Abstract
The presence of VPS33B in tumors has rarely been reported. Downregulated VPS33B protein expression is an unfavorable factor that promotes the pathogenesis of lung adenocarcinoma (LUAD). Overexpressed VPS33B was shown to reduce the migration, invasion, metastasis, and chemoresistance of LUAD cells to cisplatin (DDP) in vivo and in vitro. Mechanistic analyses have indicated that VPS33B first suppresses epidermal growth factor receptor (EGFR) Ras/ERK signaling, which further reduces the expression of the oncogenic factor c-Myc. Downregulated c-Myc expression reduces the rate at which it binds the p53 promoter and weakens its transcription inhibition; therefore, decreased c-Myc stimulates p53 expression, leading to decreased epithelial-to-mesenchymal transition (EMT) signal. NESG1 has been shown to be an unfavorable indicator of non-small-cell lung cancer (NSCLC). Here, NESG1 was identified as an interactive protein of VPS33B. In addition, NESG1 was found to exhibit mutual stimulation with VPS33B via reduced RAS/ERK/c-Jun-mediated transcription repression. Knockdown of NESG1 activated EGFR/Ras/ERK/c-Myc signaling and further downregulated p53 expression, which thus activated EMT signaling and promoted LUAD migration and invasion. Finally, we observed that nicotine suppressed VPS33B expression by inducing PI3K/AKT/c-Jun-mediated transcription suppression. Our study demonstrates that VPS33B as a tumor suppressor is significantly involved in the pathogenesis of LUAD.
Keywords: Chemoresistance, Lung adenocarcinoma, Metastasis, Nicotine, VPS33B
Introduction
Lung cancer is the most commonly occurring cancer worldwide and is the leading cause of cancer-related death.1 The number of deaths related to lung cancer alone exceeds the total number of deaths caused by the next three most prevalent cancers (colon cancer, breast cancer, and prostate cancer).2 Lung cancer is classified into two histological types: small-cell lung cancer and non-small-cell lung cancer (NSCLC). As a subtype of NSCLC, lung adenocarcinoma (LUAD) is the most common pathologic type of lung cancer and has a poor prognosis.3,4 To date, the molecular mechanisms underlying its initiation and development remain unclear.
Carcinogenesis results from an imbalance between tumor-activating genes and tumor suppressors.5, 6, 7, 8, 9, 10 The VPS33B gene is a member of the Sec-1 domain family and encodes the human ortholog of rat VPS33B, which is homologous to the yeast class C VPS33 protein.11 It is a core component of two sorting complexes: the class C core vacuole/endosome tethering complex and the homotypic fusion and vacuole protein sorting complex.12, 13, 14 Previous studies have indicated that VPS33B is correlated with renal dysfunction, cholestasis syndrome, and platelet activation.15, 16, 17 Two studies have reported that VPS33B is a tumor suppressor in hepatocellular carcinoma (HCC) and nasopharyngeal carcinoma (NPC).18,19 However, the role and molecular basis of VPS33B in tumor metastasis has yet to be determined.
NESG1 and CCDC19 expressed in nasopharyngeal epithelium and trachea has been cloned and revised in a previous investigation.4,20,21 Reduced NESG1 is an unfavorable factor that promotes the pathogenesis of nasopharyngeal carcinoma (NPC) and NSCLC.22,23 However, the molecular basis for NESG1-mediated metastatic suppression remains unclear in LUAD.
Nicotine, a major component in cigarette smoke, is extremely hazardous and causes various types of cancer, including gastric cancer, colorectal cancer and lung cancer.24, 25, 26, 27, 28, 29, 30 Approximately 90% of deaths caused by lung cancer are attributable to cigarette smoking.31 Nicotine induces lung cancer cell proliferation and angiogenesis via nicotinic acetylcholine receptors and β1-arrestin (ARRB1) or IGF2 exocytosis.32, 33, 34 Thus, nicotine is a significant factor in inducing the pathogenesis of lung cancer. However, whether nicotine modulates VPS33B has not yet been determined.
The current study demonstrates that reduced VPS33B protein promotes the pathogenesis of LUAD. VPS33B is also identified as a downstream negative regulator of nicotine. VPS33B interacts with NESG1 to modulate EGFR/Ras/ERK/c-Myc/p53-mediated EMT signal, thereby suppressing cell metastasis and chemoresistance to cisplatin (DDP) in LUAD cells. These data present the detailed mechanisms underlying the function of VPS33B as a tumor metastasis suppressor that prevents the pathogenesis of LUAD.
