Abstract
How microtubules (MT) are generated in the cell is a major question in understanding how the cytoskeleton is assembled. For several decades, γ-tubulin has been accepted as the cell’s universal MT nucleator. Although there is evidence that γ-tubulin complexes are not the sole MT nucleators, identification of other nucleation factors has proven difficult. Here, we report that the well-characterized MT polymerase XMAP215 (chTOG/Msps/Stu2p/Alp14/Dis1 homologue) is essential for MT nucleation in Xenopus egg extracts. The concentration of XMAP215 determines the extent of MT nucleation. Even though XMAP215 and γ-tubulin ring complex (γ-TuRC) possess minimal nucleation activity individually, together these factors synergistically stimulate MT nucleation in vitro. The N-terminal TOG domains 1–5 of XMAP215 bind αβ-tubulin and promote MT polymerization, while the conserved C-terminus is required for efficient MT nucleation and directly binds γ-tubulin. In sum, XMAP215 and γ-TuRC together function as the principal nucleation module that generates MTs in cells.
Microtubules (MTs) form a variety of cytoskeletal structures vital to the function of the cell, including the mitotic spindle. The first step in building these structures is the generation of MTs, yet little is known about how MTs are nucleated in the cell. MTs can assemble spontaneously in vitro from high concentrations of αβ-tubulin dimers1. In contrast, spontaneous MT formation rarely occurs in vivo. Instead, MTs are nucleated at specific sites termed MT organizing centers (MTOCs)2, 3. Essential to all MTOCs is the protein γ-tubulin, which is widely accepted as the cell’s universal MT nucleator3, 4. γ-tubulin forms a complex with γ-tubulin complex proteins (GCPs) and assembles into the γ-Tubulin Ring Complex (γ-TuRC)5–7. γ-TuRC positions an outer ring of approximately 13 γ-tubulin molecules, which are thought to interface with α-tubulin subunits and thereby provide a template for MT assembly7–10. However, this model for MT nucleation remains to be experimentally tested.
There is growing evidence that γ-TuRC alone does not account for all the MT nucleation activity in the cell. In the absence of γ-TuRC, MTs still form, albeit with significantly reduced numbers and kinetics11–13. Additionally, at least one factor of unknown identity besides γ-TuRC is needed to promote nucleation from centrosomes14. Furthermore, purified γ-tubulin complexes display low nucleation potential in vitro5, 7, 9, 14–18 that does not match the nucleation activity observed from cellular MTOCs. Together, these data suggest that the current γ-TuRC-centered model of MT nucleation is incomplete. Thus, the search for factors that generate MTs in the cell is ongoing and how MT nucleation occurs remains to be elucidated.
After their nucleation, growth of MTs is necessary for building cytoskeletal structures. The major MT polymerase in the cell, XMAP215, increases polymerization rates up to 10-fold in vitro and in vivo19–28. XMAP215 is a multi-domain protein consisting of five TOG domains at the N-terminus and a C-terminal domain23. The TOG domains bind to soluble αβ-tubulin dimers22, 29, 30 and are essential for the polymerase activity of XMAP21524. Recently, TOG domains 4 and 5 have been suggested to interact with αβ-tubulin incorporated in the MT lattice31. While primarily characterized as a polymerase, XMAP215, similar to a number of other MT-associated proteins, promotes spontaneous MT assembly27, 29, 32 and re-growth from MT templates in vitro33. However, mass balance model of the mitotic spindle suggested that XMAP215 only enhances growth of individual MTs and does not influence their nucleation26. Therefore, it remains unclear whether XMAP215 is involved in nucleating MTs in the cytoplasm, because assays to visualize nucleation events in the cell are lacking. Furthermore, if XMAP215 were to function as a MT nucleator, its relationship with the universal nucleator γ-TuRC needs to be established and their mechanism determined27, 34, 35.
In this study, we demonstrate that XMAP215 is a bona-fide MT nucleation factor that cooperates with γ-TuRC. Using Xenopus egg extracts to resolve individual nucleation events, we show that XMAP215 determines the extent of MT nucleation in the cytoplasm. XMAP215 and γ-TuRC operate synergistically to generate MTs in vitro, and XMAP215’s C-terminus is required for this cooperation. We further demonstrate that XMAP215 directly interacts with γ-tubulin via its C-terminus. Thus, XMAP215 is a major MT nucleation factor that functions with γ-TuRC to give rise to the MT cytoskeleton.
Results
XMAP215 is required for microtubule nucleation
To investigate how MT structures are built via MT nucleation, we used a system that allows visualization of individual MT nucleation events in the cytoplasm. Specifically, we studied what role the polymerase XMAP215 plays in generating branched, fan-like MT structures in Xenopus egg extracts induced by the dominant active form of the small GTPase Ran (RanQ69L)36. We visualized MT nucleation from pre-existing MTs using time-lapse total internal reflection microscopy with Cy5-labeled MTs and mCherry-labeled end-binding protein 1 (EB1) to highlight the growing MT plus-ends (Supplementary Fig. 1A).
To assess XMAP215’s role, we first added recombinant XMAP215 to Xenopus egg extracts, which contains approximately 120 nM endogenous XMAP21526. We expected the branched structures to appear larger due to longer and faster growing MTs, but less dense due to unaltered level of MT nucleation26. We quantified the number of MTs by detecting each plus-tip as the readout for MT nucleation. Surprisingly, XMAP215 increased the number of MTs in a concentration-dependent manner (Supplementary Fig. 1A–B, Supplementary Video 1). Addition of 480 nM XMAP215 increased MT numbers up to 30-fold, while the growth speed of MTs remained nearly constant (Supplementary Fig. 1B–C). These results show that XMAP215 has a dramatic effect on MT generation.
XMAP215 increases nucleation events, but is it required to generate MTs? To assess this, we immunodepleted XMAP215 from Xenopus extracts (Fig. 1A). Remarkably, MT nucleation was completely abolished in the absence of XMAP215 (Fig. 1B). This was confirmed by observing the reaction up to an hour, throughout the entire sample (2000 fields of view), and across tens of extract preparations. This phenotype has previously been observed only by immunodepletion of γ-tubulin, the universal MT nucleator36. To verify the specificity of this effect, we added back purified XMAP215, which rescued the branched structures with comparable MT count to the control-depleted extracts (Fig. 1A–B, Supplementary Fig. 1D). Proteins such as TACC3, α-tubulin, augmin, TPX2 and γ-TuRC were not depleted (Supplementary Fig. 1E). In summary, XMAP215 is required for generating MTs in Xenopus egg extracts.
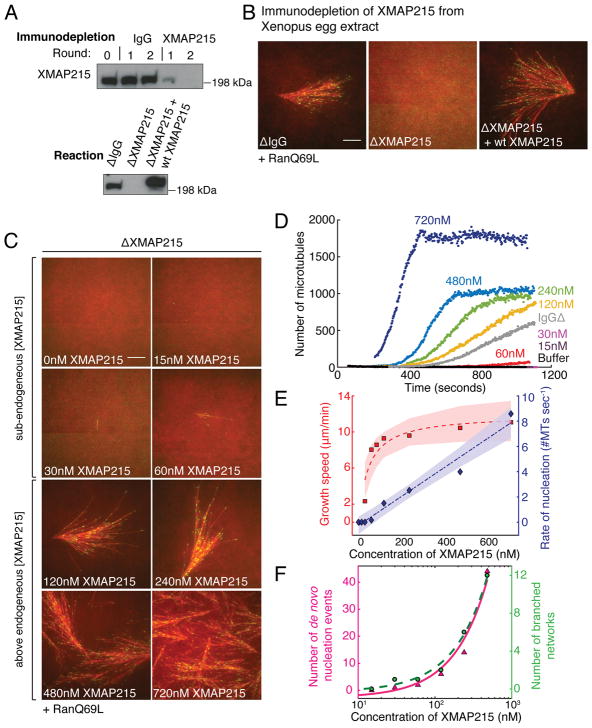
Figure 1. XMAP215 stimulates microtubule nucleation in Xenopus egg extract.
(A–B) Western blot analysis of XMAP215 or IgG immunodepletion from Xenopus extracts and add-back of wild-type XMAP215-GFP, probed using antibody against TOG12 domain. Branching MT nucleation in Xenopus extracts in the presence of 5.5μM RanQ69L. EB1-mCherry (pseudo-colored as green) and Cy5-labeled tubulin (red) was added. XMAP215-GFP was added back at 85nM (equivalent to 120nM in IgG-depleted extract). Representative images are displayed at 20 minutes of the reaction. Scale bar, 10μm. The experiments were repeated at least three times with independent extract preparations.
(C) Increasing concentration of XMAP215-GFP added back to immunodepleted extract with 5.5μM RanQ69L. Representative images are displayed at 480 seconds. Scale bar, 10μm. See Supplementary Video 2. The experiment was repeated four times with independent extract preparations.
(D) EB1 comets in the entire field of view were detected, counted and plotted with time. The analyses in (D–F) were repeated at least thrice with independent extract preparations.
