Abstract
Background: Colorectal cancer (CRC) is the third leading cause of cancer-related deaths. 5-Fluorouracil (5-FU)-based chemotherapy has always been the first-line treatment. However, development of 5-FU resistance seriously affects its curative effect. The aim of this study was to elucidate the molecular mechanisms of 5-FU resistance through miR-106a-5p in CRC. Methods: Colorectal cancer tissues were collected to analyze miR-106a-5p and TGFβR2 expressions by qPCR. Functional experiments for evaluating cell survival and metastasis were conducted to observe the biological effects of miR-106a-5p and TGFβR2. The cell survival rate was calculated using an MTT assay; the metastasis was confirmed with a Transwell invasion assay and Western blotting, which we used to measure the expression levels of the epithelial-mesenchymal transition (EMT) markers E-cadherin and vimentin. The combination of miR-106a to TGFβR2 was predicted using Targetscan, and confirmed through the construction of the luciferase reporter plasmid pGL3-basic. The interplay between miR-106a-5p and TGFβR2 was tested with qPCR and Western blotting. A Spearman rank analysis was employed to verify the correlation of miR-106a-5p and TGFβR2 expressions. Results: MiR-106a-5p was up-regulated and TGFβR2 was down-regulated in 5-FU resistant CRC tissues and HT-29 cells. MiR-106a-5p promoted cell survival and suppressed the apoptosis rate and caspase 3 activity. Additionally, cell invasion was promoted by miR-106a-5p overexpression in the HT-29 cells and was inhibited by miR-106a-5p knockdown in the 5-FU resistant HT-29 cells; miR-106a-5p overexpression contributed to migration by increasing vimentin expression and by decreasing E-cadherin expression in the HT-29 cells; miR-106a-5p functioned by directly binding to TGFβR2. The TGFβR2 knockdown conferred chemoresistance of 5-FU and metastasis in 5-FU resistant HT-29 cells, and TGFβR2 overexpression reduced cell survival, invasion numbers, vimentin expression, and increased the cell apoptosis rate and caspase 3 activity in 5-FU resistant HT-29 cells. Also, miR-106a-5p negatively regulated TGFβR2 in a linear correlation way in the CRC tissues. Conclusion: The up-regulation of miR-106a-5p contributes to the pathomechanism of colorectal cancer by promoting 5-FU resistance and metastasis via inhibiting target TGFβR2. Our findings provide new promising ways for the clinical application of the TGFβR2-miR-106a axis in clinical chemotherapy for 5-FU resistant colorectal cancer.
Keywords: MiR-106a-5p, 5-FU resistance, TGFβR2, metastasis, colorectal cancer
Introduction
Colorectal cancer (CRC), the third leading cause of cancer-related mortality and morbidity, is one of the most common cancers in the gastrointestinal tract [1]. The chemotherapeutant, 5-fluorouracil (5-FU), which causes cytotoxic damage, is the basis of standard chemotherapy for CRC [2]. 5-FU-based chemotherapy, including combined therapy and adjuvant therapy, is largely used in the medical care of various kinds of cancers. However, the clinical responses to 5-FU vary greatly, and chemoresistance is a major reason for CRC therapy failure [3,4]. The mechanisms associated with 5-FU resistance and metastasis have attracted the attention of several researchers in recent years.
MicroRNAs (miRNAs), a group of endogenous small noncoding RNAs which are 21-25 nucleotides, suppress gene expression by inducing target mRNA degradation and/or blocking translation. Statistics show that miRNAs regulate the expression of > 60% of human protein-encoding genes [5]. The aberrant expression of certain miRNAs has been observed in an array of human cancer types [6,7], and miRNAs are thought to serve important roles in tumorigenesis [8,9]. MiR-106a is a member of the miR-17 family [10], which includes miR-17, miR-20, miR-92b, and miR-106. It is generally recognized that the members of the miR-17 family are typical oncogenes [11,12]. MiR-106a, located on the human X chromosome [13], overexpresses among various tumor tissues, especially in digestive system neoplasms [14]. It was reported that miR-106a is up-regulated in radiation-resistant cells in prostate cancer [15]. Recently, studies showed that miR-106a takes part in 5-FU-resistance [16], metastasis [17], and apoptosis [17-19] in CRC cells. However, the mechanism of the resistant CRC cells by miR-106a remains unclear.
One of the key reasons for the aggressiveness of malignant tumors is attributed to a tumor cell remodeling process called epithelial-to-mesenchymal transition (EMT) [20]. Mechanistically, EMT is characterized by the gaining of mesenchymal phenotypes [21,22] including increased motility, invasiveness, and chemoresistance. Transforming growth factor-beta (TGF-β) is a multifunctional cytokine secreted into the tumor microenvironment which primarily promotes the EMT process [23]. TGF-β signaling is initiated by the binding of the TGF-β ligand to its receptor (TGFβR2). Numerous studies have focused on miRNA/TGFβR2 to reveal its pathogenesis in many cancers. However, the role of miR-106a-5p/TGFβR2 has not been clarified in CRC.
In this study, we examined the up-regulated miR-106a-5p link to 5-FU resistance in colorectal cancer tissues and HT-29 cells. MiR-106a-5p overexpression contributes to 5-FU resistance by promoting cell survival and inhibiting apoptosis, and facilitates invasion and EMT in HT-29 cells. What’s more, miR-106a-5p negatively regulates TGFβR2 by directly binding in a linear correlation manner. These results indicate that the TGFβR2-miR-106a axis is a novel molecular mechanisms of 5-FU resistance, suggesting a promising application of miR-106a-5p in clinical chemotherapy for 5-FU resistant colorectal cancer.
Materials and methods
Recruiting patients and acquiring specimens
Fifty-six tumor specimens, containing 32 5-FU sensitive tissues and 24 5-FU resistant tissues were separated from CRC patients who underwent surgery at the Second Affiliated Hospital of Zhejiang Chinese Medical University (Xinhua Hospital of Zhejiang Province). The specimens were stored at -80°C or in liquid nitrogen immediately. The patients included 36 males and 20 females, ranging in age from 40 to 80 years. Informed consent was obtained from all individual participants included in the study, and the study was approved by the ethics committees of the Second Affiliated Hospital of Zhejiang Chinese Medical University (Xinhua Hospital of Zhejiang Province). We extracted total RNA and protein from the specimens.
Cells and cell culture
It was reported that miR-106a expression is the highest in HT-29 cells among several types of CRC cells [19]. The colon adenocarcinoma HT-29 cells were purchased from ATCC (Beijing, China) and used in in vitro experiments. Routinely, the cells were seeded in plastic flasks and cultured in ATCC-formulated McCoy’s 5a Medium Modified and supplemented with 10% (v/v) fetal bovine serum (FBS, Thermo Fisher Scientific, Waltham, MA, USA) and 0.5% (v/v) penicillin/streptomycin in a humidified atmosphere containing 5% (v/v) CO2 at 37°C. The cells were supplied with fresh medium every second day and digested with 0.25% trypsin-0.53 mM EDTA (Invitrogen, Carlsbad, CA, USA) when the confluence was about 100%.