Materials and methods
Immunohistochemistry
Two paraffin-embedded tissue arrays with different lung adenocarcinoma samples were purchased from Shanghai Outdo Biotech. Co., Ltd. For the use of these clinical materials for research purposes, prior consent was obtained from the patients, along with approval from the Ethics Committee of Shanghai Outdo Biotech. Co., Ltd. The tissue arrays were deparaffinized, and antigen retrieval was performed in citrate buffer for 3 min at 100 °C. Endogenous peroxidase activity and nonspecific antigens were blocked with a peroxidase blocking reagent, followed by incubation with primary antibody overnight at 4 °C. The antibody dilutions and sources are listed in Table S3. After washing, the sections were incubated with a biotin-labeled secondary antibody and were subsequently incubated with streptavidin-conjugated horseradish peroxidase. The peroxidase reaction was developed using a 3,3-diaminobenzidine (DAB) chromogen solution in a DAB buffer substrate (Maixin, Fuzhou, China). The sections were visualized with DAB and counterstained with hematoxylin, mounted in a neutral gum, and analyzed by bright-field microscopy.
To evaluate the VPS33B staining, a semiquantitative scoring criterion was used in which both the staining intensity and the percentage of positive cells were recorded. A staining index (ranging from 0 to 7) was obtained from the intensity of VPS33B staining (0 = negative, 1 = weakly positive, 2 = moderately positive, 3 = strongly positive) multiplied by the proportion of immunopositive tumor cells (<10% = 1, 10% to <50% = 2, 50% to <75% = 3, ≥75% = 4). A score of 6 or greater was classified as indicating VPS33B overexpression. For scoring, two independent pathologists were blinded to the clinicopathological information.
Cell culture
The lung cancer cell lines A549 and H1975 were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and maintained in RPMI 1640 medium supplemented with 10% newborn calf serum (ExCell, Shanghai, China). A549-DDP cell lines were constructed in-house via progressively increasing the concentration of DDP. The cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2.
Lentivirus production and infection
Lentiviral particles carrying the VPS33B cDNA and GFP vector were constructed by GeneChem (Shanghai, China). A549 and H1975 cells were infected with lentiviral vectors, and the levels of VPS33B were measured using reverse transcriptase qPCR and Western blot analysis.
RNA isolation, reverse transcription, and qRT-PCR
RNA isolation, reverse transcription, and qRT-PCR were performed in LUAD cell lines according to the instructions provided by TAKARA, Co., Ltd. The specific qPCR primers for VPS33B, c-Jun, p53, c-Myc and NESG1 are listed in Table S1. All experiments were repeated at least three times.
In vitro cell migration and invasion assays
For cell migration assays, 1 × 105 cells in a 100 μL of serum-free medium were seeded onto a fibronectin-coated polycarbonate membrane inserted in a Transwell apparatus (Corning, USA). On the lower surface, 500 μL of RPMI 1640 medium containing 10% fetal bovine serum was added as a chemoattractant. After incubating the cells for 10 h at 37 °C in a 5% CO2 atmosphere, Giemsa-stained cells adhering to the lower surface were counted under a microscope in five predetermined fields (100×). All assays were repeated independently at least three times. The procedure for the cell invasion assays was similar to that of the cell migration assay, except that the Transwell membranes were precoated with 24 μg/mL Matrigel (R&D Systems, USA).
In vivo metastasis in nude mice
For in vivo metastasis assays, 50 μL of A549 and H1975 cells (5 × 106) stably overexpressing VPS33B (or an equal number of their respective control cells) were injected under the liver capsule of each mouse (5 mice per group). All mice were sacrificed 4 weeks later. The liver and lungs were subjected to fluorescence image detection, which visualized primary tumor growth and metastatic lesion formation. The mice were maintained in a barrier facility with high-efficiency particulate air (HEPA)-filtered racks and fed with an autoclaved laboratory rodent diet. All animal studies were conducted in accordance with the principles and procedures outlined in the Southern Medical University Guide for the Care and Use of Animals.
