to immobilized HaloTag-IFNAR2 or C-terminally truncated HaloTag-IFNAR2 (R2Δ375) in Hela cells. As negative controls, HeLa cells were transfected with cytosolic mEGFP or TagRFP-T, respectively. n: number of cells analyzed in two independent experiments. Significance was quantified using the two-sample Kolmogorov-Smirnov test. *** P < 0.001, n.s. not significant.
STAT2-USP18 interaction regulates ternary complex assembly of the type I IFN receptor
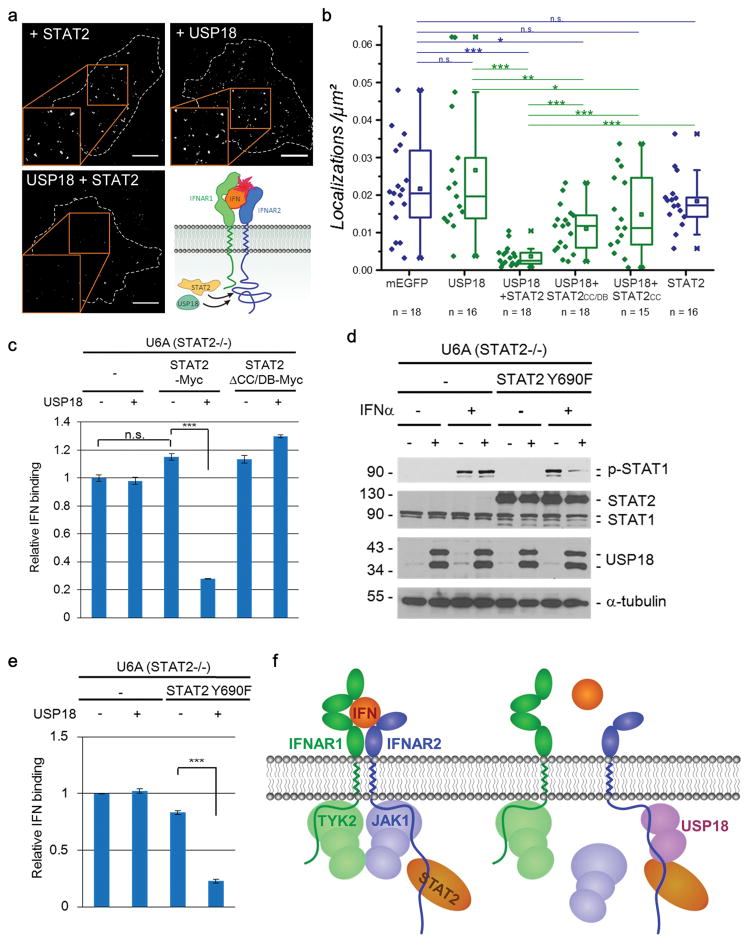
We recently reported that human USP18 negatively regulates the binding affinity of type I IFNs to their cell surface receptor, thus reducing the responsiveness of IFN-primed cells to subsequent IFN stimulation 22,23. Quantitative single molecule dimerization assays revealed that USP18 interferes with the assembly of the ternary IFN/IFNAR1/IFNAR2 complex, which explains the loss in ligand binding affinity 22. To explore whether the STAT2-USP18 interaction is important for the effect of USP18 on ligand binding and ternary complex formation, ligand binding assays in U6A cells were performed. To this end, we quantified at a single molecule level the binding of IFNα2 M148A labeled with DY647 (DY647IFNα2 M148A). This ligand requires simultaneous interaction with both IFNAR1 and IFNAR2 for binding to the cell surface receptor and thus indirectly probes ternary complex formation 22,43. In contrast to previous experiments performed in HeLa or IFNAR2-deficient U5A cells 22, no substantial difference in the amount of DY647IFNα2 M148A bound to the cell surface receptor was detected in U6A cells that express USP18 (Fig. 6a and 6b). This experiment suggests impaired negative regulation by USP18 in absence of STAT2. By contrast, complementation by co-expression of STAT2 significantly decreased the number DY647IFNα2 M148A on the cell surface as expected for effective desensitization by USP18. Complementation with transiently transfected STAT2, without USP18, had no effect on ligand binding. For both, CC-DB and CC fragments of STAT2 a significant support of USP18-mediated negative regulation of ternary complex formation, confirming the relevance of the STAT2-USP18 interaction for inhibiting IFNAR assembly. These results corroborate that the presence of both proteins, STAT2 and USP18, is required for USP18-mediated inhibition of ternary complex formation at the plasma membrane.
Figure 6. STAT2-USP18 interaction regulates ternary complex assembly of the Type I IFN receptor.
(a) Binding of DY647IFNα2 M148A bound to the IFNAR at the surface of U6A cells expressing STAT2-TagRFP-T (top-left), USP18-mEGFP (top-right) or both (bottom-left). The images are superimpositions of single molecule localizations from 100 consecutive frames. Scale bars: 10 μm. Representative images of 13–18 cells analyzed for each condition.
(b) Comparison of the density of DY647IFNα2 M148A bound to cell surface IFNAR of U6A cells expressing STAT2, USP18 or both proteins. Furthermore, protein with only STAT2 CC domain or with only CC and DB domains of STAT2 were also used in the assay. As a control, localizations of DY647IFNα2 M148A on the surface of U6A cells transfected with mEGFP were quantified. Data were acquired in two independent experiments, n indicates number of cells analyzed for each condition. Significance was quantified using the two-sample Kolmogorov-Smirnov test. *** P < 0.001; **P < 0.01; * P < 0.05, n.s. not significant.
(c) Relative amount of FITC-labeled IFNα bound to the surface of U6A cells was examined by flow cytometry. Cells are transduced to express the indicated constructs. Data are normalized to the mean fluorescence intensity (MFI) of U6A cells in the absence of STAT2 and USP18.
(d) U6A cells, stably transduced with control (−) or C-terminally Myc-tagged STAT2 Y690F mutant (STAT2 Y690F), were infected with MIP (−) or MIP-USP18 (+) retrovirus. With or without IFNα (1000 U/ml) treatment for 15 minutes, cell lysates were analyzed by Western blotting.
(e) Relative amount of FITC-labeled IFNα bound to the surface of U6A cells was examined by flow cytometry. Data are normalized to the MFI of U6A cells in the absence of STAT2 and USP18. Data of figure 6 (c) and (e) are presented as mean ± S.E.M. for three independent experiments. ***P < 0.001., n.s. not significant.