Development of 5-FU resistant HT-29 cells (HT-29-5-FU)
Commonly, highly metastatic cancer cells exhibit a drug-resistant phenotype [24,25]. To establish the drug-resistant cell subline, the HT-29-5-FU, HT-29 cells were exposed to stepwise increases of 5-FU (Sigma-Aldrich, St Louis, MO, USA) concentrations from 10 to 100 μm. When no significant cell deaths were noted after the 5-FU treatment, the cells were checked by cell survival assay in the presence of 5-FU. 50% inhibitive concentration (IC50) values of HT-29 and HT-29-5-FU were counted for the resistance index (RI). RI is the rate of HT-29-5-FU IC50/HT-29 IC50.
Total RNA isolation and quantitative real-time PCR (qPCR)
The total RNA of the CRC tissues and the HT-29 cells was extracted using Trizol (Dingguo, Beijing, China) according to manufacturer’s protocol. For TGFβR2, the RNA was reverse transcribed into cDNA using the PrimeScript RT PCR Kit (Takara, Dalian, China); for the miR-106-5p, the first-strand cDNA synthesis was performed by TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific). The expression of TGFβR2 and miR-106-5p was calculated using the SYBR Premix Dimmer Eraser Kit (Takara) on an ABI PRISM 7500 (Applied Biosystems). The miR-106a primer sequence was: 5’-GGAAAAGTGCTTACAGTGCAGGTAG-3’. The expression of miR-106a was normalized to that of U6 (U6: forward primer: 5’-GTCGTATCCAGTGCAGGGTCCGAGGT-3’; reverse primer: 5’-GCACTGGATACGACAAAATATGGAAC-3’). TGFβR2 primers sequence: 5’-CCGCTGCATATCGTCCTGT-3’ (forward primer); 5’-AGTGGATGGATGGTCCTATTACA-3’ (reserve primer). And, the expression of TGFβR2 was normalized to that of GAPDH (GAPDH: forward primer: 5’-AAGGTGAAGGTCGGAGTCAA-3’; reverse primer: 5’-AATGAAGGGGTCATTGATGG-3’).
All experiments were performed at least in triplicate. The relative quantification of gene expression was performed by the 2-ΔΔCt method.
Total protein extraction and Western blotting
The expressions of TGFBR2 and the EMT markers, E-cadherin and vimentin, were screened using Western blotting. The total protein was lysed with a RIPA regent (Beyotime, Shanghai, China) and the concentration was measured using a Bradford assay (Bio-Rad, CA, USA). 20 μg total protein was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore-Sigma, Billerica, MA, USA). After blocking with 5% nonfat milk for 1 hour at room temperature, the blots were incubated with primary antibodies (TGFβR2: Abcam, ab186838, 1:250; E-cadherin: Abcam, ab1416, 1:50; Vimentin, Abcam, ab92547, 1:5000; GAPDH: Abcam, ab8245, 1:5000) at 4°C overnight with gentle shaking. The blots reacted with the second antibody at room temperature for 2 h. Finally, the bands were measured using an ECL kit (Beyotime). The bands were quantified using Image J, and GAPDH was the loading control.
Cell transfection
Plasmid pEGFP and pSilencer 2.1-U6 hygro were purchased from Bio Vector (Beijing, China). TGFBR2 overexpression in the 5-FU resistant HT-29 cells was achieved by the construction and transfection of the recombined plasmid pEGFP-TGFβR2. TGFβR2 knockdown in the HT-29 cells was obtained by the construction and transfection of the recombined plasmid pSilencer-TGFBR2. The MiR-106a-5p knockdown and overexpression were provided by Gene Pharma (Shanghai, China). MiRNAs were transfected at 50 nm and the plasmid DNA was transfected at 2 μg for 48 hours using the Lipofectamine 2000 transfection reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The sequences of miR-106a-5p were: 5’-AAAAGUGCUUACAGUGCAGGUAG-3’; anti-miR-106a-5p: 5’-CUACCUGCACUGUAAGCACUUUU-3’.
Transwell invasion assay
Cell invasion assays were performed using Matrigel-coated plates (invasion assay 24-well Transwell inserts with 8 μm pores) (BD, Jiangsu, China). The cells in the 200 serum-free medium were loaded into the upper Transwell chamber (8.0-lm pore size, BD Biosciences, Franklin Lakes, NJ, USA) for the invasion assay. The chambers were incubated in media with 10% FBS in the bottom chambers for 48 h. Cells that migrated and invaded to the reverse side of chamber inserts were fixed and stained with methanol and 0.1% crystal violet. Finally, the stained cells were counted under a microscope. The experiments were independently carried out in triplicate.
Cell survival assay
The cells were seeded and cultured at a density of 5 × 104 cells/well in a 96-well plate overnight. The cells were subsequently exposed to various concentrations of 5-FU for 48 h. The cell survival assay was measured by a 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. With the addition of 20 μl of MTT dye in 200 mL of phosphate-buffered saline (PBS) (5 mg/mL) per well for 4 hours, the formazan crystals were dissolved in 150 μl dimethyl sulfoxide (DMSO, Dingguo) and the absorbance was measured at 490 nm using a SpectraMax M3 microplate reader (Molecular Devices, LLC, CA, USA). The experiments were undertaken in quadruplicate.
Flow cytometry
The cells were cultured in 6-well plates at 2 × 105 cells per well in a complete culture medium. After reaching 90% confluence, the cells were pretreated with a serum-free medium for 8-12 h in order to synchronize the cells. Then, the cells were exposed to a complete medium containing 5-FU for another 48 h. The apoptosis assays were performed using an Annexin V-FITC apoptosis detection kit (Beyotime) according to the manufacturer’s protocol. In short, the treated cells were collected and re-suspended in 1 × binding buffer at a concentration of 1 × 106 cells per ml. A 100 μl cell suspension was mixed with 5 μl annexin V-FITC and 5 μl propidium iodide (PI) for 15 min at room temperature in the dark, followed by the addition of a 400 μl binding buffer. The samples were analyzed by fluorescence-activated cell sorting (FACSAria, BD).