(E) Growth speed of MTs was obtained by tracking all EB1 comets observed over during the experiment. No MTs nucleated below 30nM XMAP215, and growth speed was not measured. At 30nM, growth speed was measured manually as 2.3±0.8 μm/min (mean ± s.d.; n=25). For 60nM and above, growth speed was computed via image analysis: 60nM–8.0±1.9 (n=1470), 85nM–8.5±1.8 (n=18190), 120nM–9.3±2.0 (n=45090), 240nM–9.6±2.2 (n=59297), 480nM–10.5±2.4 (n=79381) and 720nM–11.1±2.7 μm/min (n=147008). n represents the number of growth speed measurements obtained from consecutive frames of all tracks. Mean speed (red squares) versus concentration was fit to Michaelis-Menten kinetics (dashed red). Rate of nucleation (blue diamonds) was measured as the slope of linear region of nucleation kinetics. Rate of nucleation versus concentration was regressed to straight line (dashed blue). 95% confidence intervals of the fits are shaded.
(F) De novo nucleation events and branched networks that emerged were counted manually for each reaction (magenta triangles and green circles respectively). Linear fit to each is plotted as solid and dashed curves. x-axis displayed in log scale, where 0nM concentration cannot be shown (green circles).
See Supplementary Figs. 1 and 9, Supplementary Video 1, and Supplementary Table 2 for source data.
Characterization of XMAP215’s microtubule nucleation activity
To further characterize its role, we investigated the effect of XMAP215 concentration in MT nucleation and polymerization processes. We added increasing concentrations of XMAP215 to immunodepleted extracts and evaluated a range of XMAP215 concentrations: from sub-endogenous levels (15 nM) to 6-fold the endogenous concentration (720 nM). No MTs formed at 15 nM XMAP215 and only a few MTs were visible at 30 nM (Fig. 1C–D, Supplementary Video 2). From 60 to 720 nM XMAP215, MT nucleation increased dramatically and the measured rate of MT nucleation varied linearly with XMAP215 concentration, without reaching saturation (Fig. 1E). We simultaneously measured the rate of polymerization or growth speed of MTs, which saturated at twice the endogenous concentration (240 nM), as previously reported26.
In summary, MT nucleation and polymerization by XMAP215 demonstrates different characteristics. First, no saturation limit for MT nucleation was observed for the range of XMAP215 concentrations tested (Supplementary Fig. 1A–B, Fig. 1E), yet the MT polymerization rate saturates near the endogenous concentration (Supplementary Fig. 1C, Fig. 1E). Second, a minimum concentration of XMAP215 (30 nM) is required for nucleating MTs, while XMAP215 promotes MT growth even at low concentrations24, 26. These results support that XMAP215 not only functions as a polymerase, but also promotes MT nucleation in the cytoplasm.
XMAP215 is a general microtubule nucleation factor
These findings led us to investigate whether XMAP215 increases only branching MT nucleation events, or generates MTs independently, which we define as de novo nucleation events. The number of de novo nucleation events and consequently, the branched MT networks formed, increased linearly with XMAP215 concentration (Fig. 1F), suggesting that XMAP215 functions as a general MT nucleation factor. To directly characterize this, XMAP215-depleted extracts were used in the absence of RanQ69L, thereby not inducing branching MT nucleation, and increasing concentration of XMAP215 was added (Fig. 2A, Supplementary Video 3). Remarkably, de novo MT nucleation events increased by 150-fold between 60 nM and 720 nM of XMAP215 (Fig. 2B). The occurrence of branching nucleation events was negligible (Fig. 2B). As before, no MTs formed at XMAP215 concentration below 30 nM and the polymerization rate saturated at 240 nM (Supplementary Fig. 2A). Additionally, we verified that XMAP215 stimulates de novo nucleation in the absence of the branching factors TPX2 and augmin36 (Fig. 2C–E, Supplementary Video 4). Thus, XMAP215 functions as a general MT nucleation factor in the cytoplasm. Notably, MT nucleation by XMAP215 was indistinguishable in extracts depleted of TPX2 or augmin, excluding a specific synergy between TPX2 and XMAP215 in this assay that was previously observed in vitro27.
Figure 2. XMAP215 is required for all microtubule nucleation events.
(A) De novo (or MT-independent) nucleation events were observed with increasing XMAP215 concentration added back to XMAP215-depleted extract without RanQ69L. Images displayed at 700 seconds of the reaction. Scale bar, 10 μm. See Supplementary Fig. 2A–B, Supplementary Video 3. The experiment was repeated twice with independent extract preparations, along with more than three additional experiments were performed where fewer concentration points were evaluated.
(B) Total number of MT-independent (de novo) and MT-dependent nucleation events that occurred until 600 seconds of the reaction were counted and plotted against XMAP215 concentration. Data for MT-independent nucleation events versus XMAP215 concentration was fitted to a linear curve (solid green). XMAP215 concentration is plotted on log scale. Few MT-dependent nucleation events were also observed (red circles). The analysis was repeated twice with independent extract preparations. See Supplementary Table 2 for source data.
(C) Immunoblot for TPX2 and augmin depletion corresponding to (D–E). Augmin was immunodepleted using anti-HAUS1 antibodies, and depletion was assessed by western blotting for two subunits - HAUS1 and HAUS6. The experiments in (C–E) were repeated twice with independent extract preparations, along with two supporting experiments performed. See Supplementary Fig. 9 for unprocessed blot.
(D) XMAP215-GFP was added to TPX2 and augmin immunodepletion at 360 nM in addition to endogenous protein in the presence of 5.5 μM RanQ69L. Equal volume of buffer was added for control reactions. Representative images at 20 minutes at displayed. Scale bar, 10 μm. See Supplementary Video 4.
(E) EB1 comets in the entire field of view were tracked using the analysis procedure described in Methods. Number of EB1 tracks were counted and plotted over time.
To further understand the relationship between XMAP215’s nucleation and polymerization activities, we designed an experiment to specifically observe MT polymerization by XMAP215 in Xenopus extracts. Instead of observing endogenous MT nucleation, we added pre-formed MT seeds stabilized by GMPCPP to XMAP215-depleted extracts. Subsequently, wild-type XMAP215 was added back to initiate MT growth (Supplementary Fig. 2B–E). In absence of XMAP215, polymerization was suppressed, resulting in infrequent EB1 blinking events on one end of MT seeds (roughly 20% seeds; Supplementary Fig. 2C,E). When 7.5 nM of XMAP215 was added back, MT growth immediately occurred from a large proportion of seeds (nearly 70%). At 15 nM XMAP215, MTs polymerized from all seeds. In contrast, no MT nucleation events were observed in the absence of seeds at XMAP215 concentrations below 30 nM (Figs. 1, 2A–B). Therefore, by titrating XMAP215 in Xenopus egg extracts, we find regimes where polymerization from MT plus-end occurs while nucleation does not (Figs. 1 and 2A–B, Supplementary Fig. 2B–E), and where polymerization saturates while nucleation continuously increases (Supplementary Fig. 1A–C, Figs. 1C–F and 2A–B).
γ-TuRC is required for microtubule nucleation by XMAP215
Our results show that XMAP215 is essential for MT generation, similar to the universal nucleator γ-TuRC, yet is γ-TuRC required for nucleation by XMAP215? To address this, we depleted γ-TuRC from Xenopus extracts and induced MT nucleation by RanQ69L, while XMAP215 and proteins required for branching MT nucleation were not depleted (Supplementary Fig. 3A). Almost no MTs formed with endogenous XMAP215 level in the absence of γ-TuRC (Fig. 3A). Surprisingly, both de novo and branching nucleation events were severely reduced even upon addition of excess XMAP215 to γ-TuRC-depleted extracts (Fig. 3A–B, Supplementary Fig. 3B–C; Supplementary Video 5). By specifically assaying de novo MT nucleation in Xenopus extracts as before (Fig. 2A), we further verified that XMAP215 promoted negligible de novo nucleation without γ-TuRC (Supplementary Fig. 3D–E). These results clearly demonstrate that both XMAP215 and γ-TuRC are required for all MT nucleation events in Xenopus extracts.
Figure 3. γ-TuRC is required for microtubule nucleation by XMAP215.
(A) Branching MT nucleation was induced in control and γ-tubulin depleted extract with 5.5 μM RanQ69L. XMAP215-GFP was added at 120 nM and 240 nM concentration in excess of the endogenous protein. Representative images are displayed at 1100 seconds of the reaction. Scale bar, 10 μm. See Supplementary Video 5. The experiment was repeated with three independent extract preparations, including verification with XenC antibody for γ-TuRC immunodepletion5, 11.
(B) Number of MT-independent and MT-dependent nucleation events observed was tabulated with excess XMAP215 concentration. Nucleation events were counted until 800 seconds for each reaction. The few MTs that emerged in γ-tubulin depleted extracts were counted manually (both MT-independent and MT-dependent nucleation events). For IgG depletion, total number of MTs (EB1 comets) in the field of view was counted using image analysis, while de novo nucleation events were counted manually. The number of MT-dependent nucleation events was calculated by subtracting MT-independent nucleation events from the total number of MTs.
See Supplementary Fig. 3. The analyses were repeated twice with experiments performed on independent extract preparations, with one additional supporting set of results with anti-XenC immunodepletion.
XMAP215 and γ-TuRC synergistically nucleate microtubules in vitro
Because we found that XMAP215 and γ-TuRC are both required for nucleation in Xenopus extract, we investigated their effect on MT nucleation in vitro. We first purified γ-TuRC via affinity purification from Xenopus egg extracts. All components of γ-TuRC were present as confirmed by mass spectrometry (Supplementary Fig. 4A), while proteins such as XMAP215, TPX2, α-tubulin, or TACC3 were not detected (Supplementary Fig. 4B). Furthermore, the characteristic ring shape of γ-TuRC was observed by electron microscopy (Fig. 4A). Affinity-purified γ-TuRC was further purified by sucrose gradient centrifugation, where γ-TuRC fractionated at the expected size (Fig. 4B).