(f) Model of USP18-STAT2 regulating IFN response.
Furthermore we analyzed the effective cell surface binding affinity of FITC-labeled IFNα to cell surface IFNAR by flow cytometry. FITC-labeled IFNα showed concentration-dependent binding to 2fTGH cells, but did not bind to IFNAR2-deficient U5A cells (Supplementary Fig. 6a and 6b). FITC-labeled IFNα had similar biological activity to non-labeled IFNα (Supplementary Fig. 6c). These results demonstrate that FITC-labeled IFNα is suitable for testing the IFN-receptor interaction. We ectopically expressed USP18, STAT2, and STAT2ΔCC/DB, which loses its USP18-binding ability (Fig. 3), in U6A cells. USP18 decreased IFNα binding to U6A cells only in the presence of full length STAT2 but not STAT2ΔCC/DB (Fig. 6c), supporting the notion that interaction of STAT2 and USP18 is important for the type I IFN ligand-receptor binding. Phosphorylation of tyrosine (Y) 690 in STAT2 is not only critical for ISGF3 formation and ISG regulation, but also for STAT2-mediated STAT1 phosphorylation 44. Accordingly, expression of the STAT2 mutant Y690F in U6A cells did not enhance IFNα-induced STAT1 phosphorylation (Fig. 6d). Since the STAT2-USP18 interaction is not affected by this mutation (Supplementary Fig. 6d), USP18 expression still reduced phosphorylation of STAT1 in STAT2 Y690F-expressing U6A cells stimulated with IFNα (Fig. 6d). Importantly, expression of USP18 also decreased the level of IFNα binding to STAT2 Y690F-expressing U6A cells (Fig. 6e). Together, these results establish that the interaction between USP18 and STAT2 mediates the inhibitory effect of USP18 on type I IFN receptor assembly and signaling. Our data suggest that USP18 is recruited to IFNAR2 via its interaction with the STAT2 CC and DB domains to allow an additional interaction of USP18 with the membrane-proximal domain of IFNAR2 (Fig. 6f), which probably causes JAK1 dissociation 21.
Targeted disruption of the STAT2-USP18 interaction enhances IFN-dependent response
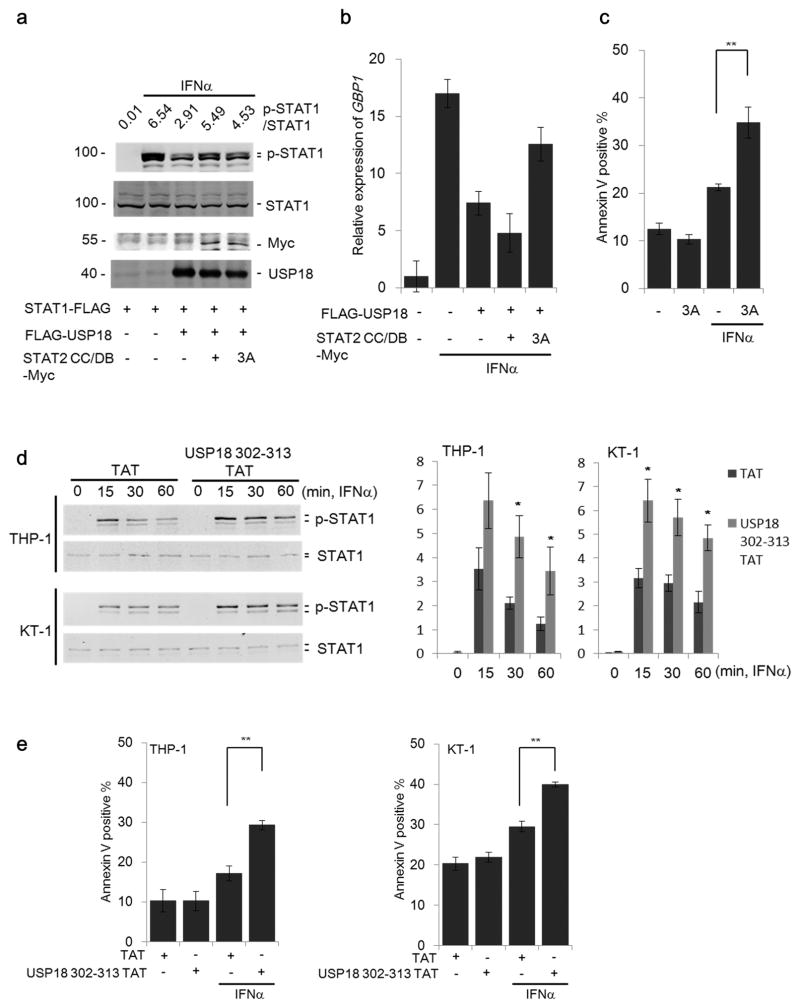
IFNs modulate crucial immune responses during pathogen infection and against malignant cells, which are effectively abrogated by expression of USP18. Therefore, we next examined whether a peptide comprising the STAT2 CC and DB domains can block the USP18-STAT2 interaction and thus maintain IFN signaling responses in presence of USP18. To this end, STAT1, USP18, and the STAT2 CC/DB fragment were co-expressed in HEK 293T cells (Fig. 7a). As expected, STAT2 CC/DB counteracted the negative effect of USP18, resulting in increased STAT1 phosphorylation.
Figure 7. Inhibiting negative feedback regulation of USP18 by targeting its interaction with STAT2.
(a) 293T cells were co-transfected with plasmids encoding STAT1-FLAG, FLAG-USP18 and either STAT2 CC/DB-Myc or the mutant STAT2 CC/DB L227A R409A K415A-Myc (3A). Following treatment with IFNα (1000 U/ml) for 15 minutes as indicated cell lysates were collected and immunoblotted with indicated antibodies. The ratio of p-STAT1/total STAT1 was quantified by LI-COR Odyssey system.
(b) Cells indicated in Fig. 7a were treated with IFNα (1000 U/ml) for 12 hours and then expression of GBP1 was analyzed by RT q-PCR. Data represents mean ± S.D. for two independent experiments.
(c) THP-1 cells transduced with either MIP control (−) or MIP-STAT2 CC/DB 3A (3A) were treated with IFNα (1000 U/ml) for 48 hours and then Annexin V positive cells were analyzed by flow cytometry. Data represents mean ± S.E.M. for three independently generated stable cell lines. **P < 0.01.