Caspase 3 activity assay
The cysteine-requiring aspartate protease (Caspase) family plays vital role in the apoptosis process [26]. Caspase 3, the most well studied caspase in mammalian cells, has been known to be the key executor in apoptosis. In our experiment, the caspase 3 activity was examined using a Caspase-3 Activity Assay Kit (Beyotime) according to the manufacturer’s instructions. The cells were cultured in 96-well plates and treated with 5-FU for 48 h. In brief, the treated cells were lysed with a lysis buffer (100 μl/well) for 15 min on ice, followed by washing with cold HBSS. After incubating the mixture composed of a 10 μl cell lysate, 80 μl reaction buffer and 10 μl of 2 mM caspase 3 substrate at 37°C for 4 h, the caspase 3 activity was quantified in the samples with a SpectraMax M3 microplate reader (Molecular Devices) at an absorbance of 405 nm. The experiments were carried out in triplicate.
Dual-luciferase reporter assay
The putative target prediction of miR-106a-5p was performed using web server tools: www.Targetscan.org. The potential binding sites on the human TGFBR2 gene were mutated and cloned by the PCR method into plasmid pGL3-basic, which expressed the luciferase reporting gene, to develop the overexpression of TGFBR2 mutation (TGFBR2-Mut) and TGFBR2 wild type (TGFBR2-Wt). The cells were plated in a 24-well plate at 1 × 104 cells/well, followed by co-transfection with either 20 ng of TGFBR2-Mut or TGFBR2-Wt and 20 nm of anti-miR-106a-5p in HT-29-5-FU cells for 48 h, and 20 nm of miR-106a-5p in HT-29 cells for 48h. The cells were collected to measure the relative luciferase by using the Dual-luciferase Reporter Assay System (Promega, WI, USA). The experiments were carried out in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS 17.0. The values given were the mean ± SEM. The P-values were determined for experimental versus control treatments using a two-tailed Student’s t-test, *P < 0.05. The correlations between the TGFβR2 and miR-106a-5p expressions were evaluated by Spearman rank analysis.
Results
The 5-FU-resistance role of miR-106a-5p in CRC tissues and cells
We examined the level of miR-106a-5p, both in 5-FU-resistant CRC patient specimens (n = 24) and in 5-FU-sensitive specimens (n = 32). As Figure 1A shows, the miR-106a-5p was apparently up-regulated in the 5-FU-resistant tissues. Next, we established the 5-FU-resistant HT-29 cells and measured cell survival when they were exposed to different concentrations of 5-FU for 48 h. The MTT assay (Figure 1B) showed a generally higher cell survival of the HT-29-5-FU cells than the control HT-29 cells. According to the IC 50 value, 15 μm 5-FU on the HT-29 cells and 60 μm 5-FU on the HT-29-5-FU cells were retained for further experiments. Additionally, we measured the miR-106a-5p expression levels in cultured HT-29 cells and HT-29-5-FU cells (Figure 1C). Compared with the CRC tissues, the miR-106a-5p expression was significantly higher in the HT-29-5-FU cells, still. These results showed that the miR-106a-5p was up-regulated in the 5-FU-resistant CRC tissues and HT-29 cells.
Figure 1.
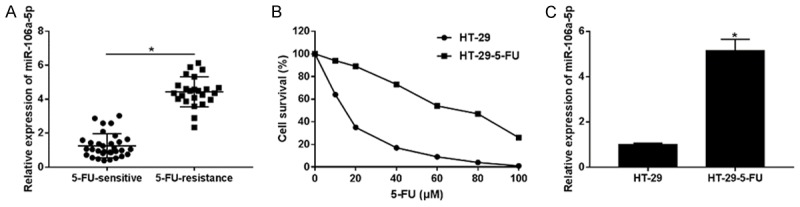
The 5-FU-resistance role of miR-106a-5p in CRC tissues and cells. A. The expressions of miR-106a-5p were compared among 5-FU-resistant CRC patient specimens (n = 24) with 5-FU-sensitive specimens (n = 32). MiR-106a-5p was up-regulated in 5-FU-resistant tissues. B. The survival of HT-29 cells and HT-29-5-FU resistant cells treated with 0, 10, 20, 40, 60, 80, 100 μm of 5-FU for 48 h. The IC 50 values of HT-29 and HT-29-FU were 14.09 and 66.26 μm respectively. The MTT assay showed the higher cell survival of the HT-29-5-FU cells. C. The mRNA expressions of miR-106a-5p in the HT-29 cells and the HT-29-5-FU resistant cells were examined by qPCR. The statistical analysis results were expressed as the mean ± SEM. *P < 0.05 as compared with control group.
MiR-106a-5p regulated 5-FU-resistance in HT-29 cells
Considering the up-regulation of miR-106a-5p in the 5-FU-resistant CRC tissues and HT-29 cells, we investigated the role of miR-106a-5p in the 5-FU-resistant HT-29 cells. First, we knocked down miR-106a-5p in HT-29-5-FU and overexpressed miR-106a-5p in the HT-29 cells. The relative expressions of miR-106-5p were examined by qPCR respectively. As shown in Figure 2A and 2B that anti-miR-106-5p defensed miR-106a-5p expression in the HT-29-5-FU cells, and miR-106a-5p overexpression promoted miR-106a-5p expression in the HT-29-5-FU cells. The cell survival of HT-29-5-FU cells (Figure 2C) was impaired by the transfection of anti-miR-106a-5p, whereas miR-106a-5p overexpression rescued and promoted the cell survival of the HT-29 cells (Figure 2D). The apoptosis rate was also calculated by flow cytometry. In contrast to cell survival, more HT-29-5-FU cell apoptosis (Figure 2E) was detected because of anti-miR-106a-5p, and less HT-29 cell apoptosis (Figure 2F) was detected because of miR-106-5p. Similar results were achieved in the caspase 3 activity assay (Figure 2G and 2H). These results show that miR-106-5p is likely to promote cell survival and inhibit cell apoptosis, contributing to 5-FU resistance in HT-29 cells.
Figure 2.