MTT cytotoxicity assay
DDP (QiluPharmo Co., Ltd, Jinan, China) was resuspended in PBS (0.5 mg/mL) and stored at −20 °C. Drug sensitivity was determined by MTT assay. The cells were seeded in 96-well plates in 100 μL of RPMI-1640 medium supplemented with 10% FBS at 2 × 103 cells/well. Once attached, the cells were treated with 2.5, 5, 10, 20, or 40 μM DDP (0.5 mg/mL) and incubated at 37 °C in 5% CO2 for 48 h. Experiments were conducted three times.
In vivo DDP sensitivity experiment
To establish an LADC mouse model, 6 × 105 VPS33B-overexpressing A549 cells or their controls were injected intraperitoneally into nu/nu mice (n = 40) aged 4 weeks. Tumors were allowed to grow for 3 d, and animals were randomized into four groups, treated with either normal saline (NS) or DDP injected intraperitoneally every 3 d (NC + NS, NC + DDP, VPS33B + NS, and VPS33B + DDP, n = 10/group). The survival time of the nude mice was observed, and survival curves were analyzed by Kaplan–Meier analysis.
Transient transfection with small-interfering RNAs and plasmids
siRNAs for VPS33B and NESG1 were designed and synthesized at RiboBio Inc. (Guangzhou, China). The sequences of each siRNA, mimic and inhibitor are listed in Table S2. The VPS33B and NESG1 cDNA was constructed (in-house) in pCMV vectors and contained an HA or Myc tag, respectively. The c-Jun, c-Myc and p53 pcDNA3.1 plasmids were purchased from Vigene Biosciences (Shandong, China). A549 and H1975 cells were plated in 6-well and 96-well plates (Nest, Biotech, China) at 30%–50% confluence 24 h prior to transfection. Subsequently, siRNAs, miRNAs, or the plasmids with different genes were respectively transfected or co-transfected at a working concentration of 100 nM using the TurboFect™ siRNA Transfection Reagent (Fermentas, Vilnius, Lithuania) and Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, Waltham, USA) according to the protocol provided by the manufacturers. Cells were collected after 48–72 h for further experiments.
Western blot analysis, reagents, and antibodies
Western blot analyses were performed as described in a previous study, with primary antibodies against VPS33B, EGFR, NESG1, PI3K, p-PI3K, AKT, p-AKT, ERK, p-ERK, K-Ras, c-Myc, E-cadherin, N-cadherin, Vimentin, Snail, and p53. β-actin and GAPDH were used as loading controls. Dilutions and the sources of antibodies are listed in Table S3. Images were captured using Minichemi Chemiluminescence Imaging System, Beijing Sage Creation Science Co., Ltd., China.
Luciferase reporter assay
To evaluate the effects of c-Jun on NESG1 and VPS33B promoters, as well as c-Myc on the p53 promoter activity, fragments containing the c-Jun binding sites in NESG1 and VPS33B promoters, as well as the c-Myc binding sites in the p53 promotor, were cloned into the pGL3-Basic luciferase reporter vector. In addition, the c-Jun binding site mutation vectors and c-Myc binding site mutation vectors were constructed. These vectors, the c-Jun plasmid, and the c-Myc plasmid were co-transfected into A549 and H1975 cells. The luciferase activity of these promoters was examined 48 h after transfection.
Co-Immunoprecipitation (Co-IP)
Co-IP was conducted using the Pierce Co-Immunoprecipitation Kit (Thermo Scientific, USA) according to the instructions provided by the manufacturer. Total proteins were extracted and quantified. A total of 3000 μg of protein in 200 μL of supernatant was incubated with 10 μg of anti-HA and anti-MYC for 12 h at 4 °C. The resin was washed and the sample was eluted in sample buffer, followed by boiling for 10 min at 100 °C. Immune complexes were subjected to Coomassie Brilliant Blue staining and Western blot analysis.
Confocal laser scanning microscopy
A549 cells were co-transfected with the VPS33B plasmid carrying an HA-tag and the NESG1 plasmid carrying a myc-tag in a six-well plate. The cells were cultured overnight and then fixed with 3.5% paraformaldehyde and permeabilized with 0.2% Triton X-100 at room temperature. The cells were incubated with anti-HA and anti-myc antibodies for 30–45 min at 37 °C. After incubation for 30–45 min 37 °C with secondary antibody, the coverslips were mounted onto slides using a mounting solution containing 0.2 mg/mL DAPI. Images were captured by Zeiss LSM 800 laser confocal microscope.