(d) THP-1 and KT-1 cells were treated with TAT or USP18 aa 302-313 TAT peptide. Five hours after peptide treatment, IFNα (1000 U/ml) was added for the indicated time and cells were analyzed by Western blotting. The ratio of p-STAT1/total STAT1 from three independent experiments was quantified (right panel). Data are presented as mean +/c S.E.M. of three independent experiments. *P < 0.05.
(e) TAT- or USP18 aa 302-313 TAT peptide-treated THP-1 and KT-1 cells were incubated with IFNα (1000 U/ml) for 48 hours and then Annexin V positive cells were analyzed by flow cytometry. Data are presented as mean +/− S.E.M. of three independent experiments. **P < 0.01.
It is known that the CC domain of STAT2 interacts with IRF9 and that the DB domain of STAT2 is essential for activation of ISG transcription 45,46. Although expression of STAT2 CC/DB can successfully disrupt the STAT2-USP18 negative feedback interaction, it may also compete with wild-type STAT2 in the formation of the ISGF3 complex and in the binding to ISRE promoter regions upon nuclear translocation. To investigate these downstream biological functions, we generated a triple mutant within STAT2 CC/DB, carrying the exchanges L227A, R409A, and K415A (further designated as STAT2 CC/DB 3A). The single L227A point mutation substantially reduced interaction of STAT2 CC/DB with IRF9 (Supplementary Fig. 7a). Furthermore, it has been reported that the residues R409 and K415 are important for nuclear translocation 47. STAT2 CC/DB 3A is therefore expected to be primarily cytosolic and to lack its ability to bind to IRF9. Yet, STAT2 CC/DB 3A retained its ability to disrupt the USP18 inhibitory effect on STAT1 phosphorylation (Fig. 7a, right lane). In addition, STAT2 CC/DB 3A, but not STAT2 CC/DB, partially blocked the effect of USP18 on transcription of ISGs, such as GBP1 and IFIT1 (Fig. 7b and Supplementary Fig. 7b). Both IFNα and IFNβ are known to promote apoptosis of several cancer cell lines 48. IFNα or IFNβ treatment induced apoptosis in the human myeloid cell line THP-1, and this effect was significantly enhanced upon addition of STAT2 CC/DB 3A (Fig. 7c and Supplementary Fig. 7c).
Since we identified the critical region for the USP18 interaction with STAT2 (Supplementary Fig. 3c), we also examined whether a peptide comprising USP18 aa 302-313 could have a similar effect as STAT2 CC/DB domains. IFN-induced STAT1 phosphorylation in THP-1 and KT-1 cells treated with this peptide was enhanced and prolonged as compared to the control (Fig. 7d). Consistent with these results, we observed enhanced GBP-1 expression and increased apoptosis in THP-1 and KT-1 cells treated with the USP18 aa 302-313 peptide (Fig. 7e and Supplementary Fig. 7d). Taken together, our results demonstrate that, by interfering with the USP18-STAT2 interaction, USP18-mediated inhibition of the type I IFN signaling can be suppressed. This implicates that the STAT2-USP18 interaction interface could be a useful drug target for enhancing type I IFN responses.
Discussion
Type I IFNs are involved in a variety of different processes of innate and adaptive immune responses 49,50, which are coordinated by a highly fine-tuned regulatory signaling network 1. Unraveling the molecular and cellular determinants governing this network will be imperative for a better understanding and for therapeutically manipulating immunological responses in a variety of disease contexts. A critical role of negative regulators in type I IFN signaling is emerging15, which have been targeted by inhibitors to enhance IFN response: PKD2 exerts negative feedback via IFNAR1 51, but the PKD inhibitor CID755673 only slightly prolonged the IFN response. SOCS1 and SOCS3 inhibit tyrosine phosphorylation and nuclear translocation of STAT1 by binding to JAKs 52. However, inhibition of SOCS1 only transiently enhances the IFN response, since it is expressed in the early phase after type I IFN treatment and undetectable later on 53. Additionally, SOCS1 affects not only type I but also type II IFN signaling. In contrast, inhibition of USP18 leads to enhanced type I IFN signaling at the early stage 21 and produces a prolonged response at the later stage 27,54. Therefore, we aimed to explore the specific mechanism of signal inhibition by the STAT2-USP18 negative feedback interaction.
STAT2 is well known as an unique effector of type I and type III IFN signaling not only by being an integral component of the ISGF3 complex responsible for the induction of ISGs, 55,56, but also by positively regulating STAT1 phosphorylation 38,57. The biological significance of STAT2 in type I IFN signaling has been further corroborated by the study of STAT2-deficient humans and mice, which become immune compromised and are vulnerable to viral infection 39,58. However, for the first time, we here demonstrate that USP18 requires STAT2 for exerting its inhibitory effect on IFN signaling. USP18 reduced IFN binding and receptor dimerization as well as JAK1 phosphorylation only when STAT2 was present. These observations established the key role of STAT2 in USP18-mediated inhibition of IFN signaling and moreover suggest that the increased STAT2 levels induced by IFN signaling may further enhance negative feedback by USP18.
In order to fulfill its inhibitory function, USP18 needs to be recruited to the receptor 21. Previously, we reported that USP18 interferes with the recruitment of IFNAR1 to the IFN-IFNAR1-IFNAR2 ternary complex, by an unknown mechanism 22. Here, we have demonstrated that STAT2 acts as an adaptor for recruiting USP18 to IFNAR2 via its membrane-distal constitutive binding site for STAT2. Binding of STAT2 and USP18 to IFNAR2 is synergistic, in line with the previous observation that the STAT2-IFNAR2 interaction was strengthened by USP18 33. Hence, through stabilizing the interaction between STAT2 and IFNAR2, USP18 may also negatively regulate the STAT phosphorylation process by decreasing the STAT phosphorylation turnover rate and the activation of the ISGF3 complex. We assume that recruitment of USP18 to IFNAR2 via STAT2 promotes the otherwise weak interaction of USP18 with a membrane-proximal site of IFNAR2. We previously have shown this interaction to compete with JAK1 binding, thus effectively reducing ternary complex formation and signal activation at the plasma membrane, which probably are the main inhibitory functions of USP18 21–23. Here, we found that STAT2 interacted with the N- as well as the C-terminal regions of USP18. Consistent with previous results 21, our experiments confirmed that the N- and C-terminal regions of USP18 directly interact with IFNAR2. Taken together, our data suggest that USP18 simultaneously interacts with IFNAR2 via STAT2 in the membrane distal region and directly in the membrane-proximal region (Fig. 6f).