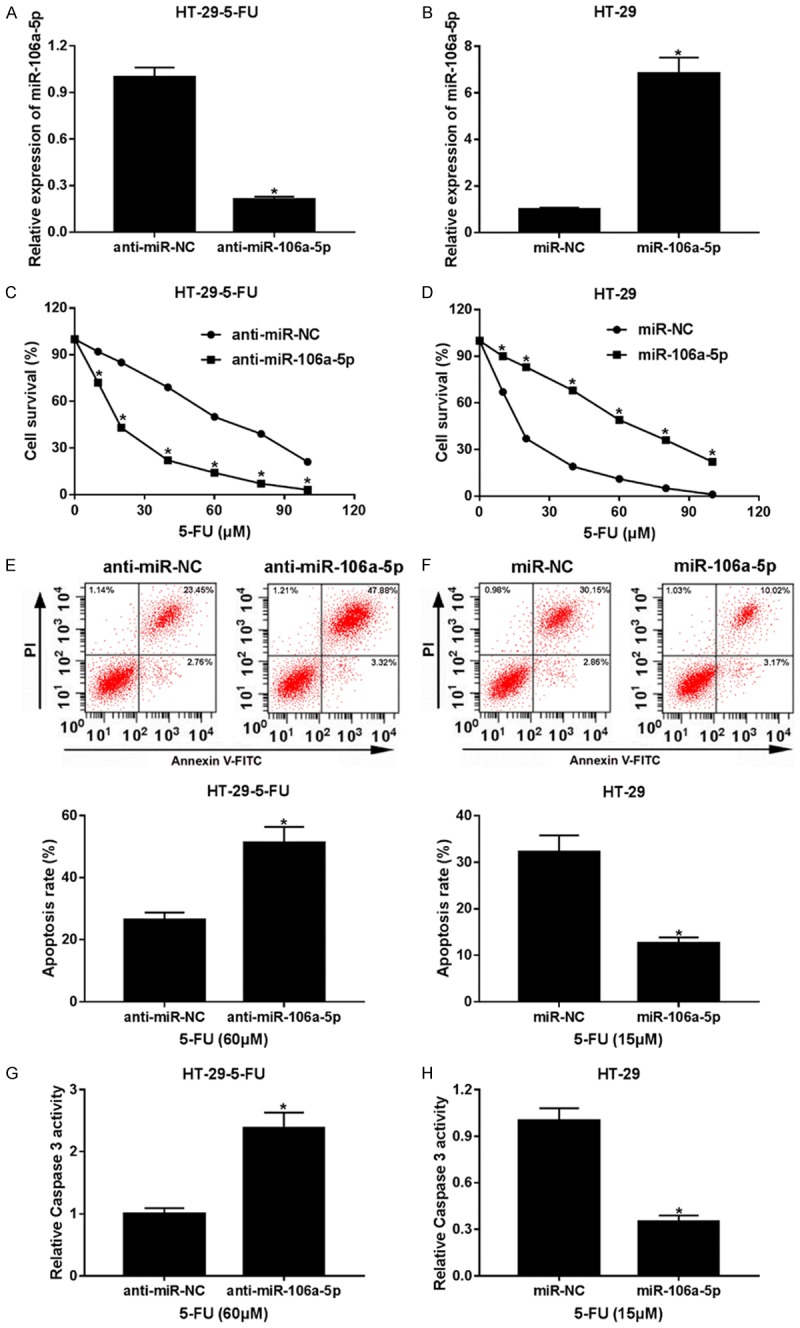
MiR-106a-5p regulated 5-FU-resistance in the HT-29 cells. (A) Knockdown of miR-106-5p in HT-29-5-FU cells. (B) Overexpression of miR-106-5p in the HT-29 cells. After transfection of anti-miR-106-5p and miR-106-5p into the cells, the relative expressions of miR-106-5p were examined by qPCR respectively. The cell survival of HT-29-5-FU cells (C) transfected of the anti-miR-106-5p and HT-29 cells (D) transfected of miR-106-5p was measured by MTT assay. The apoptosis of HT-29-5-FU cells (E) overexpressed of anti-miR-106-5p and HT-29 cells (F) overexpressed of miR-106-5p was detected by flow cytometry, and the rates of the apoptotic cells were counted and analyzed. 15 μm 5-FU was for the HT-29 cells, and 60 μm 5-FU was for the HT-29-5-FU cells. The caspase 3 activity of the anti-miR-106-5p overexpression HT-29-5-FU cells (G) and the miR-106-5p overexpression of the HT-29 cells (H) were monitored by a Caspase 3 Activity Assay Kit. Statistical analyses results are shown as mean ± SEM. *P < 0.05, compared with control group.
MiR-106a-5p promoted metastasis of HT-29 cells
In consideration of the protective effect of miR-106a-5p in HT-29-5-FU cells, we conducted further experiments to figure out the role of miR-106a-5p in cell invasion. A Transwell invasion assay was done, and the expressions of the EMT markers, E-cadherin and vimentin, were analyzed. More cell invasion numbers (Figure 3A), higher vimentin expression, and lower E-cadherin expression (Figure 3C) were noted in the HT-29-5-FU cells. The effects of miR-106a-5p on cell invasion and EMT in the HT-29 cells were determined. As shown in Figure 3B and 3D, miR-106a-5p knockdown weakens invasion and vimentin expression in HT-29-5-FU cells; while miR-106a-5p overexpression motivated invasion and vimentin expression in HT-29 cells. The above data suggests a protective effect of miR-106a-5p on cell metastasis and 5-FU resistance in HT-29 cells.
Figure 3.
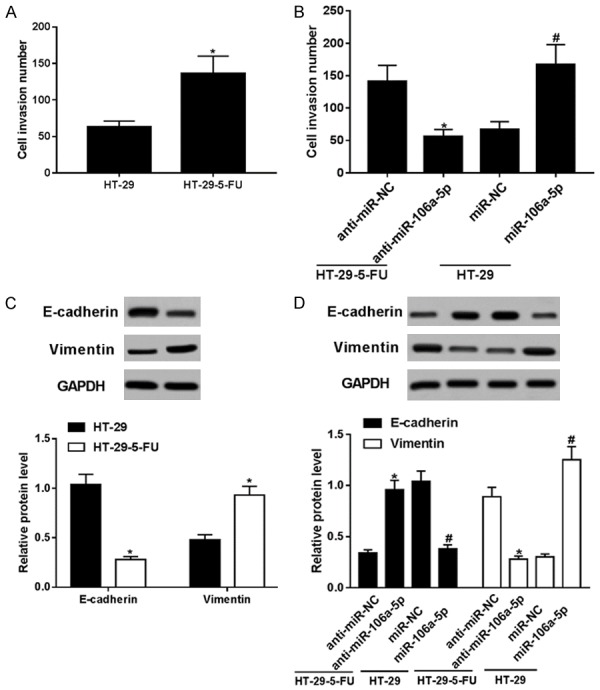
Cell invasion and EMT was promoted by miR-106a-5p in the HT-29 cells. The cell invasion number (A) and the expressions of the EMT markers, E-cadherin and vimentin, (C) in the HT-29-5-FU cells were tested using a Transwell invasion assay and Western blotting. The effects of miR-106a-5p knockdown/overexpression on cell invasion (B) and EMT markers expression (D) in HT-29-5-FU/HT-29 cells were determined. All data above were represented as the mean ± SEM. *P < 0.05, compared with control groups.