Figure 4. XMAP215 stimulates microtubule nucleation by γ-TuRC.
(A) Negative stain electron microscopy shows 25 nm diameter ring structures characteristic of the γ-tubulin ring complex. Scale bar, 100 nm. The experiment was repeated thrice, with at least three supporting experiments with sucrose-gradient fractionated γ-TuRC, all showing distinct γ-TuRC ring structures.
(B) The peptide-eluted γ-TuRC was fractionated by sucrose gradient centrifugation. Fractions were analyzed by Silver-stained SDS-PAGE (top) and immunoblot (bottom) with antibodies against γ-tubulin, GCP5 and GCP4. These components were observed to peak in fractions 5 and 6, at the expected size for intact γ-TuRC. The bands for GCP6, 5, 4, 3, 2, γ-tubulin, NEDD1 and Mzt2 can clearly be seen in fractions 5 and 6 using silver staining. Representative image is displayed, and the experiment was repeated more than three times. See Supplementary Fig. 9 for unprocessed scans.
(C) Combination of purified γ-TuRC and XMAP215 promotes MT nucleation in vitro. Purified γ-TuRC at 250–400 pM added to αβ-tubulin with GTP promotes MT nucleation as compared to control (elution from IgG beads, upper left panel). Addition of recombinant XMAP215, at concentrations from 5 nM to 130 nM, promotes low levels of MT nucleation, with bundling and increased MT length seen at high concentrations. Addition of both XMAP215 and γ-TuRC together causes a significant increase in the number of MTs nucleated, greater than simply adding the MTs generated by each component independently. Representative fields of MTs are shown, nucleation assays were repeated with at least 3 independent γ-TuRC purifications. Scale bar, 10 μm.
(D) Fluorescent intensity was quantified as the readout of amount of polymerized tubulin for 10 fields of view for each reaction condition. Data from three independent γ-TuRC preparations was pooled and displayed as mean±s.d. n=30 fields of view analyzed per reaction. The XMAP215+γ-TuRC reactions had greater fluorescent intensity as compared to control. A significant increase between microtubule mass can be seen starting from 35nM XMAP215 when combined with γ-TuRC.
See Supplementary Fig. 4.
MT nucleation was analyzed in vitro by incubating soluble αβ-tubulin with purified γ-TuRC and XMAP215 independently or together (Fig. 4B). At minimal αβ-tubulin concentration of 10–12 μM, γ-TuRC (250–400 pM) or XMAP215 alone nucleated few MTs above the buffer control. Addition of γ-TuRC to increasing concentrations of XMAP215 (5–130 nM) resulted in a continuous increase in the amount of MTs observed (Fig. 4C). Remarkably, low concentration of XMAP215 (5–30 nM) together with γ-TuRC resulted in 6 to 10-fold increase in the number of MTs generated when compared to the sum of MTs generated by XMAP215 and γ-TuRC individually (Supplementary Fig. 4C). With higher XMAP215 concentrations, even more prominent increase in MT generation was observed when γ-TuRC was present (Fig. 4C). However, due to emergence of long, bundled MTs, individual MTs could not be counted and total MT fluorescence intensity across all reactions was measured instead (Fig. 4D). These measurements demonstrate that XMAP215 and γ-TuRC together induce more MT generation than the sum of MT mass produced by either component individually across the range of XMAP215 concentration (Fig. 4C–D, Supplementary Fig. 4D). Similar results were obtained with the gradient-fractionated γ-TuRC (Supplementary Fig. 4E). This in vitro reconstitution agrees with our results in Xenopus extracts (Fig. 1–2). Thus, XMAP215 and γ-TuRC synergistically promote MT nucleation both in vitro and ex vivo.
TOG domains of XMAP215 are essential for microtubule nucleation
We next asked what regions of XMAP215 are required for MT nucleation activity in the cytoplasm. We generated versions of XMAP215 lacking TOG domains from the N-terminus or missing its C-terminus. All constructs were tested for their nucleation activity in Xenopus extracts following depletion of endogenous XMAP215. Branching MT nucleation was stimulated by TPX2’s C-terminal half in addition to RanQ69L as described previously36, which allows for robust measurements of low nucleation levels by some XMAP215 constructs.
Deleting the TOG1 domain (TOG2-CT) caused significant reduction in MT nucleation (Fig. 5A, Supplementary Fig. 5A–B; Supplementary Video 6), and further deletion of TOG2 (TOG3-CT) or TOG domains 1–4 (TOG5-CT) completely abrogated MT generation. Smaller branched structures were generated when the C-terminal domain of XMAP215 was removed (TOG1-5), while further truncation from the C-terminus preserving TOG regions 1–4 additionally reduced the nucleation activity of the protein (Fig. 5B–C, Supplementary Video 7). These results indicate that the entire protein is needed for full MT nucleation activity, with the TOG domains being essential for retaining a minimal level of nucleation.
Figure 5. Microtubule nucleation by XMAP215 protein constructs in Xenopus egg extract.
Branching MT nucleation activity of XMAP215 constructs. All proteins were added back to XMAP215-depleted extract at final concentration of 120 nM in the presence of 5.5 μM RanQ69L and 1 μM GST-TPX2 α3-7. All experiments and analyses were repeated with at least three independent extract preparations.
(A) Representative images are displayed for N-terminal deletion constructs at 560 seconds of the reaction. Scale bar, 10 μm. See Supplementary Fig. 5A–B and Supplementary Video 6.
(B) Representative images are displayed for C-terminal deletion constructs at 480 seconds of the reaction. Scale bar, 10 μm. See Supplementary Video 7.
(C) Number of EB1 comets were detected, counted and plotted with time for protein constructs displayed in (B).
(D) Branching MT nucleation activity of tubulin-binding mutant constructs of TOG1-5 protein. Representative images are displayed at 650 seconds of the reaction. Scale bar, 10 μm. See Supplementary Video 8.
(E) Number of EB1 comets were detected, counted and plotted with time for protein constructs displayed in (D).
XMAP215’s TOG domains 1 and 2 linked to a MT-binding region has been reported to recapitulate the minimal polymerase activity24. This protein construct (TOG12-Kloop) did not have any discernable nucleation activity, suggesting that the minimal polymerization domains are not sufficient to promote MT nucleation.
Because TOG domains interact with αβ-tubulin to promote polymerization30, we tested whether disrupting αβ-tubulin binding to TOGs by mutating two residues in each TOG domain24 affects the nucleation activity of XMAP215. This mutation in all TOGs within the full-length XMAP215 and TOG1-5 construct completely abolished MT nucleation activity (Figure 5D–E, Supplementary Fig. 5C–D, Supplementary Video 8). Mutating residues in TOGs 1–3 individually dramatically reduced the nucleation activity of TOG1-5, with the more N-terminally situated TOGs having a greater effect, matching their described function in polymerization24. In contrast, mutation of TOG4 had a minor effect, while mutating TOG5 did not significantly impair MT nucleation. Interestingly, the loss of TOG5 domain caused significant reduction in MT nucleation (Fig. 5B–C, compare TOG1-5 with TOG1-4) while mutating TOG5 did not, suggesting that TOG5’s role may not involve its interaction with αβ-tubulin dimers. Lastly, maintaining the αβ-tubulin binding of TOGs 1 and 2 (TOG1-5 3–5AA) did not restore nucleation, demonstrating that TOG1 and TOG2 alone are not able to promote MT nucleation. Taken together, the interaction between αβ-tubulin and XMAP215’s TOG domains, in particular by the N-terminally situated TOGs, is important for MT nucleation activity.
C-terminus of XMAP215 is required for efficient microtubule nucleation
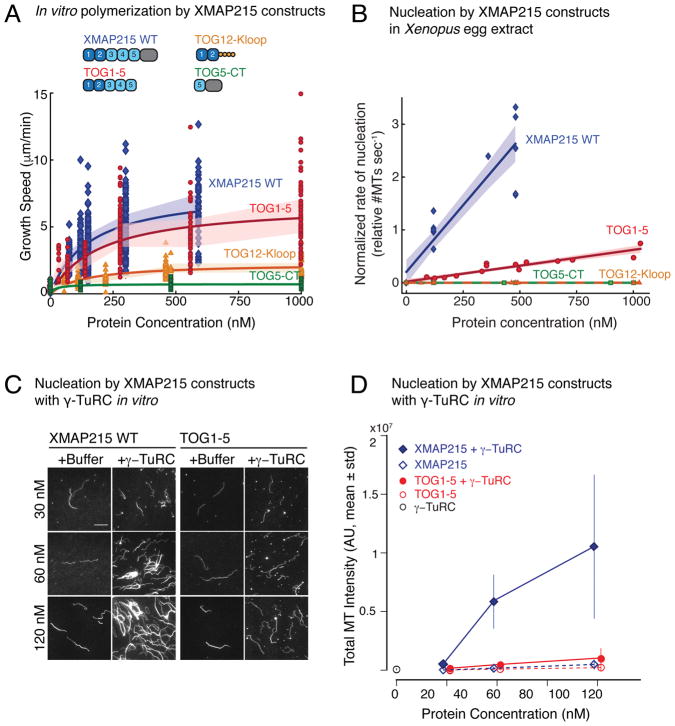
To compare how XMAP215’s domains contribute to MT polymerization versus MT nucleation, we chose three constructs, namely the N-terminal TOG domains 1–5 (TOG1-5), the construct containing TOG5 and the C-terminus (TOG5-CT) and the minimal polymerase (TOG12-Kloop). First, we performed in vitro MT polymerization from stabilized MT seeds in the presence of these constructs. Wild-type XMAP215 promoted MT plus-end growth in a concentration-dependent manner (Fig. 6A, Supplementary Fig. 6A), as described previously23, 24, 26, 37. TOG1-5 strongly promoted MT polymerization to an extent similar to the full-length protein. The maximum MT polymerization rate by the minimal polymerase TOG12-Kloop was 2–3 fold lower than wild-type XMAP215, as previously reported24. Most importantly, the C-terminal construct TOG5-CT did not increase MT polymerization rate even at high protein concentrations. In summary, XMAP215’s N-terminal TOG domains constitute the MT polymerizing region of the protein, and its C-terminus is neither required for nor affects the growth rate of MT plus-ends.