Both the CC and DB domains of STAT2 are critical for the interaction with USP18 and for USP18-mediated negative feedback regulation of type I IFN signaling, as measured by type I IFN binding and STAT phosphorylation. Interestingly, these domains are also critical for the STAT2 interaction with IFNAR2, which can explain the binding synergy and moreover tightly couples STAT2 and USP18 functions. Indeed, we could demonstrate that a construct containing only the STAT2 CC/DB has an inhibitory effect on the function of USP18 and thus increases STAT1 phosphorylation. Notably, our study uncovers that a USP18 302-313 peptide can disrupt the USP18-STAT2 interaction interface. Both peptides significantly enhanced IFN-triggered responses. It should be noted that, while this inhibitory effect is strong, it is not complete, indicating either the existence of other factors involved or that there is a room for improving these inhibitors. Further studies will be required for unraveling the STAT2 CC/DB and USP18 302-313 inhibitory effect on USP18 function including a structural analysis using purified proteins when they become available. Thus, novel strategies to manipulate negative feedback by USP18 by means of small molecule PPI modulators, which are currently emerging 59, can be envisaged.
Importantly, USP18 has a negative role not only in type I but also in type III IFN signaling 60, which also involves STAT2. Therefore, a similar mechanism of negative regulation by USP18 and STAT2 in type III IFN signaling can be expected. Indeed, preliminary data from our laboratory show that USP18 can bind to type III IFN receptor IL-28RA, and has no inhibitory effect on type III IFN signaling in U6A cells (Arimoto et al. unpublished data). Although our current functional peptide studies presented in figure 7 still lack in vivo validation, it is tempting to speculate that the tight functional linkage of STAT2 and USP18 has evolved to warrant efficient control of ISGF3-based gene expression. Design of modulators for controlling the USP18-STAT2 interaction could therefore yield the ability to enhance or diminish type I and type III IFN responses in therapeutic settings.
Online Methods
Yeast two-hybrid screen
cDNA encoding full length human USP18 (wild-type and C64A mutated form) were cloned into the LexA DNA-binding domain plasmid derived from pBTM116 and used as bait in yeast two-hybrid screens of a human placental cDNA library (complexity of 106 colonies) as previously described 61. A total of 11 independent clones displaying similarity to STAT2 proteins were isolated.
Yeast two-hybrid assay
To confirm specific interactions, the following yeast two-hybrid vectors were created in this work, which employ the Gal4 DNA-binding and activation domains: pGBD-C1 (vector), control), and DNA-binding domain fusions pJJH1722 (pGBD-STAT2) and pJJH1719 (pGBD-USP18); as well as pGAD424A (vector), control), and pJJH1721 (pGAD-STAT2) and pJJH1720 (pGAD-USP18) for expression of activation domain fusion proteins. Complete sequences of these plasmids are available upon request. For two-hybrid analyses with these constructs the S. cerevisiae strains pJ69-4a and pJ69-4alpha were used 62. 3 μl of overnight cultures from strains pJ69-4a/pJ69-4alpha carrying two-hybrid plasmids with the indicated coding sequences were spotted onto selective media for plasmid maintenance and incubated for 10 days.
Plasmid construction
Human STAT2 and its mutant cDNAs were cloned into pcDNA3.1 vector. Human STAT2 was also cloned into pCAG and GST (6p-1) vector. Murine Stat2 was cloned into MSCV-IRES-Puro (MIP) retroviral vector. shRNA plasmids (pLKO.1 vectors) for mouse Stat2 (TRCN0000081538-0000081542) and control were purchased from Dharmacon.
Human USP18 and its mutant cDNAs were cloned into pcDNA3.1, pCMV7.1 3 × FLAG, pEGFP-C1, and MIP vector. Murine Usp18 was cloned into pCX4-bsr retroviral vector. Plasmid encoding human IFNAR2 in the pcDEF3 vector was kindly provided by Dr. Sergei Kotenko. Human IFNAR2 was also cloned into pCAG and pEBG vector.
Monomeric GFP, human STAT2-mEGFP and human mEGFP-USP18 fusion constructs (kind gift from Sylvie Urbé, Liverpool 63) were cloned into the plasmid vector pSEMS-26m (Covalys) for expression in mammalian cells. Monomeric EGFP was obtained by the A206K mutation within the EGFP sequence of pEGFP-N1 (Clontech). An artificial transmembrane domain (TMD) with the sequence ASALAALAALAALAALAALAALAKSSRL (ALA7) (as described by 64) extracellular fused to HaloTag (Promega) and mTagBFP (obtained from Vladislav Verkhusha, New York 65) and intracellularly fused to human STAT2 was cloned into pDisplay™ (Invitrogen). For cloning of pSEMS-HaloTag®-IFNAR2, the gene coding for the HaloTag followed by the genes of full length IFNAR2 or IFNAR2Δ375 without the N-terminal signal sequences were inserted into pDisplay (Invitrogen). The constructs including the Igκ signal sequence from the pDisplay vector were transferred by restriction with EcoRI and NotI into pSems-26m. The genes for mTagBFP, HaloTag and IFNAR2 were inserted into pDisplay™ –HaloTag to generate the construct pDisplay-HaloTag-mTagBFP-IFNAR2.
Cell culture and primary bone marrow cells from Usp18−/− mice
HEK293T (ATCC), Hela (ATCC), MDA-MB-231 (kindly provided by Dr. David Cheresh, previously), Stat2−/− MEFs (kindly provided by Dr. Adolfo Garcia-Sastre), and U series (2fTGH, U1A, U2A, U4A, U5A, and U6A) (kindly provided by Dr. George Stark) cells were grown in DMEM medium supplemented with glutamine, penicillin/streptomycin, and 10% FBS. THP-1 (ATCC) and KT-1 cells (kindly provided by Dr. Shigeru Fujita, previously) were grown in RPMI medium supplemented with glutamine, penicillin/streptomycin, and 10% FBS. Ba/F3 cells were grown in RPMI1640 medium supplemented with 15% FBS and 5% conditional media from WEHI-3B cells (kindly provided by Dr. Carrie Dolman, previously). Bone marrow cells from Usp18−/− 66 mice were grown in RPMI1640 medium supplemented with 20% FBS and IL3 and SCF conditional media. Hela and U series cells are routinely tested for mycoplasma. All the procedures for Usp18−/− mice experiments were approved by the UCSD institutional animal care and use committee
Transfection, and lentivirus or retrovirus infection
Transfection was conducted by using PEI (Polyethylenimine) 67. For the retrovirus or lentivirus production, 293T cells were co-transfected with plasmids encoding viral vectors and packaging vectors pCL-10A1 for human or Ecopac for murine cells. Viral particles were collected 48 h after transfection, filtered with 0.45 μm sterile filter. For the retrovirus or lentivirus infection, spin infection (2000g, 3hours, 30°C; Allegra X12R (Beckman Coulter)) in the presence of polybrene (8ug/ml) was performed.