Chromatin immunoprecipitation assay
A ChIP assay was performed as previously described using the ChIP Assay Kit38,39 (Millipore, Catalog: 17–371) to determine whether c-Jun combined with the NESG1 and VPS33B promoters and whether c-Myc combined with the p53 promoter in A549 and H1975 cells. Crosslinked DNA was sheared to 200–1000 base pairs in length with sonication and then subjected to immunoselection with an anti-c-Jun antibody. Finally, qPCR was conducted to measure the enrichment of DNA fragments in the putative transcription binding sites in these gene promoters on the basis of the specific primers. The specific primers are listed in Table S1.
Electrophoretic mobility shift assay (EMSA)
The binding activity on the promoter region of p53, NESG1, and VPS33B were detected using the EMSA Kit (Roche, Switzerland) according to the instructions provided by the manufacturer. The probes used are listed in Table S4. Preformed c-Jun-recognized probes (Biosense Bioscience Co., Ltd., Guangzhou, China) were used as a positive control, and samples without nucleoprotein were used as negative controls. For competition experiments, a 100-fold specific oligonucleotide competitor (unlabeled wild-type or mutant gene probes) was added to the binding mixture 10 min before the labeled probe was added. Visualized bands were analyzed using a BioSens Gel Imaging System (BIOTOP, China). The EMSA analysis was performed at Biosense Bioscience Co., Ltd. (Guangzhou, China).
Yeast two-hybrid screening
NESG1 bait screening was conducted using the Mate&Plate™ Library-Universal Human and Screening Kit (Clontech) according to the guidelines provided by the manufacturer. The NESG1 bait constructed in the DNA-binding domain (DBD)-containing pGBKT7 vector was transformed into the yeast haploid strain Y2H. Y2H was mated with yeast Y187 cells (which contain a human cDNA library) and formed diploid yeast colonies. The DNA was isolated from individual colonies after selection on media lacking leucine, tryptophan, histidine, and adenine (SD/-Trp/-Leu/-His/-Ade). The isolated yeast DNA was transformed into electrocompetent DH5α cells. NESG1 interacting partners were determined by sequence analysis using the pGADT7-Rec3′ internal sequencing primer (upstream 5′-AGGCTGAGCTGCGAAAAAG-3′) and BLAST searches against the human genome (NCBI: http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Nicotine-treated LUAD cells
A549 and H1975 cells were seeded (2 × 105) in RMPI 1640 medium. Nicotine (Sigma–Aldrich; Merck KGaA, Darmstadt, Germany) was used to treat these cells at concentrations of 0.1, 1, 10, and 100 μM at 37 °C for 72 h. Additionally, cells were treated with 10 μM nicotine for different periods of time (48, 60, 72, 132, and 144 h).
Statistical analysis
Statistical analyses were performed using SPSS ver. 20.0 (SPSS Inc. Chicago, IL, USA) and GraphPad Prism v5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The data were expressed as the mean ± SD of at least three independent experiments. Differences were considered statistically significant at P < 0.05 by Student's t-test for two groups, one-way ANOVA for multiple groups, and parametric generalized linear model with random effects. Chi-squared testing was used to examine the correlation of VPS33B expression with clinical features. A survival analysis was performed using the Kaplan–Meier method. All statistical tests were two-sided, and asterisks indicate statistical significance.
Results
Reduced VPS33B protein expression acts as an unfavorable prognostic factor
Immunohistochemical staining was conducted to determine VPS33B protein expression in 155 LUAD cases with prognosis information (and in 42 lung tissues). The results showed the expression of cytoplasmic VPS33B in LUAD and their matched lung tissues (Fig. 1A). VPS33B protein expression in LUAD tissues was significantly downregulated relative to the control lung tissues (P = 0.001) (Table 1). Reduced VPS33B protein expression also exhibited a significantly positive correlation with the overall survival time of the patient (Fig. 1B) (P = 0.016). These data demonstrated that reduced VPS33B protein is an unfavorable factor promoting LUAD pathogenesis.
Figure 1.