Figure 6. C-terminus of XMAP215 is required for microtubule nucleation with γ-TuRC.
(A) Polymerization from stabilized seeds was performed with several XMAP215 constructs: wild-type XMAP215, TOG1-5, TOG12-Kloop, TOG5-CT. Growth speed was measured from kymographs and plotted against protein concentration. Number of kymographs analysed (n): Buffer (n=21); XMAP215-WT: 75nM (n=43), 120nM (n=62), 150nM (n=141), 300nM (n=131), 590nM (n=53); TOG1-5: 35nM (n=12), 70nM (n=24), 140nM (n=57), 280nM (n=84), 560nM (n=49), 1000nM (n=59); TOG12-Kloop: 55nM (n=21), 110nM (n=51), 220nM (n=44), 460nM (n=70), 1000nM (n=32); TOG5-CT: 120nM (n-53), 480nM (n=74), 1000nM (n=46). Shaded region represents s.d. at individual concentrations. Michaelis-Menten fit to all measurements versus concentration is displayed (solid curves).
(B) Branching MT nucleation activity of XMAP215 constructs in (A) was observed in Xenopus egg extracts. Proteins were added to XMAP215-depleted extract at specified final concentration along with 5.5μM RanQ69L and 1μM GST-TPX2 α3-7. Rate of MT nucleation was measured from linear region of nucleation curves (representative plot for TOG1-5 shown in Supplementary Fig. 6B). Rate of nucleation by 120nM wild-type XMAP215 was normalized to 1 in each experimental set with Xenopus extracts. Where more than one reaction at 120nM wild-type XMAP215 was performed, their average was set to 1. Data was pooled from the following independent extract preparations: wild-type XMAP215(8), TOG1-5 protein(4), TOG12-Kloop(3), and TOG5-CT(2). Normalized rate of nucleation versus protein concentration was fit to straight line, with shaded region representing 95% confidence interval. See Supplementary Video 9.
(C–D) Purified γ-TuRC was incubated with wild-type XMAP215 or TOG1-5 to promote MT nucleation in vitro. Addition of 30–120nM wild-type XMAP215 promotes synergistic MT nucleation with γ-TuRC, while TOG1-5 shows minimal synergy with γ-TuRC. Representative fields of MTs are displayed. Experiments were repeated with two independent γ-TuRC purifications. Scale bar, 10μm.
(D) Quantification of total fluorescent intensity. All data from two independent γ-TuRC preparations was pooled and plotted as mean±s.d., except 30nM TOG1-5 condition, which was performed with one γ-TuRC preparation. γ-TuRC (n=44); XMAP215: 30nM (n=20), 60nM (n=50), 120nM (n=43); TOG1-5: 30nM (n=20), 60nM (n=39), 120nM (n=44); γ-TuRC+XMAP215 30nM (n=20), 60nM (n=45), 120nM (n=39); γ-TuRC+TOG1-5: 30nM (n=20), 60nM (n=41), 120nM (n=41). n represents the number of fields of view analyzed.
See Supplementary Table 1 for source data of Figs. 6B–D, and Supplementary Fig. 6.
To thoroughly compare the nucleation activity to polymerization by these domains, we repeated the branching MT nucleation assay in Xenopus extracts as a function of XMAP215 concentration. As before, upon depletion of endogenous XMAP215 followed by add-back of increasing concentrations of wild-type XMAP215, an increase in the rate of MT nucleation was observed (Fig. 6B, Supplementary Fig. 6B; Supplementary Video 9). Although add-back of TOG1-5 forms MT networks, the rate of MT nucleation was significantly reduced. Even at high TOG1-5 concentrations (500 nM – 1 μM), the rate of MT nucleation remained lower than at endogenous concentrations of full-length XMAP215 (120 nM). Neither addition of up to 1 μM of the C-terminal construct (TOG5-CT) nor the minimal polymerase (TOG12-Kloop) restored MT nucleation. The side-by-side comparison of these constructs in MT polymerization and nucleation assays (Fig. 6A–B) demonstrates that XMAP215’s C-terminus does not directly promote polymerization, yet is required for rapid MT nucleation in Xenopus extracts.
To further analyze the requirement of XMAP215’s C-terminus in nucleating MTs, we performed in vitro MT nucleation assay with purified γ-TuRC and increasing concentrations of TOG1-5 protein (Fig. 6C). Similar to full-length XMAP215, TOG1-5 alone displays low nucleation activity. Combining γ-TuRC and TOG1-5 resulted in slightly increased MT nucleation when compared to γ-TuRC alone (Fig. 6C, Supplementary Fig. 6C), yet TOG1-5 induced lower MT nucleation with γ-TuRC than the wild-type XMAP215. At higher protein concentrations, the difference in nucleation activity of full-length XMAP215 and TOG1-5 was more pronounced and TOG1-5 shows a dramatically reduced effect on MT generation when combined with γ-TuRC (Fig. 6D). Together, these results demonstrate that the polymerizing region of XMAP215, the N-terminal TOG1-5, is not sufficient to promote MT nucleation similar to the full-length XMAP215 and γ-TuRC, and that the conserved C-terminus is required for robust MT nucleation.
XMAP215 and γ-TuRC components interact in Xenopus egg extract
Because γ-TuRC and XMAP215 cooperate to generate MTs, we hypothesized that these components may interact in Xenopus egg extracts. Indeed, components of γ-TuRC co-immunoprecipitate with XMAP215 (Supplementary Fig. 7A) and reciprocally, endogenous XMAP215 was co-precipitated with γ-TuRC (Supplementary Fig. 7B), confirming an interaction between these components in egg extracts. Notably, the branching factor TPX2 did not co-precipitate in either experiments, while augmin did not co-precipitate with XMAP215, verifying the specificity of this interaction. We note that in Drosophila S2 cells, XMAP215 and augmin were co-immunoprecipitated38. This could be due to system-specific differences, such as the species, cell-cycle state, or presence of MTOCs that are absent from Xenopus extracts.
It is noteworthy that αβ-tubulin was not co-immunoprecipitated with endogenous XMAP215 (Supplementary Fig. 7A). This was surprising because XMAP215 and αβ-tubulin directly interact in vitro23, 24. We determined that XMAP215 and αβ-tubulin interact in vitro using size exclusion chromatography only under low salt buffer 23, 24 (Supplementary Fig. 7C, 75 mM NaCl). This interaction was not maintained near the physiological salt concentration (150 mM NaCl), suggesting that XMAP215’s binding to αβ-tubulin is transient in the cytoplasmic environment of Xenopus extracts.
XMAP215’s C-terminus directly binds to γ-tubulin
To further characterize the interaction between XMAP215 and γ-TuRC, we specifically tested binding between XMAP215 and γ-tubulin, the most abundant component of γ-TuRC. We performed size exclusion chromatography with purified human γ-tubulin and XMAP215. To prevent formation of γ-tubulin filaments at several hundred nM concentration39, we used a low molar ratio of γ-tubulin to XMAP215. In this setup, γ-tubulin alone elutes in fractions H–K (Fig. 7A) and partially oligomerizes in the chromatography buffer (compare to Supplementary Fig. 8A). Interestingly, in the presence of XMAP215, γ-tubulin shifts toward an earlier elution volume corresponding to XMAP215’s elution, demonstrating its direct interaction with γ-tubulin. This binding was verified using in vitro immunoprecipitations (Supplementary Fig. 8B). Notably, this interaction occurs even at the low γ-tubulin concentrations used in these experiments (140 nM in Fig. 7A, 80 nM in Supplementary Fig. 8B), which are within the cytoplasmic γ-tubulin concentrations40. Mutating the residues essential for αβ-tubulin interaction in TOG domains did not abolish γ-tubulin binding (Supplementary Fig. 8C). Next, we identified the XMAP215 domain responsible for interacting with γ-tubulin. The N-terminal truncation containing TOGs 1-4, or TOGs 1-5, or the protein containing TOGs 1 and 2 repeated twice did not interact with γ-tubulin (Fig. 7A, Supplementary Fig. 8C). Surprisingly, the C-terminal construct containing TOG5 and the C-terminal domain (TOG5-CT) strongly shifted γ-tubulin to an earlier elution volume, similar to full-length XMAP215 (Fig. 7A). These results demonstrate that XMAP215 directly binds to γ-tubulin, potentially in an oligomeric form, via its C-terminal domain.
Figure 7. XMAP215 interacts with γ-tubulin via its C-terminal domains.