Reagents and antibodies
Antibodies were commercially purchased as followed; anti-phospho-JAK1 (Tyr1022/1023) (Cell Signaling), anti-JAK1 (Cell Signaling), anti-phospho-STAT1 (Tyr 701) (Cell Signaling), anti- STAT1 (Cell Signaling), anti-STAT2 (Santa Cruz), anti-green fluorescent protein (anti-GFP), and anti-tubulin (Sigma). Antibodies against FLAG (anti-FLAG; M2), Myc (9E10), and hemagglutinin (HA) (12CA5 or 3F10) were purchased from Sigma, Santa Cruz, and Roche, respectively. Anti-IFNAR1 antibody was provided from Biogen Idec. Anti-IFNAR2 FITC was purchased from Sino Biological Inc. Anti-USP18 antibody is described as before 68. Recombinant human IFNβ was provided from Biogen Idec. Recombinant human IFNα, mouse IFNβ, and human IFNλ were purchased from Peprotech. Recombinant IFNα2 and the mutant IFNα2-M148A used in cellmicropatterning and single molecule assays was produced in E. coli and purified as described before 69. For site-specifically fluorescent labeling, IFNα2 and the mutant IFNα2-M148A fused to an N-terminal ybbR-tag were produced in E. coli and conjugated with DY 647 as described previously 70. A degree of labeling >90% was obtained as determined by UV/Vis spectroscopy.
The RGD peptide Ac-CGRGDS-COOH was custom synthesized by Coring System Diagnostix, Gernsheim/Germany. Poly-L-lysine (PLL) hydrobromide (Mw: 15,000–30,000 g/mol) was purchased from Sigma Aldrich. Homobifunctional dicarboxy-PEG (COOH-PEG3000-COOH, Mw of PEG: 3000 g/mol) was from Rapp Polymere, Tübingen/Germany. Poly-L-lysine graft modified with Methoxy-PEG (Mw: 2000 g/mol) (PLL-PEG-OMe) was purchased from SuSoS AG, Dübendorf/Switzerland. HaloTag®-O2-amine ligand (HTL) was purchased from Promega. Dimethylformamide (DMF), N-Ethyldiisopropylamine (DIPEA), N,N′-Diisopropylcarbodiimide (DIC), N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride (EDC) were purchased from Sigma Aldrich.
Synthesis of functionalized poly-L-lysine-graft-(polyethylene glycol) copolymer (PLL-PEG derivatives, PLL-PEG-HTL and PLL-PEG-RGD) was carried out as described recently 71.
RNA isolation and qRT-PCR analysis
RNA was extracted with Trizol (Thermo Fisher Scientific). For qRT–PCR analyses, equal amounts of RNA were reverse-transcribed by qScript (Quanta Biosciences) and the resulting cDNA templates were subjected to qRT-PCR using KAPA SYBR FAST universal qPCR kit (Kapa Biosystems, Inc. Wilmington, MA) and CFX96 thermal cycler (BIO-RAD).
Primer sequences for are as follows;
RT- Isg15 -Fw; GAC TAA CTC CAT GAC GGT G
RT- Isg15 -Rev; AAC TGG TCT TCG TGA CTT G
RT- Gbp1 -Fw; GGA GGC CAT TGA GGT CTT CAT
RT- Gbp1 -Rev; CAA AGG CAT CTC GTT TGG CT
RT- Cxcl9 -Fw; TCCTTTTGGGCATCATCTTCC
RT- Cxcl9 -Rev; TTTGTAGTGGATCGTGCCTCG
RT- Irf9 -Fw; GCCTTTGCCCCATCCCCATCTC
RT- Irf9 -Rev; CCCCTGGCCCTGGAAGTACTGG
RT- Ifit1 -Fw; TGGCGACCTGGGGCAACTGTG
RT- Ifit1 -Rev; TGGGCTGCCTGTTTCGGGATGTC
RT- Igtp -Fw; CGCCTCATCAGCCCGTGGTCTAA
RT- Igtp -Rev; TGCCATTGCCAGAGTCCCCAGTC
RT-GBP1 -Fw; CCAGTTGCTGAAAGAGCAAGAGA
RT-GBP1 -Rev; TCCCTCTTTTAGTAGTTGCTCCTGTT
RT- IFIT1 -Fw; AAGGCAGGCTGTCCGCTTA
RT- IFIT1 -Rev; TCCTGTCCTTCATCCTGAAGCT
Western blotting
Western blotting was performed as previously described in detail 72. All samples were denatured in 1 × sample buffer [50 mM Tris-HCl (pH 6.8), 2% SDS, 2-mercaptethanol, 10% glycerol, and 1% bromophenol blue] for 5 min at 100 °C. Cells were lysed in RIPA buffer composed of 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% SDS, 1% Nonidet P-40 and 0.5% sodium deoxycholate. To analyze immune-complexes for co-immunoprecipitation assay, cells were lysed in binding buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA and 0.5% Nonidet P-40 for coimmunoprecipitation assays. The cell lysates were centrifuged (15,000 × rpm) at 4°C for 5 min. All lysis buffers in this study contain proteinase and phosphatase inhibitors (Roche). Soluble fractions were precleared by Protein G-Sepharose at 4°C for 15 min. Precleared cell lysates were immunoprecipitated for 1~4 hours with the indicated antibodies. Immunocomplexes were adsorbed to the protein G-Sepharose and, after three washes, were eluted by boiling for 5 min. FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2-agarose (Sigma). All assays were performed two to four times and representative blots were presented. All source data for the western blots are available in the supplementary data set 1. For the quantification, signals were detected with the LI-COR Odyssey system.