MiR-106a-5p is down-regulated TGFβR2 by potentially binding in HT-29 cells
HT-29 cells benefited from miR-106a-5p on cell invasion and 5-FU resistance. TGFβR2 exists extensively among cancer proliferation, invasion, progression and drug-resistance [27,28]. Accordingly, we hypothesized that miR-106a-5p displayed functions through regulating TGFβR2 in HT-29 cells. Targetscan revealed that there were potential target binding sites between miR-106a-5p and TGFβR2 3’UTR. As shown in Figure 4A, miR-106a-5p potentially binds to the TGFβR2 3’UTR 268-275 GCACUUU region; we mutated the target sites to CGUGAAA. Then, we constructed the recombination plasmid pGL3-basic-TGFβR2-Mut. After co-transfection, the cells were collected and analyzed using a dual-luciferase reporter assay. The luciferase activity declined in the HT-29 cells’ co-expression of TGFβR2-Wt and miR-106a-5p (Figure 4C) and was enhanced in the HT-29-5-FU cells’ co-expression of TGFβR2-Wt and anti-miR-106a-5p (Figure 4B). The effect of miR-106a-5p on the TGFβR2 expressions, both at the mRNA (Figure 4D) and protein levels (Figure 4E and 4F), was disclosed as follows. The TGFβR2 level was lower in the HT-29-5-FU cells, compared with the level in the HT-29 cells. The level of TGFβR2 was raised by anti-miR-106a-5p in the HT-29-5-FU cells and reduced by miR-106a-5p in the HT-29 cells. The quantification of TGFβR2 protein expression was normalized by Image J (Figure 4F). This evidence shows that TGFβR2 can be down-regulated by miR-106a-5p and reversed by miR-106a-5p knockdown in HT-29 cells, by directly targeting the binding.
Figure 4.

MiR-106a-5p down-regulated TGFβR2 by potentially binding in HT-29 cells. (A) Prediction of the potential target binding sites between miR-106a-5p and TGFβR2 3’ UTR using Targetscan. The cells were collected and analyzed with a dual-luciferase reporter assay. (B) The co-transfection of anti-miR-106a-5p with plasmids overexpressed of TGFβR2 and TGFβR2-Mut in HT-29-5-FU cells, respectively. The luciferase activity of co-expressed TGFβR2-Wt and anti-miR-106a-5p was higher. (C) The transfection of miR-106a-5p into TGFβR2/TGFβR2-Mut overexpression HT-29 cells. The luciferase activity of co-expressed TGFβR2-Wt and miR-106a-5p was lower. Detection of the expression of TGFβR2 in HT-29-5-FU cells expressed of anti-miR-106a-5p and HT-29 cells expressed of miR-106a-5p, both in mRNA (D) and protein (E) level. The relative TGFβR2 expression level was lower in the HT-29-5-FU cells, compared with the level in HT-29 cells. The expression of TGFβR2 was raised by anti-miR-106a-5p in the HT-29-5-FU cells and reduced by miR-106a-5p in the HT-29 cells. (F) The quantification of TGFβR2 protein expression was normalized by Image J. All data shown represented the mean ± SEM. *P < 0.05, compared with control groups.
TGFβR2 regulated 5-FU resistance and invasion in HT-29 cells
The overexpression of TGFβR2 in HT-29-5-FU cells (Figure 5A), helped estimate cell survival (Figure 5C), invasion (Figure 5I) and apoptosis (Figure 5E and 5G). TGFβR2 was up-regulated about 7-fold in the HT-29-5-FU cells. TGFβR2 overexpression decreased cell survival at different concentrations of 5-FU and increased the apoptosis rate, and caspase 3 activity was induced by 60 μm 5-FU in the HT-29-5-FU cells. In addition, TGFβR2 overexpression reduced the cell invasion numbers. The knockdown of TGFβR2 in HT-29 cells (Figure 5B), left the following to be determined: cell survival (Figure 5D), invasion (Figure 5J), and apoptosis (Figure 5F and 5H). TGFβR2 was down-regulated about 10-fold more than the control in the HT-29 cells. TGFβR2 promoted cell survival at different concentrations of 5-FU and attenuated the apoptosis rate, and caspase 3 activity was induced by 15 μm 5-FU in the HT-29 cells. Moreover, TGFβR2 down-regulation increased the cell invasion numbers. This indicated that TGFβR2 acted with pro-apoptosis, anti-proliferation, and anti-invasion roles in the HT-29 cells.
Figure 5.
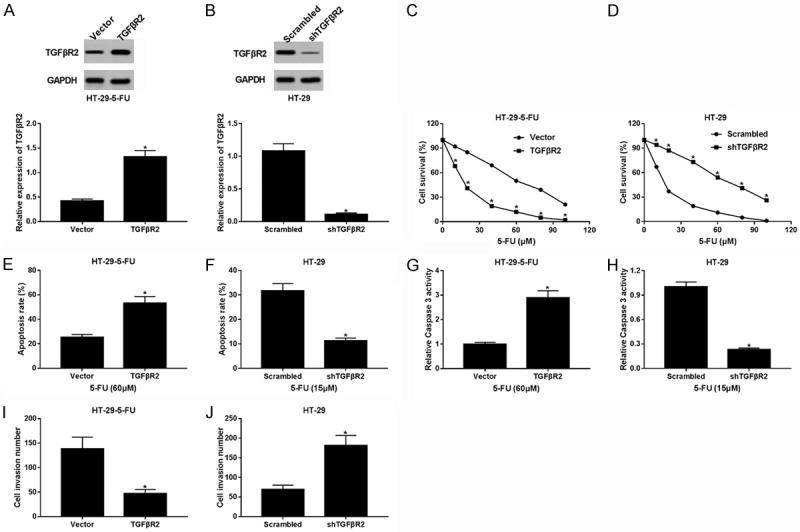
TGFβR2 regulated 5-FU resistance and invasion in colorectal cancer. TGFβR2 was overexpressed in the HT-29-5-FU cells (A) and knocked down in the HT-29 cells (B) by transfection of the reconstruction plasmid. A cell survival assay was performed to monitor the effects of TGFβR2 (C) and shTGFβR2 (D) on cell activity. The survival was decreased/increased at different concentrations of 5-FU in HT-29-5-FU cells and HT-29 cells. The apoptosis rate was recorded by flow cytometry in the HT-29-5-FU cells (E) treated with 60 μm 5-FU, and in HT-29 cells (F) treated with 15 μm 5-FU. TGFβR2 promoted the 5-FU-induced apoptosis of HT-29-5-FU cells, and shTGFβR2 rescued the apoptosis of the HT-29 cells. The caspase 3 activity induced by 5-FU was measured in HT-29-5-FU cells (G) treated with 60 μm 5-FU, and in HT-29 cells (H) treated with 15 μm 5-FU. Similarly, TGFβR2 facilitated 5-FU-stimulated caspase 3 activity in HT-29-5-FU cells, and shTGFβR2 inhibited caspase 3 activity in HT-29 cells. A Transwell invasion assay was carried out to clarify the role of TGFβR2 (I) and shTGFβR2 (J) on cell invasion. Conversely, TGFβR2 attenuated cell invasion in HT-29-5-FU cells, and shTGFβR2 contributed to cell invasion in HT-29 cells by the knockdown of TGFβR2. The data above are the mean ± SEM. *P < 0.05, compared with control groups.