(A) Size exclusion chromatography was performed with 140 nM of human γ-tubulin alone and with 1–1.4 μM XMAP215 protein constructs. 500 μl sample volume was injected into the column, 300 μl fractions were collected and alternate fractions eluted between 8.5 ml to 16.6 ml were analyzed via SDS-PAGE followed by immunoblot with γ-tubulin antibodies and GFP antibodies to detect the elution profile of γ-tubulin and GFP labeled XMAP215 proteins, respectively. Shift in γ-tubulin signal was observed to assess the region of XMAP215 that γ-tubulin interacts with. Stoke’s radii of reference proteins are marked at their peak elution: Thyroglobulin (8.6 nm), Aldolase (4.6 nm) and Ovalbumin (2.8 nm). The contrast of each image was adjusted to display the entire elution profile clearly. Each chromatography run was repeated at least twice on different days at the specified concentration, and at least one additional supporting experiment for each displayed run (more than nine supporting runs in total) was performed at slightly different protein concentrations. Void volume of the column was measured as 8.7–8.8 ml.
See Supplementary Fig. 8A–C. See also Supplementary Fig. 9 for unprocessed scans.
We next validated the region of XMAP215 that interacts with αβ-tubulin using size exclusion chromatography. The N-terminal TOG1-4 protein formed a complex with αβ-tubulin similar to wild-type XMAP215, while the C-terminal construct TOG5-CT did not bind αβ-tubulin (Fig. 8A, Supplementary Fig. 8D). Altogether, these results show that XMAP215’s N-terminus binds to αβ-tubulin, while its C-terminus interacts with γ-tubulin.
Figure 8. N-terminus of XMAP215 interacts with αβ-tubulin, and model for how XMAP215 and γ-TuRC promote microtubule nucleation.
(A) XMAP215 N-terminal TOG1-4 construct or C-terminal TOG5-CT construct (5–6 μM) and bovine αβ-tubulin (6.2 μM) were mixed and applied to size exclusion chromatography. 100 μl sample volume was injected into the column. Each chromatography run was performed once at the specified concentration. More than three additional supporting experiments were performed either with slightly different protein concentrations or using alternative protein constructs. Void volume of the column was measured as 8.7–8.8 ml. See Supplementary Fig. 8D. See also Supplementary Fig. 9 for unprocessed scans.
(B) Schematic representation for how XMAP215 and γ-TuRC could together promote MT nucleation. While γ-TuRC independently induces low MT nucleation (left), its cooperation with XMAP215 promotes efficient nucleation of MTs. Synergistic microtubule nucleation model (right): XMAP215 binds to γ-TuRC with its C-terminus and promotes the assembly of αβ-tubulin dimers onto γ-TuRC via its N-terminus.
Discussion
How MT nucleation occurs is critical for understanding how MT cytoskeletal structures that enable cell function are assembled. γ-TuRC has been thought to play a prominent role in MT nucleation since its discovery in 198941, but evidence suggested other factors might exist as well. In this study, we uncover that the major polymerase of the cell, XMAP215, is a principal MT nucleation factor that directly interacts with γ-tubulin and functions synergistically with γ-TuRC to generate MTs in the cytoplasm.
We propose several reasons why XMAP215’s role in MT nucleation has been underappreciated until now. Of primary importance, assays to resolve MT nucleation in the cytoplasm and to distinguish it from plus-end growth were previously unavailable. Additionally, compensatory nucleation mechanisms in the mitotic spindle11,26, 42, introduction of MTOCs such as centrosomes32 that contain XMAP21543, or incomplete loss of XMAP21520,44 could have masked its function in MT nucleation. Here, by developing a high-resolution assay and analyses system, stimulating nucleation without MTOCs, and via complete XMAP215-depletion, we demonstrate that XMAP215, along with γ-TuRC, is necessary for all nucleation events in Xenopus egg extracts and determines the rate of MT generation in the cytoplasm.
One widely accepted model for MT nucleation7 proposes that γ-tubulin complexes perform the entire nucleation process. In this scenario, XMAP215, acting solely as a polymerase, catalyzes the addition of αβ-tubulin dimers to the plus-end after MT nucleation has occurred26. In contrast, our study clearly demonstrates that XMAP215 and γ-TuRC function cooperatively in the nucleation process, while either factor independently supports low MT nucleation (Fig. 8B, left). What is the molecular mechanism by which γ-TuRC and XMAP215 nucleate MTs? We propose a synergistic nucleation model in which both XMAP215 and γ-TuRC are directly involved in assembling MT precursors. The C-terminal region of XMAP215 binds to γ-TuRC where the TOG domains recruit αβ-tubulin dimers and both factors together form the MT nucleation intermediates (Fig. 8B, right). The domain organization of the conserved C-terminus still remains to be characterized, but the existence of a sixth TOG domain has been speculated45, 46, which could be a potential γ-tubulin interaction site. In the N-terminus, TOG domains 1 and 2 could primarily recruit αβ-tubulin dimers, whereas the later TOG domains stabilize the newly forming MT. This is supported by recent work proposing TOG5’s interaction with lattice-incorporated αβ-tubulin31, 47. Following MT nucleation, XMAP215 could translate with the growing MT plus-end to catalyze MT polymerization. In the future, it will be important to further investigate the mechanism of how XMAP215 and γ-TuRC cooperate to nucleate a MT using single molecule techniques and structural studies.
Because MT nucleation is the rate-limiting process in constructing the cytoskeleton, activity of the principal nucleation module consisting of XMAP215 and γ-TuRC must be critically regulated in space and time to rapidly assemble complex MT structures. While γ-TuRC has been proposed to be regulated by activator proteins such as CDK5RAP215, it is feasible that XMAP215 is also regulated. For example, the localization of XMAP215 at the centrosome by TACC348 could function to concentrate XMAP215 near γ-TuRCs, or post-translational modifications19, 49 could temporally regulate its nucleation activity. With the discovery of XMAP215 and γ-TuRC’s cooperation in the MT nucleation process, we can now begin evaluating additional factors that regulate MT generation.
Methods
Purification of recombinant proteins
Wild-type XMAP215 C-terminal GFP-7xHis clone was a gift from Simone Reber26, and TOG12-Kloop construct from Per Widlund (TOG12+++24). XMAP215 1-5AA-GFP-6xHis was synthesized (Genscript) and cloned into pFastBac vector. Remaining XMAP215 constructs with C-terminal GFP-7xHis-Strep tags were cloned into pST50Tr-STRHISNDHFR vector50 using Gibson Assembly (NEB). The deletion constructs consisted of the following residues: TOG2-CT(264-2065), TOG3-CT(544-2065), TOG5-CT(1091-2065), TOG1-5(1-1460), TOG1-4(1-1090), TOG1212(1-543+1-515). Tubulin-binding mutants were derivatives of TOG1-5 or full-length XMAP215. Two residues in each TOG domain were mutated to alanine as described24. All constructs were fully sequenced.
Wild-type XMAP215-GFP and XMAP215 1-5AA were purified from Sf9 cells using Bac-to-Bac system (Invitrogen). All other constructs were expressed in E. coli Rosetta2 cells (Novagen) by inducing with 0.5–1 mM IPTG for 12–16 hours at 16 °C or 6–8 hours at 25 °C. The cells were lysed (EmulsiFlex, Avestin) and lysate was clarified by centrifugation at 13,000 rpm in Fiberlite F21-8 rotor (ThermoFisher) or at 65,000 rpm in Ti70 rotor (Beckman Coulter) for 30–45 minutes.
All proteins, except wild-type XMAP215 and TOG12-Kloop, were purified using Strep-affinity (Strep-Trap HP, GE Healthcare), followed either by gel filtration (HiLoad 16/600 Superdex, GE Healthcare) or dialyzed into storage buffer. Wild-type XMAP215 was purified using His-affinity (His-Trap, GE Healthcare), then cation-exchange (Mono S 10/100 GL, GE Healthcare). TOG12-Kloop was purified as described previously24. His-GFP was purified using His-affinity. C-terminal GFP was replaced with mCherry tag in the pET21a vector carrying EB151. EB1-mCherry was purified using His-affinity and dialyzed into CSF-XB buffer. Purifications of His-RanQ69L and GST-tagged TPX2 α3-7 were described recently52.
All proteins were dialyzed into CSF-XB buffer (100mM KCl, 10mM K-HEPES, 5mM K-EGTA, 1mM MgCl2, 0.1mM CaCl2, pH 7.7) with additional 10% w/v sucrose, flash-frozen and stored at −80°C. Protein concentrations were determined by analyzing a Coomassie-stained SDS-PAGE against known concentrations of BSA (A7906, Sigma).
Antibodies
See Supplementary Table 1 for information on antibodies used in this study.
Xenopus egg extracts, immunodepletion and immunoprecipitation experiments
CSF extracts were prepared from Xenopus laevis oocytes as described 53, 54 and either used immediately or immunodepleted51, 53. When working with Xenopus laevis, all relevant ethical regulations were followed, and all procedures were approved by Princeton IACUC. For immunodepletions, XMAP215-, TPX2-, HAUS1-, or γ-tubulin- and IgG(control)-antibody were conjugated to Protein A Dynabeads (Life Technologies #10002D) overnight. 50–150 μl extracts were subjected to two sequential rounds of immunodepletion by incubating with roughly equal volume of antibody-conjugated bead slurry for 30–40 minutes in each round, as described previously51, 53. See Supplementary Table 1 for antibody concentrations. Control immunodepletion was always performed and depletion efficiency was assessed using western blots and functional assays.