Cell micropatterning
Micropatterned surfaces were fabricated by microcontact printing. Poly(dimethylsiloxane) (PDMS) stamps was generated from basic elastomer (Sylgard 184, Dow Chemicals) mixed with curing agent (Dow Chemicals) in a 10:1 ratio, and applied to a silicon master at 80°C overnight. The silicon master containing an array of lines with a width of 5 μm, a spacing of 10 μm and a depth of 3 μm was generated by photolithography using a custom designed beam mask (nb technologies GmbH).
Standard glass coverslides for fluorescence microscopy were cleaned in a plasmacleaner for 10 minutes, followed by inking of the stamp with 0.5mg/ml PLL-g-PEG-HTL in PBS buffer for 10 minutes. For PLL-g-PEG-HTL transfer, stamps were placed onto the glass coverslides for 10 minutes to generate HTL patterns. After removing the stamps, the coverslides were incubated with a mixture of 0.002mg/ml PLL-g-PEG-RGD and 0.1mg/ml PLL-PEG-MeO in PBS buffer for 1 minute to backfill the uncoated area and to allow cell adhesion. The surface was then rinsed in MilliQ water and dried with N2.
For cellular micropatterning, cells were cultivated at 37°C and 5% CO2 in MEM supplemented with 10% fetal calf serum (MEM/FCS) 1% Hepes buffer and 1% non-essential amino acids. For transfection, cells were plated in 60 mm cell culture dishes to a density of approximately 50% confluence. One day after seeding, cells were transfected via calcium phosphate precipitation as described earlier 73. After 24–36 hours cells were plated on chemically modified cover glasses for 15–20 hours with medium containing penicillin and streptomycin (PAA). For labeling of micropatterned IFNAR, cells were incubated in presence of 10 nM fluorescently labeled interferon coupled to ATTO655 (AT655IFNα2).
Fluorescence imaging
Total internal reflection fluorescence microscopy (TIRFM) was performed using an inverted microscope microscope (Olympus IX81) equipped with a 4-line TIRF condenser (Olympus cell^tirf MITICO), a back-illuminated electron multiplied (EM) CCD camera (C9100-13, 512×512 pixel from Hamamatsu) as well as lasers at 405 nm (100 mW), 488 nm (150 mW), 561 nm (150 mW) and 640 nm (140 mW). A 60× objective with a numerical aperture of 1.49 (UAPON 60×/1.49, Olympus) or a 150× objective with a numerical aperture of 1.45 (UAPON 60×/1.45, Olympus) was used for TIRF excitation.
The excitation beam was reflected into the objective by a quad-band dichroic mirror (HC-BS R405/488/561/635, AHF) and the fluorescence was detected through a quadbandpass filter (BrightLine HC 446/523/500/677). For multicolor experiments, a QuadView (QV2, from Photometrics) equipped with suitable dichroic beamsplitters (480dcxr, 565dcxr and 640dcxr, Chroma) and emission filters (BrightLine HC 438/24, AHF, BrightLine HC 520/35 AHF, EmitterHQ 600/30, AHF and BrightLine HC 685/40, Chroma) was utilized to avoid spectral cross-talk. Data acquisition was performed with the acquisition software Olympus Xcellence rt Version 1.2.
Fluorescence recovery after photobleaching (FRAP) experiments were performed at 37°C in an incubation chamber (Olympus) and using a 150× TIRF objective with a numerical aperture of 1.49 (UAPON 150×/1.49, Olympus) for TIR excitation. A circular area with a diameter of 8 μm was bleached by 405 nm excitation for 5 s with a laser power of 7.5 mW, followed by acquisition with a cycle time of 1–5 s by a 1 mW 488 nm or 561 nm laser excitation.
Data analysis
Image analysis and image processing was performed using ImageJ (NIH, Bethesda, MD). Image processing comprises cropping, scaling, rotation as well as adjustment of brightness and contrast levels. The fluorescence contrast of patterned proteins inside vs. outside the pattern was calculated from the average fluorescence intensities of the bait and prey proteins obtained from rectangular ROIs using the “Measure” function in ImageJ. The fluorescence contrast C of the bait proteins was calculated as
where Ibait,in denotes the mean pixel intensities from selected areas inside the pattern, Ibait,out the mean pixel intensities from selected areas outside the pattern and Ibg the background intensity from the glass surface obtained from a ROI outside the cells. Cbait reflects the relative enrichment of the bait proteins and the maximal enrichment that can be achieved by the prey proteins. The contrast of the prey proteins Cprey was obtained from the background corrected mean pixel intensities from selected areas inside and outside the pattern as: . Since Cprey varies proportional to Cbait, Cprey was corrected to obtain
For quantitative analysis, data were visualized in box plots indicating data distribution of the second and third quartile (box), median (line), mean (open squares), and whiskers (1.5× interquartile range). Outliers were plotted as individual points.
For analysis of FRAP experiments, a rectangular region of interest within the bleached area of the pattern and a rectangular or circular ROI within the bleached area but outside the patterned area were chosen for obtaining intensity values per pixel over time, respectively. FRAP curves were obtained by the following equation:
The offset intensity (Foffset) was determined from a ROI outside of the cell and was substracted by all intensity values. Free cytoplasmic diffusion in living cell was consistent both inside and outside the pattern and its effect to FRAP could be determined as (FROIoutside − Foffset). Thus the unbiased fluorescence recovery inside the pattern could be obtained by a subtraction of recovery outside the pattern as: (FROIinside − Foffset) − (FROIoutside − Foffset). A normalization factor of was implemented in order to correct background photobleaching during FRAP experiments. For this purpose, a rectangular ROI in a not-bleached patterned region was assigned as a reference and the sequential intensities in this area were normalized to the original one. The recovery of the fluorescence intensity was fitted by a simple monoexponential function 74,75).
Single molecule ligand binding assay
Single molecule ligand binding experiments were performed in presence of 2 nM DY647IFNα2 M148A and after an incubation time of 10 minutes by TIRF imaging as described previously. All binding experiments were carried out using media complemented with oxygen scavenger [0.5mg ml-1 glucose oxidase (Sigma), 0.04 mgml-1 catalase (Roche AppliedScience), 5% w/v glucose, 1 μM ascorbic acid and 1μM methyl viologene] in order to minimize photobleaching 76. In order to minimize background from non-specifically adsorbed dye molecules during single molecule experiments, glass coverslips were coated with a poly-L-lysine-graft-(polyethylene glycol) copolymer functionalized with RGD as described before77. The assay was performed two times and a representative blot was presented. Localization and quantification of individual IFNα2 M148A molecules were determined by using the multiple target tracking algorithm (MTT) 78 as described previously 79.