TGFβR2 expression is negatively associated with the miR-106a-5p level in colorectal cancer tissues
MiR-106a-5p benefited HT-29 cells on cell metastasis and 5-FU resistance, probably via target binding to TGFβR2. We randomly chose two specimens of 5-FU sensitive and 2 samples of 5-FU resistant tissues, and inspected the TGFβR2 protein levels (Figure 6A). TGFβR2 was down-regulated in the 5-FU-resistant CRC tissues, both on the protein and mRNA expression levels (Figure 6B). Among the 56 specimens (Figure 6C), TGFβR2 expression, to a certain extent, had a negative linear correlation with miR-106a-5p expression. These results supported the theory that miR-106a-5p contributes to cell metastasis and 5-FU resistance, probably via target binding to TGFβR2 in CRC.
Figure 6.
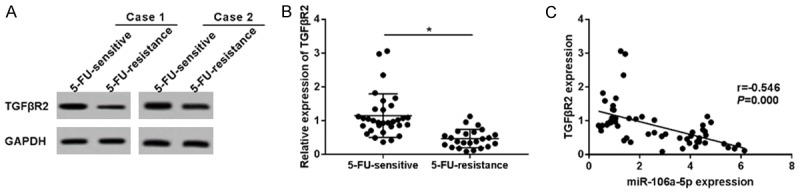
TGFβR2 expression was negatively associated with the miR-106a-5p expression level in colorectal cancer tissues. (A) Western blotting displayed TGFβR2 expressions in 2 specimens of 5-FU-sensitive and 2 samples of 5-FU-resistance, which were randomly selected. TGFβR2 was down-regulated in 5-FU-resistant CRC tissues, both on the mRNA expression level (B) and the protein level. (C) TGFβR2 expression, to a certain extent, is in a manner of negative correlation with miR-106a-5p expression in CRC tissues. Spearman rank analysis was chosen to explain the correlation. The results of the statistical analyses were expressed as the mean ± SEM. *P < 0.05 as compared with control group.
Discussion
In this study, we determined that miR-106a-5p is up-regulated and TGFβR2 is down-regulated in 5-FU resistant CRC tissues and HT-29 cells. 5-FU resistant HT-29 cells displayed more characteristics of cell survival, invasion, and EMT than the control HT-29 cells. The knockdown of miR-106a-5p impaired 5-FU resistance by reducing the high survival rate and raising the low apoptosis rate and caspase 3 activity in HT-29 cells. The overexpression of TGFβR2, similarly, reduced the resistance of 5-FU. What’s more, miR-106a-5p contributed to cell invasion and EMT, and the high 5-FU resistance and metastasis could be reversed by TGFβR2 in HT-29 cells. Notably, miR-106a-5p negatively regulated TGFβR2 in a linear correlation manner by potentially target binding. In short, it can be concluded that miR-106a-5p promotes 5-FU resistance and metastasis in colorectal cancer through the down-regulation of its target TGFβR2. These results indicate a promising application of the miR-106a-5p/TGFβR2 axis in clinical curative 5-FU-based chemotherapy in colorectal cancer.
A higher expression of miR-106a is found among various tumor tissues, especially in digestive system neoplasms [14]. Our study showed an up-regulation of miR-106a-5p in 60% of colon cancer tissues compared with the matched adjacent normal tissues. And miR-106a was significantly higher both in patients’ tumor tissues and colon cancer cell lines [29], including PKO, Lovo, HCT116, SW480, and SW620 cells, compared with the normal adjacent tissues and the colon mucosal epithelial cell line, NCM460 cells. In addition, our results indicate that miR-106a expression is the highest in HT-29 cells among several CRC cells [19]. In this study, we observed a higher expression of miR-106a-5p in 5-FU resistant CRC tissues and in HT-29 cells than in 5-FU sensitive CRC tissues and control HT-29 cells, which is consistent with the claim that miR-106a is up-regulated in radiation-resistant cells in prostate cancer [15]. Therefore, we logically speculated that miR-106a has a higher expression level in 5-FU resistant CRC tissues/HT-29 cells, compared with the 5-FU sensitive CRC tissues/HT-29 cells and the normal adjacent colon mucosal epithelial cell/cell line.
Drug-resistance is viewed as a chokepoint that restricts its effectiveness and practicality in clinical therapy. Recently, an increasing number of studies have shown that miRNAs play key role in the pathogenesis of drug-resistance [16,30]. For example, it is reported that miR-106a takes part in 5-FU-resistance, metastasis, and apoptosis in cancer cells. MiR-106a promotes proliferation and suppresses senescence in prostate cancer cells which contributed to radioresistance [15]. One study demonstrated that miR-106a protects cell viability from 5-FU-induced cytotoxicity, thus reducing the 5-FU sensitivity of CRC in SW620, HCT116 cells [16]. Rothschild [31], however, challenged the term “oncogenic” in the context of miR-106a, as it can promote lung cancer sensitivity to Src-TKIs. Coincidently, it was reported that miR-106a inhibited cell proliferation and induced apoptosis by the enhancement of caspase 9 in HCT116 and SW620 cells [18]. Our finding support the oncogenic role of miR-106a-5p, which functions as a proliferation promoter and apoptosis inhibitor. In consideration of the controversy of the role of miR-106a on cell proliferation and apoptosis in cancers, including colorectal cancer, more investigation ought to be directed towards miR-106a and its role in drug-resistance and metastasis.
The TGFβ-signaling pathway plays an important role in the pathogenesis of colorectal cancer and the inactivation of the pathway is a common event in CRC tumorigenesis. Previous studies illuminate the phenomenon that TGFβR2 as a common target of several miRNAs, including miR-211 [32], miR-17 [33,34], and miR-202 [35]. TGFβR2 exists extensively in cancer proliferation, invasion, progression and drug-resistance, and the significant correlation between high TGFβR2 gene expression and shorter disease-free survival [36]. As Ullmann reported, TGFβR2 is strongly up-regulated in SW620 cells and higher TGFβR2 protein expression in tissue sections had been noticed in 65 CRC patients as well. Here, our data showed the opposite results on TGFβR2 expressions. TGFβR2 mRNA and protein expression levels were higher in CRC tissues and HT-29 cells. However, the protein level of TGFβR2 was measured by 2 randomly selected tissue samples. Hence, our data enrich the knowledge of the effect of TGFβR2 in CRC.