The immunoprecipitation (IP) experiment of XMAP215 and reciprocal IP of γ-TuRC (Supplemental Fig. 7A–B) was performed by coupling either XMAP215, γ-tubulin or Mzt1 antibody to Dynabeads. For IP of XMAP215, 75μl of egg extract was incubated with 125μl of Dynabeads slurry for 1 hour on ice. Beads were washed thrice with 5 volumes of TBS buffer + 0.1% Tween20. For IP of γ-TuRC, 50μl of extract was incubated with 50μl of Dynabeads slurry for 1 hour at 4°C, then washed twice with 5 volumes of CSF-XB. Beads were boiled and protein content analyzed using immunoblot (chemiluminescence), imaged via X-ray film or iBright imaging system (ThermoFisher Scientific).
Microtubule nucleation assays in Xenopus egg extracts
Microtubule (MT) nucleation was assayed in Xenopus egg extracts as previously described36, 55. Briefly, all reactions were performed with 0.5mM vanadate (sodium orthovanadate, NEB) to avoid sliding of MTs. 5.5μM RanQ69L was added for branching nucleation experiments. For de novo nucleation reactions, an equal volume of buffer was added. MTs were labeled with 0.89μM Cy5-labeled porcine brain tubulin and 200nM EB1-mCherry (plus-tips). For testing activity of XMAP215 constructs (Fig. 5, 6), 1 μM GST-tagged X.l. TPX2 α3-736 was also added to decrease the time lag until MT nucleation. For all experiments, either untreated extracts were used, or endogenous XMAP215, TPX2, augmin or γ-tubulin was immunodepleted prior to use, and purified wild-type XMAP215 or XMAP215 constructs were added to the extract mixture at specified final concentrations. For Fig. 6, freshly purified proteins (TOG1-5, TOG12-Kloop, TOG5-CT) were used for comparing MT nucleation in extracts to in vitro polymerization (Fig. 6A), and the same protein purification was used in both assays. The reaction mixture was prepared, incubated on ice for 2–3 minutes, 6μl mixture was pipetted into a flow cell to start the reaction (timed as 0 seconds), and imaged for 15–20 minutes at 18–20°C in a temperature-controlled room. All reagents added to extracts were stored in CSF-XB buffer, no more than 25% volumetric dilution of extracts was performed, and reaction composition was kept constant for each experimental set. All reactions shown in individual figure panels were performed with one Xenopus extract preparation. TIRF imaging was performed with Nikon TiE microscope using 100x 1.49 NA objective. Andor Zyla sCMOS camera was used for acquisition, with the field of view of [165.1x139.3 μm] or [132.1x132.1 μm]. 2x2 binned, dual color images were acquired every 2 seconds using NIS-Elements software (Nikon). Brightness and contrast were optimized individually for display.
Analysis of microtubule nucleation in Xenopus egg extracts
Number of MTs and their growth speed in extracts was measured using a combination of custom-built and existing MATLAB software. EB1 intensity in the image sequence was homogenized by dividing a blank background image that was pre-blurred with Gaussian kernel of 20 pixels. For experiments in Figs. 1–3, immotile features were first discarded by applying temporal median filter of 6–8 frames. EB1 comets, were detected by comet detection and tracked by plus-end tracking module of uTrack56, 57. An alternative pre-processing method was devised to analyze nucleation by XMAP215 constructs (Fig. 5–6) that was insensitive to growth speed or EB1 comet size, which varied between constructs. After background correction, EB1 comets were enhanced by applying 2 pixels wide, Laplacian of Gaussian filter, followed by uTrack’s comet detection. MT nucleation curves were generated by plotting the number of EB1 comets detected or the number of EB1 tracks for each frame over time, as specified in each figure legend. For growth speed calculation, all EB1 tracks were classified into growth and forward gaps using uTrack’s plus-end dynamics classification. Growth speed was measured from consecutive frames of growth events for all tracks.
De novo nucleation events and branched MT networks (Figs. 1F, 2B) were counted manually for the entire experimental duration or until a fixed time specified in figure legends. Lifetime of MTs was measured manually for Supplementary Fig. 1D. Data regression to Michaelis-Menten kinetics or straight line was performed using curve fit function in MATLAB.
Purification of γ-TuRC from Xenopus egg extracts
γ-TuRC purification was based on previous reports5, 17, 58, 59. 10 ml of Xenopus egg extract was diluted with 5 volumes of CSF-XB+ buffer (CSF-XB buffer + 1mM GTP, 1mM DTT and protease inhibitors) supplemented with 10% w/v sucrose and spun at 3500 rpm for 10 minutes to pellet large particles. 1mM GTP was added to all buffers and all steps were performed at 4°C. The supernatant was diluted two-fold with CSF-XB+ buffer, and filtered through a 0.22μm filter. γ-TuRC was precipitated by addition of 30% polyethylene glycol (PEG) solution to final PEG concentration of 6.5%, incubated for 30 minutes, then centrifuged for 20 minutes at 17,500xg. The PEG pellet was resuspended in 20ml CSF-XB+ buffer with 0.05% NP-40, sample was centrifuged at 136,000xg for 7 minutes, and supernatant was pre-cleared with Protein A Sepharose beads (GE LifeSciences #17127901) for 20 minutes. Beads were removed, 1ml of γ-tubulin antibody (1mg/ml) was added, and sample was incubated for 2 hours with rotation. 1ml of washed Protein A Sepharose beads were then added and incubated for 2 hours. Beads containing bound γ-TuRC were centrifuged and the flow-through removed. Beads were washed thrice with 10ml of CSF-XB+ with 0.05% NP-40, with 30ml of CSF-XB+ containing 250mM KCl, 10ml of CSF-XB+ with 1mM ATP to remove heat-shock proteins, and finally with 10ml of CSF-XB+ buffer. 1ml of γ-tubulin peptide (residues 413–451) at 0.4mg/ml in CSF-XB+ buffer incubated with beads overnight. After 10 hours, additional CSF-XB+ buffer was added and fractions 1–3 (1ml each) were collected. For sucrose gradient fractionation, CSF-XB+ buffer with 10% sucrose and CSF-XB+ with 50% sucrose was layered in an ultra-clear 2.2ml tube (11x34 mm, Beckman Coulter) and continuous 10–50% sucrose gradient was made using two-step program in Gradient Master 108 machine. Peptide-eluted fractions were combined, concentrated, layered on top of the gradient and subjected to centrifugation at 200,000xg in TLS55 rotor for 4 hours. The gradient was fractionated from the top in 10–11 fractions and immunoblotted against GCP4, GCP5 and γ–tubulin to determine γ-TuRC fractions.
Negative stain electron microscopy
5μl of γ-TuRC sample was pipetted onto carbon grids and incubated at room temperature for 5 minutes. The grid was rinsed briefly in deionized water, and 1% uranyl acetate was flowed over. Uranyl acetate was wicked away and the grid was air-dried. Images were taken on CM100 TEM microscope.
Mass Spectrometry
Peptide-eluted γ-TuRC obtained was precipitated with Trichloroacetic acid (TCA), resuspended in SDS buffer, separated by PAGE and stained with Coomassie dye. Gel bands were excised and analyzed by the Mass Spectrometry Core facility at Princeton University. Gel bands were trypsin-digested and peptides analyzed on Thermo Orbitrap Elite in data dependent mode with 120,000 MS1 resolution and up to 20 MS/MS scans in ion trap. Raw files were searched using Proteome Discoverer 2.1 (Thermo Scientific) with 10ppm MS1 and 0.5Da MS2 tolerances. Caramidomethylation of cysteine was set as fixed modification, while oxidation of methionine, deamination of asparagine and conversion glutamine to pyro-glutamate (at N-termini) as dynamic modifications. Data were searched against Xenopus protein database defined in ref. 40 supplemented with common contaminant proteins. Scaffold 4.7.5 (Proteome Software) was used to validate MS/MS identifications. 1% protein FDR and at least 2 peptides were accepted as confident protein identifications.
Nucleation assay with purified γ-TuRC
Peptide-eluted γ-TuRC was diluted two-fold in BRB80 buffer (80mM K-PIPES, 1mM MgCl2, 1mM EGTA, pH 6.8) to final concentration of 3–6nM GCP4 subunit, determined by quantitative immunoblot against purified, recombinant GCP4. Gradient fractionated γ-TuRC was concentrated and exchanged into BRB80 using Amicon Ultra-4 ml 10,000 NMWL centrifugal filter unit. Bovine brain αβ-tubulin (Pursolutions) +10% Alexa-568 labeled porcine brain tubulin, BRB80 buffer and 1.5mM GTP were combined and clarified by centrifugation at 80,000 rpm (TLA100.1 rotor, Beckman Coulter) for 20 minutes at 2°C. The final tubulin concentration for each reaction ranged within 10–12μM. Microtubule nucleation was performed as described previously5, 17, 58, 60. Clarified tubulin mixture was combined with γ-TuRC with or without recombinant XMAP215 or TOG1-5 at described concentrations to 20μl final volume on ice. The solution was incubated at 37°C for 5 minutes, diluted with 80μl warm BRB80, and the reaction terminated with addition of 100μl of 2% glutaraldehyde in BRB80 buffer, then incubated at room temperature for 5 minutes. Samples were diluted 1:10 in BRB80, layered on top of 5ml cushion of 20% glycerol in BRB80 buffer in 15ml Corex tube containing custom inserts to support round, poly-lysine coated coverslips. The sample was centrifuged for 45 minutes at 25,000xg in HB-6 rotor at 4°C. Following centrifugation, MTs were fixed with ice-cold methanol, coverslip mounted in Prolong Diamond and imaged via TIRF microscopy as described above. All images from each experimental set were taken in the same imaging session with constant TIRF angle, laser power and exposure.