IFN FITC labeling and binding affinity assay
Recombinant IFNα2b (ProSpec, Cat#CYT-205) was labeled with fluorescein isothiocyanate (FITC) using the SureLINK FITC-Labeling Kit (KPL, Cat# 82-00-01), per the manufacturer’s instructions. A conjugate with an optimal molar ratio (F/P) of ~4.1 was used for binding affinity experiments. For binding affinity experiments, indicated cell lines were incubated with a saturating concentration of FITC-IFNα2b and FITC MFI was measured on a BD FACSCanto instrument with standard lasers and optical filters.
Receptors expression analysis
Infected U6A cell lines were trypsinized and incubated with mouse anti-IFNAR1 or IFNAR2-FITC antibodies. For the detection of IFNAR1, we used anti-mouse PE secondary antibody.
Peptides
THP-1 or KT-1 cells were treated with cell permeable TAT (Trans-Activator of Transcription Protein) (GRKKRRQRRRPQ) or USP18 aa 302-313 linker TAT (YELFAVIAHVGMGGGSGRKKRRQRRRPQ) (Eton Bioscience Inc., San Diego, CA) (final 10mM in 2%FBS medium) in this study.
Apoptosis assay
Apoptosis was measured by staining with Annexin V-APC and 7-AAD using Annexin V apoptosis detection kit (BD) according to the manufacturer’s protocol. Cells were analyzed by flow cytometry on a BD FACSCanto with standard lasers and optical filters. The results in the paper are the means of the percent of apoptotic cells from three independently infected (MIP or MIP-STAT2 CC/DB 3A) or peptide (TAT or USP18 aa 302-313 TAT)-treated cells.
Statistical analyses
Where applicable, statistical significance was determined by using a two-tailed Student’s t- test using the statcel2 software (OMS Ltd. Japan). A p value of < 0.05 was considered statistically significant. Changes in the interactions between different bait and prey proteins in the single molecule ligand binding assay and cell micropatterning experiments as determined by the contrast values were statistically analyzed, using the two-sample Kolmogorov-Smirnov test. The p values for the contrast values of two samples were calculated using MATLAB software. A p value of < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. A. Garcia-Sastre for Stat2−/− MEFs, Dr. G. Stark for sharing U series Cell lines, Dr. T. Akagi for providing pCX4 series vectors, Dr. S. Kotenko for pcDEF-hIFNAR2 DNA construct, Dr. D. Baker from Biogen Idec Inc. for supplying recombinant human IFNβ and Anti-human IFNAR1 antibody, Hybrigenics staff for its contribution, G. Hikade for technical support and Dr. R. Kurre for advice in fluorescence microscopy. This study was supported by NIH R01HL091549 and R01CA177305 to D.E.Z. and SFB 944 from the DFG to J.P. and J.J.H.
Footnotes
Author Contributions
KIA, SL, and SAS designed, performed, analyzed experiments and wrote the manuscript; CB, YZ, SM, JBF, SW, JJH, ZL, MY, SP, FC performed experiments or provided critical information; JP and DEZ conceived the project, designed experiments, analyzed experimental data, and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Data Availability Statement
Source data for all western blots in this study is available online. Other data supporting our findings in the study are available from corresponding authors upon reasonable request.
References
- 1.Hertzog PJ, Williams BR. Fine tuning type I interferon responses. Cytokine Growth Factor Rev. 2013;24:217–25. doi: 10.1016/j.cytogfr.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15:405–14. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 5.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sistigu A, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20:1301–9. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 7.Rieger PT. Interferon-alpha: a clinical update. Cancer Pract. 1995;3:356–65. [PubMed] [Google Scholar]
- 8.Dusheiko G. Side effects of alpha interferon in chronic hepatitis C. Hepatology. 1997;26:112S–121S. doi: 10.1002/hep.510260720. [DOI] [PubMed] [Google Scholar]
- 9.Crow YJ. Aicardi-Goutieres syndrome. Handb Clin Neurol. 2013;113:1629–35. doi: 10.1016/B978-0-444-59565-2.00031-9. [DOI] [PubMed] [Google Scholar]
- 10.Meyer O. Interferons and autoimmune disorders. Joint Bone Spine. 2009;76:464–73. doi: 10.1016/j.jbspin.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Uze G, Lutfalla G, Mogensen KE. Alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Leung S, Qureshi S, Darnell JE, Jr, Stark GR. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271:5790–4. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- 13.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature reviews Immunology. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 14.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–72. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36:150–160. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Marchetti M, et al. Stat-mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin-dependent endocytosis of IFN receptors. Mol Biol Cell. 2006;17:2896–909. doi: 10.1091/mbc.E06-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015;36:139–149. doi: 10.1016/j.it.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Liu LQ, et al. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol Cell Biol. 1999;19:3029–38. doi: 10.1128/mcb.19.4.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer H, et al. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18) Genomics. 2000;65:44–52. doi: 10.1006/geno.2000.6148. [DOI] [PubMed] [Google Scholar]
- 20.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–81. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 21.Malakhova OA, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–67. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilmes S, et al. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J Cell Biol. 2015;209:579–93. doi: 10.1083/jcb.201412049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francois-Newton V, et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One. 2011;6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meuwissen ME, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J Exp Med. 2016;213:1163–74. doi: 10.1084/jem.20151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie KJ, et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–8. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 27.Kim KI, et al. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J Immunol. 2005;175:847–54. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- 28.Honke N, et al. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol. 2012;13:51–7. doi: 10.1038/ni.2169. [DOI] [PubMed] [Google Scholar]
- 29.Honke N, et al. Usp18 driven enforced viral replication in dendritic cells contributes to break of immunological tolerance in autoimmune diabetes. PLoS Pathog. 2013;9:e1003650. doi: 10.1371/journal.ppat.1003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldmann T, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34:1612–29. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketscher L, et al. Selective inactivation of USP18 isopeptidase activity in vivo enhances ISG15 conjugation and viral resistance. Proc Natl Acad Sci U S A. 2015;112:1577–82. doi: 10.1073/pnas.1412881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yim HY, et al. Elevated Response to Type I IFN Enhances RANKL-Mediated Osteoclastogenesis in Usp18-Knockout Mice. J Immunol. 2016;196:3887–95. doi: 10.4049/jimmunol.1501496. [DOI] [PubMed] [Google Scholar]