Reduced VPS33B protein expression as an unfavorable prognosis factor in LUAD. (A) IHC demonstrated VPS33B protein expression in LUAD cells and bronchial epithelial cells: 1: High VPS33B protein expression in bronchial epithelial cells; 2 and 3: Low VPS33B protein expression in LUAD cells; 4 and 5: High VPS33B protein expression in LUAD cells. (B) Kaplan–Meier survival analysis showed prolonged overall survival for LUAD patients with high VPS33B protein expression compared to patients with low VPS33B protein expression. The log-rank test was used to calculate the P values.
Table 1.
The expression of VPS33B in lung adenocarcinoma compared to non-cancerous lung tissues.
| Group | Cases (n) | VPS33B expression |
P value | |
| High expression | Low expression | |||
| Cancer | 155 | 77 | 78 | P = 0.001 |
| Bronchial Epithelium | 42 | 33 | 9 | |
VPS33B suppresses cell metastasis and chemoresistance to DDP
To explore the biological role of VPS33B in LUAD, endogenous VPS33B expression was detected in H460 and H446 cells, whereas the other three lines (A549, SPCA1, and H1975) showed undetectable or very low levels of endogenous VPS33B; these cells were selected for subsequent experiments (Fig. S1A). Lentivirus-carrying VPS33B cDNA was injected into H1975 and SPCA1-1 cells with low VPS33B mRNA and protein expression. Quantitative polymerase chain reaction and Western blot analysis showed markedly higher VPS33B mRNA and protein expression levels in A549 and H1975 cells than in their respective empty vector control cells (Fig. S1B and C).
Transwell (Fig. 2A) (Fig. S1D) and Boyden assays (Fig. 2B) (Fig. S1E) were conducted to evaluate the effects of VPS33B on the migration and invasion ability of LUAD cells in vitro. The data indicated that overexpressed VPS33B markedly reduced migration and invasion in A549 and H1975 cells relative to their respective controls. The in vivo metastasis assay indicated that intrahepatic dissemination and lung metastasis were significantly lower in nude mice that had been injected under the liver capsule with Lv-VPS33B-GFP A549 and H1975 cells compared to their respective control groups (Fig. 2C, Fig. S1F).
Figure 2.
VPS33B suppresses LUAD migration, invasion, metastasis and DDP resistance in vivo and in vitro. (A–B). Transwell (A) and Boyden (B) assays demonstrated that A549 and H1975 cell migration and invasion were decreased after transfection with Lv-GFP-VPS33B. Student's t-test, one-way ANOVA, mean ± s.d., *P < 0.05. (C).In vivo metastasis assays indicated that intrahepatic dissemination and lung metastasis levels were reduced in Lv-GFP-VPS33B A549 and H1975 cells. (D). Survival analysis showed the cumulative overall survival time, ranked low to high, as follows: NC + NS < VPS33B + NS < NC + DDP < VPS33B + DDP, n = 10/group. Log-rank test. (E). Changes in EGFR, Ras/ERK/p-ERK, c-Jun, c-Myc, p53, E-cadherin, N-cadherin, Vimentin and Snail expression were detected by Western blot analysis in A549 and H1975 cells after transfection of Lv-GFP-VPS33B. β-Actin was used as a loading control. All experiments were repeated three times.
In a subsequent study, we observed that VPS33B A549 and H1975 cells exhibiting stable overexpression had significantly increased sensitivity to DDP in vitro (Fig. S1G). The inhibition rates 48 h after treatment with DDP at different concentrations were evaluated for cells transfected with VPS33B and its empty control. The 50% inhibitory concentration (IC50) of DDP decreased from 5.13 μM to 2.42 μM in A549 cells after VPS33B transfection. A similar IC50 reduction, from 20.25 μM to 8.83 μM, was observed in H1975 cells (Fig. S1L). These decreases were verified in vivo by applying VPS33B-overexpressing A549 xenografts in nude mice. The Kaplan–Meier survival analysis confirmed that DDP treatment (NC + DDP) or VPS33B overexpression (VPS33B + normal saline (NS)) alone extended the lifespan of the mice relative to that of the untreated normal controls (NC + NS). However, overexpressed VPS33B coupled with DDP treatment (VPS33B + DDP) markedly prolonged the survival time beyond those of the other three groups (Fig. 2D).