Number of MTs in the entire field of view was counted manually, and their total intensity was determined using a MATLAB script. Images were thresholded via Otsu method, eroded to eliminate tubulin aggregates having aspect ratio of one and a mask for MT signal was generated. Average intensity from reverse mask was subtracted from the image as background. The residual fluorescent intensity per image was summed and reported.
In vitro microtubule polymerization
Coverslips and glass slides were coated with dichlorodimethylsilane61. Biotin-labeled, Alexa-594, GMPCPP MTs were made as described61, flash frozen and stored at −80°C. Aliquots were thawed at 37°C and diluted 2000-fold in BRB80 buffer. TIRF objective was warmed to 35°C using objective heater (Bioptechs, model 150819-13) prior to experiments.
Polymerization reactions were performed similar to previous reports62. Briefly, the flow chamber was incubated with anti-biotin antibody for 5 minutes, rinsed with BRB80 buffer, incubated with 1% Pluronic F127 in BRB80 for 5–10 minutes, rinsed again with BRB80, and incubated with diluted MT seeds for 10 minutes. The slide was protected from light. Unattached seeds were remove with BRB80 buffer + Trolox mix (2.5mM PCA, 25nM PCD, 2mM Trolox)63. Attached seeds were visualized on the microscope. Polymerization mix (7.5μM tubulin, 1mg/ml κ-casein, 14.3mM BME, 1mM GTP in BRB80) was flowed in at room temperature to prime the channel, followed by XMAP215 reaction mix (XMAP215 protein at indicated concentration, 6.75μM unlabeled bovine tubulin, 0.75μM Cy5-labeled tubulin, 1mg/ml κ–casein, 14.3mM BME, 1mM GTP and Trolox mix in BRB80). The chamber was immediately placed on the objective, and imaged using the TIRF setup described above with the field of view of [132.1x132.1μm]. Single color images were obtained every 6 seconds, using 647nm excitation (Cy5-dye), for 15 minutes, starting 2 minutes after the slide placement.
MT polymerization was analyzed using ImageJ software. Maximum intensity projection of movie stacks was produced and all polymerized MTs were marked using ROI generator. The stacks were resliced using multi-kymograph plugin. Growth speed of plus-end was obtained as the slope of each kymograph.
Microtubule polymerization in Xenopus egg extracts
Coverslips were passivated with dichlorodimethylsilane61. 20μM biotin and Alexa-488-labeled, GMPCPP seeds were prepared fresh as described23, digested with 0.2mg/ml subtilisin A protease (P5380, Sigma) for 20 minutes at 37°C. The digestion was stopped with 10μM PMSF. MTs were pelleted for 20 minutes and resuspended in BRB80. Flow chambers were treated sequentially with anti-biotin antibody (10 minutes), BRB80 buffer, 500–1000-fold dilution of subtilisin-digested seeds (10 minutes), and finally with CSF-XB buffer to remove unattached seeds. XMAP215 was depleted from egg extracts, and low concentrations of recombinant XMAP215-GFP was added back, along with 0.5mM vanadate, 0.89μM Cy5-tubulin and 200nM EB1-mCherry. Extract mixture was added to chamber containing subtilisin-digested seeds (timed as 0 seconds). Growth from most MT seeds was observed immediately (within 30–60 seconds). Imaging was completed within 300 seconds due to disappearance of seeds by motors and slow degradation in extracts. TIRF imaging was performed as described above. 2x2 binned, triple channel images of Alexa-488 GMPCPP seeds (pseudocolored as blue), EB1-mCherry (pseudocolored as green), Cy5-tubulin (red) were acquired every 3.5 seconds at 18–20°C.
Growth speed was obtained by analyzing composite kymographs of EB1 and tubulin intensity, as described above. Central, most-illuminated part of movie stack was cropped [103.08x103.08μm], GMPCPP seeds were observed roughly between 90–140 seconds from the start of reaction, and composite EB1 and tubulin signal was used to visually assess if the observed GMPCPP seed polymerized. For buffer addback (0nM XMAP215), tubulin signal was not observed on all seeds, and this quantification was based on seeds where one end clearly showed EB1 spots associating, translating and dissociating in the movie stack (or EB1 blinking).
Size exclusion chromatography experiments
Size exclusion chromatography (SEC) with γ-tubulin was performed as follows. All steps were performed at 4°C. Purified, human γ-tubulin was generous gift from Michelle Moritz in γ-TB buffer [defined as 50mM K-MES pH6.6, 5mM MgCl2, 1mM EGTA, 10mM thioglycerol, 10μM GDP] with additional 500mM KCl and 10% glycerol. At high protein concentration, γ-tubulin forms large oligomers or filaments below physiological salt amounts39, therefore the salt concentration was reduced only when γ-tubulin concentration was simultaneously decreased. Small volumes (100–300μl) of XMAP215 proteins were dialyzed (69590, Life Technologies) into γ-TB buffer without salt or GDP for 2 hours. After dialysis, protein aggregates were pelleted and concentration was estimated using Bradford Reagent (Bio-Rad). γ-tubulin was diluted to 280nM in γ-TB buffer containing 220mM KCl. γ-tubulin and XMAP215 proteins were individually clarified by ultracentrifugation at 80,000 rpm in TLA 100 (Beckman Coulter) for 15 minutes, then mixed in 1:1 volume ratio and incubated on ice for 10 minutes. 500μl of the mixture was loaded onto Superdex 200 Increase 10/300 column (GE Healthcare). The final concentration loaded to the column was 140nM for γ-tubulin and between 1–1.4μM for most XMAP215 proteins, except XMAP215 FL 1–5AA at 0.7μM. The column was equilibrated with γ-TB buffer containing 85 mM KCl and chromatography was performed in this buffer.
SEC experiments with αβ-tubulin was performed similar to previous reports24. Briefly, Superdex 200 Increase 10/300 column was equilibrated with chromatography buffer. Bovine brain tubulin (Pursolutions) mixed with GTP, and XMAP215 construct were clarified individually as described above. For runs performed with the truncation constructs (Fig. 8A), final concentrations of 6.2μM αβ-tubulin, TOG1-4 GFP (5μM) and TOG5-CT GFP (6μM) were used and chromatography performed in αβ-TB buffer [25mM Tris-HCl (pH 7.5), 75mM NaCl, 1mM MgCl2, 1mM EGTA, 1mM BME] supplemented with 0.05% Tween20. For wild-type XMAP215-GFP runs (Supplementary Fig. 8D), αβ-TB buffer with 0.1% Tween20 was used for chromatography along with final 4.4μM tubulin and 4.3μM XMAP215-GFP concentration. Runs in Supplementary Fig. 7C were performed in αβ-TB buffer containing either 75mM NaCl (low salt concentration) or 150mM NaCl (physiological salt concentration), both containing 0.025% Tween20 instead, and XMAP215-GFP at 4.3μM and tubulin at 4.4μM was used. Each reactions was mixed to final 0.2mM GTP concentration, incubated on ice for 10 minutes and 100μl was loaded onto the column.
For all control chromatography runs, equal volume of corresponding buffer was used. Absorbance at 280 nm was recorded. High molecular weight gel filtration standards (Thyroglobulin, Aldolase and Ovalbumin) were purchased from GE Healthcare (Catalog #28403842) and used to estimate the Stoke’s radii of eluted proteins in the same buffer as used for corresponding SEC run64. For γ-tubulin SEC experiments, 0.3ml fractions were collected and alternate fractions eluted between 8.5ml and 16.6ml were analyzed via immunoblot against γ-tubulin and GFP to detect XMAP215-GFP constructs. Secondary antibody conjugated to 800nm IRDye (LI-COR) was used and imaged with Odyssey CLx imaging station (LI-COR). For αβ-tubulin SEC experiments, 0.2ml fractions were collected and alternate fractions between 8.5ml and 13.9ml were analyzed via SDS-PAGE, stained with Coomassie dye.
In vitro immunoprecipitation with γ-tubulin
Immunoprecipitation (IP) was performed using GFP tag on XMAP215 as the bait and γ-tubulin as the prey. Anti-GFP antibody was coupled to Protein A Dynabeads overnight. All proteins were pre-clarified via ultracentrifugation. 500nM XMAP215-GFP or GFP-tag was each mixed with 80nM γ-tubulin in pulldown buffer (γ-TB buffer from above with final 90mM KCl and 0.04% Tween20). Empty beads (buffer) were used as additional control without any GFP-tagged protein. One volume of Dynabeads was suspended in one volume each IP mixture, incubated on rotisserie for 1 hour at 4°C, then washed twice with 5 volumes of pulldown buffer. Protein content on the beads was analyzed by immunoblot against γ-tubulin and GFP.