- 33.Lochte S, Waichman S, Beutel O, You C, Piehler J. Live cell micropatterning reveals the dynamics of signaling complexes at the plasma membrane. J Cell Biol. 2014;207:407–18. doi: 10.1083/jcb.201406032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 35.Ketscher L, Knobeloch KP. ISG15 uncut: Dissecting enzymatic and non-enzymatic functions of USP18 in vivo. Cytokine. 2015;76:569–71. doi: 10.1016/j.cyto.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–12. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rani MR, et al. Characterization of beta-R1, a gene that is selectively induced by interferon beta (IFN-beta) compared with IFN-alpha. J Biol Chem. 1996;271:22878–84. doi: 10.1074/jbc.271.37.22878. [DOI] [PubMed] [Google Scholar]
- 38.Leung S, Qureshi SA, Kerr IM, Darnell JE, Jr, Stark GR. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol. 1995;15:1312–7. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hambleton S, et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A. 2013;110:3053–8. doi: 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang E, Wen Z, Haspel RL, Zhang JJ, Darnell JE., Jr The linker domain of Stat1 is required for gamma interferon-driven transcription. Mol Cell Biol. 1999;19:5106–12. doi: 10.1128/mcb.19.7.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen VP, et al. Stat2 binding to the interferon-alpha receptor 2 subunit is not required for interferon-alpha signaling. J Biol Chem. 2002;277:9713–21. doi: 10.1074/jbc.M111161200. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Leung S, Kerr IM, Stark GR. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17:2048–56. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piehler J, Roisman LC, Schreiber G. New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J Biol Chem. 2000;275:40425–33. doi: 10.1074/jbc.M006854200. [DOI] [PubMed] [Google Scholar]
- 44.Improta T, et al. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci U S A. 1994;91:4776–80. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272:20070–6. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 46.Brierley MM, Fish EN. Functional relevance of the conserved DNA-binding domain of STAT2. J Biol Chem. 2005;280:13029–36. doi: 10.1074/jbc.M500426200. [DOI] [PubMed] [Google Scholar]
- 47.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J Biol Chem. 2001;276:16447–55. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 48.Chawla-Sarkar M, Leaman DW, Borden EC. Preferential induction of apoptosis by interferon (IFN)-beta compared with IFN-alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin Cancer Res. 2001;7:1821–31. [PubMed] [Google Scholar]
- 49.Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–35. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen HM, et al. Critical role for constitutive type I interferon signaling in the prevention of cellular transformation. Cancer Sci. 2009;100:449–56. doi: 10.1111/j.1349-7006.2008.01051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng H, Qian J, Baker DP, Fuchs SY. Tyrosine phosphorylation of protein kinase D2 mediates ligand-inducible elimination of the Type 1 interferon receptor. J Biol Chem. 2011;286:35733–41. doi: 10.1074/jbc.M111.263608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–65. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 53.Sarasin-Filipowicz M, et al. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol. 2009;29:4841–51. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou W, et al. Microarray analysis reveals that Type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem Biophys Res Commun. 2007;356:193–9. doi: 10.1016/j.bbrc.2007.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 56.Brierley MM, Marchington KL, Jurisica I, Fish EN. Identification of GAS-dependent interferon-sensitive target genes whose transcription is STAT2-dependent but ISGF3-independent. FEBS J. 2006;273:1569–81. doi: 10.1111/j.1742-4658.2006.05176.x. [DOI] [PubMed] [Google Scholar]
- 57.Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 59.London N, Raveh B, Schueler-Furman O. Druggable protein-protein interactions--from hot spots to hot segments. Curr Opin Chem Biol. 2013;17:952–9. doi: 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Burkart C, et al. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-lambda and elevated secretion of Cxcl10. EMBO Mol Med. 2013;5:967–82. doi: 10.1002/emmm.201201864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colland F, et al. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–32. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Urbe S, et al. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Molecular Biology of the Cell. 2012;23:1095–1103. doi: 10.1091/mbc.E11-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roder F, Birkholz O, Beutel O, Paterok D, Piehler J. Spatial organization of lipid phases in micropatterned polymer-supported membranes. J Am Chem Soc. 2013;135:1189–92. doi: 10.1021/ja310186g. [DOI] [PubMed] [Google Scholar]
- 65.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–24. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ritchie KJ, et al. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 2002;16:2207–12. doi: 10.1101/gad.1010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burkart C, et al. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-lambda and elevated secretion of Cxcl10. EMBO Mol Med. 2013;5:967–82. doi: 10.1002/emmm.201201864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piehler J, Roisman LC, Schreiber G. New structural and functional aspects of the type I interferon-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J Biol Chem. 2000;275:40425–33. doi: 10.1074/jbc.M006854200. [DOI] [PubMed] [Google Scholar]
- 70.Waichman S, et al. Functional immobilization and patterning of proteins by an enzymatic transfer reaction. Anal Chem. 2010;82:1478–85. doi: 10.1021/ac902608a. [DOI] [PubMed] [Google Scholar]
- 71.Wedeking T, et al. Spatiotemporally Controlled Reorganization of Signaling Complexes in the Plasma Membrane of Living Cells. Small. 2015 doi: 10.1002/smll.201502132. [DOI] [PubMed] [Google Scholar]
- 72.Arimoto K, et al. Plakophilin-2 promotes tumor development by enhancing ligand-dependent and -independent epidermal growth factor receptor dimerization and activation. Mol Cell Biol. 2014;34:3843–54. doi: 10.1128/MCB.00758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muster B, et al. Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS One. 2010;5:e11910. doi: 10.1371/journal.pone.0011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–95. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 76.Vogelsang J, et al. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew Chem Int Ed Engl. 2008;47:5465–9. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- 77.VandeVondele S, Voros J, Hubbell JA. RGD-grafted poly-L-lysine-graft-(polyethylene glycol) copolymers block non-specific protein adsorption while promoting cell adhesion. Biotechnol Bioeng. 2003;82:784–90. doi: 10.1002/bit.10625. [DOI] [PubMed] [Google Scholar]
- 78.Serge A, Bertaux N, Rigneault H, Marguet D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods. 2008;5:687–94. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 79.Wilmes S, et al. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J Cell Biol. 2015;209:579–93. doi: 10.1083/jcb.201412049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.