The loss of an in vivo assay to validate the 5-FU resistance promotion of miR-106a-5p is a pity. For in vivo tumorigenicity assays, we are going to suspend 1 × 106 cells in 100 μl of PBS, which will be injected subcutaneously into the flanks of nude mice. Generally, tumor size will be measured at 5-day intervals and tumors should be harvested and photographed after 40 days and individually weighed after the mice were anesthetized. In this study, we found that miR-106a-5p inversely regulates TGFβR2 by target binding, and the preliminary data of a luciferase report assay validated our speculation. Meanwhile, further confirmatory tests should be launched to obtain more convincing evidence. In general, RNA immunoprecipitation assays and RNA pull down assays are employed to confirm the results of software prediction and luciferase report assays. MiRNA-mRNA-lncRNA interactions [37] have been shown to play critical regulatory roles in cancer biology, and miR-106a has been reported to be sponged by lncRNAs, including LINC01133 [38], H19 [39], LINC00657 [40] and FER1L4 [29] to be involved in cancers, such as gastric cancer, melanoma, and hepatocellular carcinoma. Did miR-106a-5p display the effects of the promotion of 5-FU-resistence in CRC through lncRNA? More functional experiments regarding lncRNA/miR-106a-5p are needed to answer this question in the near future.
Commonly, highly metastatic cancer cells exhibit a drug-resistant phenotype, and drug-resistant cancer cells express high levels of miR-106a-5p [15], indicating that miR-106a-5p could be a potential biomarker to diagnose and predict malignant tumor and cancer cell metastasis. Located in the human X chromosome, miR-106a exists in various tumor tissues, with an especially high expression level in digestive system neoplasms, and can be detected in plasma and feces. Furthermore, plasma miRNAs [41,42] and fecal miRNAs [43] have been proven as biomarkers to screen and predict clinical outcomes in colorectal cancer. High levels of miR-106a in plasma are associated with a lack of response in metastatic colorectal cancer (mCRC); it is up-regulated in tumor and stool samples from patients with CRC or adenomas, and it is up-regulated in tumors of mCRC patients compared to CRC patients without metastases and patients treated with 5-Fu/oxaliplatin [41]. Interestingly, plasma miR-106a-5p was significantly increased in postoperative blood samples compared with the matched preoperative ones [44]. Overall, miR-106a is a key mediator and a desired biomarker in CRC.
All in all, we put forward the idea that miR-106a-5p contributes to the resistance of 5-FU by inhibiting TGFβR2 in colorectal cancer. And miR-106a-5p functions by target binding to TGFβR2. Our findings provide a new understanding on the molecular mechanism of 5-FU resistance in colorectal cancer, suggesting that the clinical application of the miR-106a/TGFβR2 axis in clinical chemotherapy for 5-FU resistant colorectal cancer.
Acknowledgements
This work was supported by the Natural Science Foundation of Zhejiang Province. (Grant no. Y14H160140).
Disclosure of conflict of interest
None.
References
- 1.Liu S, Zheng R, Zhang M, Zhang S, Sun X, Chen W. Incidence and mortality of colorectal cancer in China, 2011. Chin J Cancer Res. 2015;27:22–28. doi: 10.3978/j.issn.1000-9604.2015.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong M, Zheng W, Li H, Li X, Ao L, Shen Y, Liang Q, Li J, Hong G, Yan H, Cai H, Li M, Guan Q, Guo Z. Multi-omics landscapes of colorectal cancer subtypes discriminated by an individualized prognostic signature for 5-fluorouracil-based chemotherapy. Oncogenesis. 2016;5:e242. doi: 10.1038/oncsis.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Li X, Cen C, Ai X, Lin C, Hu G. The long non-coding RNA ENST00000547547 reduces 5-fluorouracil resistance of colorectal cancer cells via competitive binding to microRNA-31. Oncol Rep. 2018;39:217–226. doi: 10.3892/or.2017.6082. [DOI] [PubMed] [Google Scholar]
- 4.Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, Wang CX. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- 5.Paydas S, Acikalin A, Ergin M, Celik H, Yavuz B, Tanriverdi K. Micro-RNA (miRNA) profile in Hodgkin lymphoma: association between clinical and pathological variables. Med Oncol. 2016;33:34. doi: 10.1007/s12032-016-0749-5. [DOI] [PubMed] [Google Scholar]
- 6.Huang G, Du MY, Zhu H, Zhang N, Lu ZW, Qian LX, Zhang W, Tian X, He X, Yin L. MiRNA-34a reversed TGF-β-induced epithelial-mesenchymal transition via suppression of SMAD4 in NPC cells. Biomed Pharmacother. 2018;106:217–224. doi: 10.1016/j.biopha.2018.06.115. [DOI] [PubMed] [Google Scholar]
- 7.Lee JY, Yun SJ, Jeong P, Piao XM, Kim YH, Kim J, Subramaniyam S, Byun YJ, Kang HW, Seo SP, Kim J, Kim JM, Yoo ES, Kim IY, Moon SK, Choi YH, Kim WJ. Identification of differentially expressed miRNAs and miRNA-targeted genes in bladder cancer. Oncotarget. 2018;9:27656–27666. doi: 10.18632/oncotarget.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol. 2014;20:11727–11735. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xuan Y, Yang H, Zhao L, Lau WB, Lau B, Ren N, Hu Y, Yi T, Zhao X, Zhou S, Wei Y. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett. 2015;360:89–105. doi: 10.1016/j.canlet.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Li C, Zhang R, Gao X, Qu X, Zhao M, Qiao C, Xu J, Li J. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Gruszka R, Zakrzewska M. The oncogenic relevance of miR-17-92 cluster and its paralogous miR-106b-25 and miR-106a-363 clusters in brain tumors. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19030879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan W, Li Y, Lim SG, Tan TM. miR-106b-25/miR-17-92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol. 2014;20:5962–5972. doi: 10.3748/wjg.v20.i20.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Zhou Y, Xia T, Zhou X, Huang Z, Zhang H, Zhu W, Ding Q, Wang S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res Treat. 2018;170:257–270. doi: 10.1007/s10549-018-4757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang S, Wu WK, Li X, Wong SH, Wong N, Chan MT, Sung JJ, Yu J. Stratification of digestive cancers with different pathological features and survival outcomes by MicroRNA expression. Sci Rep. 2016;6:24466. doi: 10.1038/srep24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoey C, Ray J, Jeon J, Huang X, Taeb S, Ylanko J, Andrews DW, Boutros PC, Liu SK. miRNA-106a and prostate cancer radioresistance: a novel role for LITAF in ATM regulation. Mol Oncol. 2018;12:1324–1341. doi: 10.1002/1878-0261.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Y, Chen X, Liu Z, Tian X, Huo Z. miR-106a reduces 5-fluorouracil (5-FU) sensitivity of colorectal cancer by targeting dual-specificity phosphatases 2 (DUSP2) Med Sci Monit. 2018;24:4944–4951. doi: 10.12659/MSM.910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao H, Xia G, Wang C, Zhong F, Liu L, Zhang D. miR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7. Med Mol Morphol. 2017;50:76–85. doi: 10.1007/s00795-016-0150-7. [DOI] [PubMed] [Google Scholar]