Statistics and reproducibility
All nucleation or polymerization experiments with Xenopus egg extracts were reproduced with at least three independent experiments, unless stated otherwise, with extracts freshly-prepared from eggs laid by different animals on different days. Representative results are displayed, or all data was reported, as specified in individual figure legends. Similar results were seen in all replicates that were performed. For in vitro polymerization, at least two independent set of experiments were performed on different days with different protein purifications, where at least two replicates were completed for each purification. All results were pooled and reported, and no difference was observed between different experimental sets. Nucleation experiments with purified γ-TuRC were repeated at least thrice, unless stated otherwise. Each replicate consisted of an independent preparation of γ-TuRC from different extract preparations. All results from the replicates showed similar results, and were pooled and reported. All chromatography runs with γ-tubulin were repeated at least twice on different days with freshly prepared buffers and multiple protein purifications. Similar results were seen with all runs performed. Chromatography runs with αβ-tubulin were performed once at the exact buffer and concentration specified, with supporting runs performed in slightly varied conditions on different days with freshly prepared buffers and multiple protein purifications, as specified in figure legends. Immunoprecipitation experiments in Xenopus extracts or in vitro were performed two-four times as indicated in respective figure legends. All replicates showed similar results and representative experiment was reported. No statistical tests were performed or reported.
Life Sciences Reporting Summary
Further information on experimental design is available in Life Sciences Reporting Summary.
Code availability
MATLAB-based custom-built software in conjunction with open source software (uTrack, Danuser lab) was used for detecting and tracking EB1 particles. For other measurements and data analysis, MATLAB-based custom-built scripts were used. All codes are available from the authors upon request.
Data availability
Source data for figs. 1E, 1F, 2B, 6B, 6D, and supplemental figs. 2D, 2E, 4C, 4E and 6C have been provided as Supplemental Table 2. All data supporting the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Joshua Shaevitz for help with image analysis, Howard Stone for insightful discussions, Michelle Moritz for providing γ-tubulin protein, Simone Reber and Per Widlund for providing clones of XMAP215, Michelle Moritz, Fred Hughson and Simone Reber for critical reading of the manuscript, and Petry lab members for discussions. Proteomics experiments were carried out by the Mass Spectrometry Core at Princeton University. A.T. thanks the Image Analysis course at MBL for useful discussions. This work was supported by NIH New Innovator Award, Pew Scholars Program in the Biomedical Sciences, David and Lucile Packard Foundation (all to S.P.), American Heart Association predoctoral fellowship 17PRE33660328 (to A.T.) and NIH post-doctoral fellowship 1F32GM119195-01 (to R.S.K.).
Abbreviations List
- MT
Microtubule
- MTOC
Microtubule organizing center
- γ-TuRC
Gamma-tubulin (γ-tubulin) and Gamma-tubulin ring complex
- GCP
Gamma-tubulin complex protein
Footnotes
Supplementary Information includes nine figures, nine videos and two tables.
Author contributions
A.T. and S.P. conceived the project. A.T., R.S.K. and S.P. designed experiments. A.T. and R.S.K. generated and characterized the reagents and tools. A.T. and R.S.K. performed and analyzed all experiments. A.T., R.S.K. and S.P. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Voter WA, Erickson HP. The kinetics of microtubule assembly. Evidence for a two-stage nucleation mechanism. J Biol Chem. 1984;259:10430–10438. [PubMed] [Google Scholar]
- 2.Petry S, Vale R. Microtubule nucleation at the centrosome and beyond. Nature Publishing Group. 2015;17:1089–1093. doi: 10.1038/ncb3220. [DOI] [PubMed] [Google Scholar]
- 3.Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 4.Wiese C, Zheng Y. Microtubule nucleation: γ-tubulin and beyond. Journal of Cell Science. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 6.Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 7.Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by γ-tubulin complexes. Nature Publishing Group. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moritz M, Braunfeld MB, Guénebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 9.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caudron N. Microtubule Nucleation from Stable Tubulin Oligomers. Journal of Biological Chemistry. 2002;277:50973–50979. doi: 10.1074/jbc.M209753200. [DOI] [PubMed] [Google Scholar]
- 11.Groen AC, Maresca TJ, Gatlin JC, Salmon ED, Mitchison TJ. Functional overlap of microtubule assembly factors in chromatin-promoted spindle assembly. Molecular Biology of the Cell. 2009;20:2766–2773. doi: 10.1091/mbc.E09-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannak E, et al. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegansis γ-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers GC, Rusan N, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Molecular Biology of the Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. The Journal of Cell Biology. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y, Liu P, Sze S, Dai C, Qi R. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. The Journal of Cell Biology. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, Choi Y, Qi R. NME7 is a functional component of the γ-tubulin ring complex. Molecular Biology of the Cell. 2014;25:2017–2025. doi: 10.1091/mbc.E13-06-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oegema K, et al. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kollman JM, et al. Nat Struct Mol Biol. 2015:1–8. [Google Scholar]
- 19.Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tournebize R, et al. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- 21.Popov AV, et al. XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J. 2001;20:397–410. doi: 10.1093/emboj/20.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Bassam J, Van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widlund PO, et al. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proceedings of the National Academy of Sciences. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Bassam J, et al. Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Molecular Biology of the Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reber SB, et al. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nature Cell Biology. 2013;15:1116–1122. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- 27.Roostalu J, Cade N, Surrey T. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol. 2015;17:1422–1434. doi: 10.1038/ncb3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 29.Slep K, Vale R. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayaz P, Ye X, Huddleston P, Brautigam C, Rice L. A TOG: γ-tubulin Complex Structure Reveals Conformation-Based Mechanisms for a Microtubule Polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrnes AE, Slep KC. TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J Cell Biol. 2017;216:1641–1657. doi: 10.1083/jcb.201610090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popov AV, Severin F, Karsenti E. XMAP215 is required for the microtubule-nucleating activity of centrosomes. Current Biology. 2002;12:1326–1330. doi: 10.1016/s0960-9822(02)01033-3. [DOI] [PubMed] [Google Scholar]
- 33.Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat Cell Biol. 2015;17:907–916. doi: 10.1038/ncb3188. [DOI] [PubMed] [Google Scholar]
- 34.Ohi R, Zanic M. Ahead of the Curve: New Insights into Microtubule Dynamics. F1000Res. 2016;5 doi: 10.12688/f1000research.7439.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roostalu J, Surrey T. Microtubule nucleation: beyond the template. Nat Rev Mol Cell Biol. 2017;18:702–710. doi: 10.1038/nrm.2017.75. [DOI] [PubMed] [Google Scholar]
- 36.Petry S, Groen A, Ishihara K, Mitchison T, Vale R. Branching Microtubule Nucleation in Xenopus Egg Extracts Mediated by Augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanic M, Widlund PO, Hyman AA, Howard J. Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat Cell Biol. 2013;15:688–693. doi: 10.1038/ncb2744. [DOI] [PubMed] [Google Scholar]
- 38.Bucciarelli E, et al. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr Biol. 2009;19:1839–1845. doi: 10.1016/j.cub.2009.09.043. [DOI] [PubMed] [Google Scholar]
- 39.Moritz M, Rice LM, Agard DA. Microtubule Nucleation. Wiley-VCH; Weinheim: 2004. [Google Scholar]
- 40.Wuhr M, et al. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- 42.Srayko M, Kaya A, Stamford J, Hyman AA. Developmental Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 44.Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hood F, et al. J Cell Biol. 2013;202:463–478. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess SG, Bayliss R, Pfuhl M. Solution NMR assignment of the cryptic sixth TOG domain of mini spindles. Biomol NMR Assign. 2015;9:411–413. doi: 10.1007/s12104-015-9620-4. [DOI] [PubMed] [Google Scholar]
- 47.Fox J, Howard A, Currie J, Rogers S, Slep K. The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Molecular Biology of the Cell. 2014;25:2375–2392. doi: 10.1091/mbc.E13-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- 49.Vasquez RJ, Gard DL, Cassimeris L. Cell Motil Cytoskeleton. 1999;43:310–321. doi: 10.1002/(SICI)1097-0169(1999)43:4<310::AID-CM4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 50.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Petry S, Pugieux C, Nedelec F, Vale R. Proceedings of the National Academy of Sciences. 2011;108:14473–14478. doi: 10.1073/pnas.1110412108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alfaro-Aco R, Thawani A, Petry S. Structural analysis of the role of TPX2 in branching microtubule nucleation. J Cell Biol. 2017;216:983–997. doi: 10.1083/jcb.201607060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nature protocols. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 54.Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 55.King M, Petry S. The Mitotic Spindle. Methods in Molecular Biology. Vol. 1413. Humana Press; New York, NY: 2016. [Google Scholar]
- 56.Applegate KT, et al. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, Wong ML, Alberts B, Mitchison T. Reconstituting the Cytoskeleton. 1998;298:218–228. doi: 10.1016/s0076-6879(98)98021-1. [DOI] [PubMed] [Google Scholar]
- 59.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 60.Choi Y, Qi RZ. Reconstituting the Cytoskeleton. 2014;540:119–130. [Google Scholar]
- 61.Gell C, et al. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 62.Zanic M. Measuring the Effects of Microtubule-Associated Proteins on Microtubule Dynamics In Vitro. Methods Mol Biol. 2016;1413:47–61. doi: 10.1007/978-1-4939-3542-0_4. [DOI] [PubMed] [Google Scholar]
- 63.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le Maire M, Aggerbeck LP, Monteilhet C, Andersen JP, Moller JV. The use of high-performance liquid chromatography for the determination of size and molecular weight of proteins: a caution and a list of membrane proteins suitable as standards. Anal Biochem. 1986;154:525–535. doi: 10.1016/0003-2697(86)90025-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figs. 1E, 1F, 2B, 6B, 6D, and supplemental figs. 2D, 2E, 4C, 4E and 6C have been provided as Supplemental Table 2. All data supporting the findings of this study are available from the corresponding author upon request.