Knockdown of VPS33B protein expression via a specific small interfering RNA (siRNA) in VPS33B-overexpressed A549 and H1975 cells showed that silencing VPS33B reverses its suppressive effect on EGFR protein expression (Fig. S2A), cell migration and invasion, as determined by Transwell (Fig. S2B) and Boyden assays (Fig. S2C) in LUAD cells.
To clarify the molecular mechanism by which VPS33B functions as a tumor suppressor related to LUAD, the key regulators of the cell cycle and epithelial-to-mesenchymal (EMT) signals were analyzed by Western blot. VPS33B overexpression was found to downregulate c-Myc and c-Jun expression; in addition, it enhanced p53 expression in A549 and H1975 cells. In the EMT pathway, overexpressed VPS33B suppressed Snail, N-cadherin, and Vimentin and upregulated E-cadherin. We also observed that the EGFR, p-ERK, and K-Ras levels decreased in VPS33B-overexpressing A549 and H1975 cells, but total ERK remained unchanged in these cells (Fig. 2E). Taken together, these data demonstrated that VPS33B acts as a metastasis-suppressor by modulating EGFR/Ras/ERK, p53,c-Myc,c-Jun, and EMT pathways.
C-Myc directly suppresses p53
To explore whether c-Myc directly suppresses p53, c-Myc was first knocked down by siRNA (Fig. 3A). The data showed that the p53 mRNA expression level was markedly increased in A549 and H1975 cells (Fig. 3B). Notably, c-Myc was a potential binding transcription factor to the p53 promoter, as predicted using the PROMO program and UCSC online database (Fig. 3C). The ChIP and EMSA assays indicated that c-Myc was bound to the p53 promoter (Fig. 3D–F). We then found that c-Myc transduction markedly decreased the luciferase activity of the p53 promoter relative to that of its control group (Fig. 3G). These results confirmed that c-Myc directly suppresses p53 expression by binding to its promoter.
Figure 3.
c-Myc directly suppresses p53 (A) qPCR was used to confirm the effectiveness of silencing c-Myc in A549 and H1975 cells; Student's t-test, Mean ± s.d., **P < 0.01. (B) p53 was found to be upregulated in the c-Myc-silenced A549 andH1975 cells by qPCR. Student's t-test, Mean ± s.d., **P < 0.01. (C) Bioinformatics analysis revealed the promoter regions of p53 with a putative c-Myc binding site. (D-E). qPCR (D) and gel electrophoresis (E) confirmed the amplification of c-Myc-binding sites after ChIP using an antibody against c-Myc. IgG antibody was used as the negative control. Student's t-test, Mean ± s.d., **P < 0.01. (F) EMSA assay confirmed c-Myc binding to p53 promoter in A549 and H1975 cells. Labeled wild-type probe was incubated without (lane 1) or with (lane 4) cell nuclear proteins in the absence or presence of unlabeled probe (lane 2–3). Unlabeled wild-type probe (lane 2) and mutant c-Myc probe (lane 3) were used to compete with c-Myc binding, each at 100-fold excess. Supershift assay (lane 5) was performed using an anti-c-Myc antibody. (G) Luciferase reporter assay demonstrated the luciferase activities of the wild-type and Mut p53 promoter in A549 and H1975 cells transfected with c-Myc plasmid. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01.
EGFR antagonizes VPS33B to modulate Ras/ERK/c-Myc/p53
To investigate the role of EGFR in VPS33B-mediated suppression, EGFR was transfected into VPS33B-overexpressing LUAD cells. Increased EGFR levels restored the expression of the Ras/ERK/c-Myc signal and reduced p53 protein expression (Fig. S3A). Elevated EGFR increased the combination of c-Myc with the p53 promoter (Fig. S3B) and reduced p53 mRNA expression (Fig. S3C). Finally, overexpressed EGFR antagonized the VPS33B-mediated repression of migration and invasion in LUAD cells (Fig. S3D and E). These data indicated that EGFR antagonizes VPS33B to modulate Ras/ERK/c-Myc/p53 in LUAD.