- 18.Huang Q, Ma Q. MicroRNA-106a inhibits cell proliferation and induces apoptosis in colorectal cancer cells. Oncol Lett. 2018;15:8941–8944. doi: 10.3892/ol.2018.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Y, Huo Z, Song X, Chen X, Tian X, Wang X. mir-106a regulates cell proliferation and apoptosis of colon cancer cells through targeting the PTEN/PI3K/AKT signaling pathway. Oncol Lett. 2018;15:3197–3201. doi: 10.3892/ol.2017.7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung SH, Kim SM, Lee CE. Mechanism of suppressors of cytokine signaling 1 inhibition of epithelial-mesenchymal transition signaling through ROS regulation in colon cancer cells: suppression of src leading to thioredoxin up-regulation. Oncotarget. 2016;7:62559–62571. doi: 10.18632/oncotarget.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed N, Abubaker K, Findlay J, Quinn M. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Curr Cancer Drug Targets. 2010;10:268–278. doi: 10.2174/156800910791190175. [DOI] [PubMed] [Google Scholar]
- 22.Thierauf J, Veit JA, Hess J. Epithelial-to-mesenchymal transition in the pathogenesis and therapy of head and neck cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen L, Qu X, Ma Y, Zheng J, Chu D, Liu B, Li X, Wang M, Xu C, Liu N, Yao L, Zhang J. Tumor suppressor NDRG2 tips the balance of oncogenic TGF-beta via EMT inhibition in colorectal cancer. Oncogenesis. 2014;3:e86. doi: 10.1038/oncsis.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matejka VM, Finek J, Kralickova M. Epithelial-mesenchymal transition in tumor tissue and its role for metastatic spread of cancer. Klin Onkol. 2017;30:20–27. doi: 10.14735/amko201720. [DOI] [PubMed] [Google Scholar]
- 25.Guzman EA, Johnson JD, Linley PA, Gunasekera SE, Wright AE. A novel activity from an old compound: manzamine a reduces the metastatic potential of AsPC-1 pancreatic cancer cells and sensitizes them to TRAIL-induced apoptosis. Invest New Drugs. 2011;29:777–785. doi: 10.1007/s10637-010-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan TJ, Han LH, Cong RS, Liang J. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai) 2005;37:719–727. doi: 10.1111/j.1745-7270.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Ballikaya S, Schonig K, Ball CR, Glimm H, Kopitz J, Gebert J. Transforming growth factor beta receptor 2 (TGFBR2) changes sialylation in the microsatellite unstable (MSI) colorectal cancer cell line HCT116. PLoS One. 2013;8:e57074. doi: 10.1371/journal.pone.0057074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellatt AJ, Mullany LE, Herrick JS, Sakoda LC, Wolff RK, Samowitz WS, Slattery ML. The TGFβ-signaling pathway and colorectal cancer: associations between dysregulated genes and miRNAs. J Transl Med. 2018;16:191. doi: 10.1186/s12967-018-1566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, Yan D. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323–1332. doi: 10.1111/cas.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, Liu QY, Wang H, Ju YH, Li WH, Li YF. Microarray analysis of circular rna expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. doi: 10.1155/2017/8421614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothschild SI, Gautschi O, Batliner J, Gugger M, Fey MF, Tschan MP. MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Cancer. 2017;107:73–83. doi: 10.1016/j.lungcan.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Chu TH, Yang CC, Liu CJ, Lui MT, Lin SC, Chang KW. miR-211 promotes the progression of head and neck carcinomas by targeting TGFβRII. Cancer Lett. 2013;337:115–124. doi: 10.1016/j.canlet.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Qu Y, Zhang H, Duan J, Liu R, Deng T, Bai M, Huang D, Li H, Ning T, Zhang L, Wang X, Ge S, Zhou L, Zhong B, Ying G, Ba Y. MiR-17-5p regulates cell proliferation and migration by targeting transforming growth factor-β receptor 2 in gastric cancer. Oncotarget. 2016;7:33286–33296. doi: 10.18632/oncotarget.8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai N, Hu L, Xie Y, Gao JH, Zhai W, Wang L, Jin QJ, Qin CY, Qiang R. MiR-17-5p promotes cervical cancer cell proliferation and metastasis by targeting transforming growth factor-beta receptor 2. Eur Rev Med Pharmacol Sci. 2018;22:1899–1906. doi: 10.26355/eurrev_201804_14712. [DOI] [PubMed] [Google Scholar]
- 35.Mody HR, Hung SW, Pathak RK, Griffin J, Cruz-Monserrate Z, Govindarajan R. miR-202 diminishes TGFβ receptors and attenuates TGFβ1-induced EMT in pancreatic cancer. Mol Cancer Res. 2017;15:1029–1039. doi: 10.1158/1541-7786.MCR-16-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullmann P, Rodriguez F, Schmitz M, Meurer S, Qureshi-Baig K, Felten P, Ginolhac A, Antunes L, Frasquilho S, Zügel N, Weiskirchen R, Haan S, Letellier E. The miR-371~373 cluster represses colon cancer initiation and metastatic colonization by inhibiting the TGFBR2/ID1 signaling axis. Cancer Res. 2018;78:3793–3808. doi: 10.1158/0008-5472.CAN-17-3003. [DOI] [PubMed] [Google Scholar]
- 37.Wu Q, Guo L, Jiang F, Li L, Li Z, Chen F. Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER- breast cancer cell lines. J Cell Mol Med. 2015;19:2874–2887. doi: 10.1111/jcmm.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the wnt/β-catenin pathway. Mol Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luan W, Zhou Z, Ni X, Xia Y, Wang J, Yan Y, Xu B. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J Cancer Res Clin Oncol. 2018;144:531–542. doi: 10.1007/s00432-018-2582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu B, Cai H, Zheng R, Yang S, Zhou Z, Tu J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol. 2017;92:34–42. doi: 10.1016/j.biocel.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Kjersem JB, Ikdahl T, Lingjaerde OC, Guren T, Tveit KM, Kure EH. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol. 2014;8:59–67. doi: 10.1016/j.molonc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulivi P, Canale M, Passardi A, Marisi G, Valgiusti M, Frassineti GL, Calistri D, Amadori D, Scarpi E. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phua LC, Chue XP, Koh PK, Cheah PY, Chan EC, Ho HK. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among Asians. Oncol Rep. 2014;32:97–104. doi: 10.3892/or.2014.3193. [DOI] [PubMed] [Google Scholar]
- 44.Komatsu S, Ichikawa D, Tsujiura M, Konishi H, Takeshita H, Nagata H, Kawaguchi T, Hirajima S, Arita T, Shiozaki A, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33:271–276. [PubMed] [Google Scholar]