VPS33B interacts with NESG1 to suppress EGFR/RAS/ERK/c-Jun
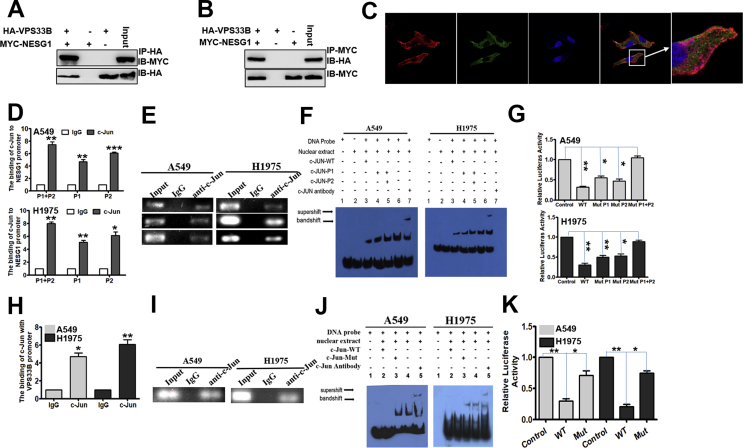
To explore whether NESG1 interacts with VPS33B, the yeast two-hybrid (Y2H) system was used to identify the interactive protein of NESG1. The coding sequence (CDS) of the NESG1 gene (Fig. S4A) was cloned into a pGBKT7 Y2H bait expression vector (Fig. S4B). PCR was used to identify the successful recombinants of pGBKT7-NESG1 (Fig. S4C). The recombinant bait plasmid exerted no self-activating effect and showed no toxicity to the Y2H cells (Fig. S4D). The human cDNA library in yeast Y187 cells was mated with pGBKT7-NESG1 Y2H cells (Fig. S4E) and further screened in the SD/-Ade/-His/-Leu/-Trp(QDO)/X-a-Gal/AbA plate (Fig. S4F and G). Finally, 30 positive clones were obtained (Fig. S3H) and were analyzed by sequencing. Four potential NESG1-interacting proteins, including RNF2, VPS33B, ENKUR, and CCDC65, were identified. In subsequent co-immunoprecipitation experiments, we confirmed the interactive combination of NESG1 with VPS33B (Fig. 4A and B) but not with RNF2, ENKUR, or CCDC65 (data not shown). Confocal laser scanning microscopy verified the cytoplasm and vesicles colocalization between NESG1 and VPS33B in LUAD cells (Fig. 4C). Overexpressed VPS33B upregulated the expression of NESG1 mRNA (Fig. S4I). Similarly, overexpressed NESG1 upregulated the expression of VPS33B mRNA (Fig. S4J). Bioinformatic assays indicated the presence of binding sites for c-Jun in the promoter regions of NESG1 and VPS33B (Fig. S4K). Increased c-Jun expression significantly reduced VPS33B and NESG1 mRNA expression (Fig. S4L). Moreover, ChIP, EMSA, and luciferase activity assays revealed that c-Jun combined with the NESG1 (Fig. 4D–G) and VPS33B (Fig. 4H–K) promoters and reduced the activity of these promoters. Transfecting EGFR cDNA into VPS33B-overexpressing LUAD cells markedly restored K-Ras/ERK/c-Jun signaling and thus downregulated NESG1 protein expression (Fig. S4M). These results demonstrated that VPS33B interacts with NESG1 to reduce EGFR/RAS/ERK/c-Jun signal.
Figure 4.
VPS33B interacts with NESG1 to suppress EGFR via RAS/ERK/c-Jun. (A–B) Western blotting confirmed the interaction of VPS33B and NESG1 after Co-IP in 293T cells transfected with HA-VPS33B and MYC-NESG1. (C) VPS33B co-located with NESG1 in the cytoplasm and vesicles, as detected by laser confocal assay. (D–E) qPCR (D) and gel electrophoresis (E) confirmed the amplification of the c-Jun-binding sites of NESG1 after ChIP using an antibody against c-Jun. IgG antibody was used as the negative control. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01. (F) EMSA assay confirmed c-Jun binding to NESG1 promoter in A549 and H1975 cells. Labeled wild-type probe was incubated without (lane 1) or with (lane 5) cell nuclear proteins in the absence or presence of unlabeled probe (lanes 2–4). Unlabeled wild-type probe (lane 2) and mutant c-Jun probe (lane 3 and 4) were used to compete with c-Jun binding, each at 100-fold excess. Supershift assay (lane 6) was performed using an anti-c-Jun antibody. (G) Luciferase reporter assay demonstrated the luciferase activities of the wild-type and Mut NESG1 promoter in A549 and H1975 cells transfected with c-Jun plasmid. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01. (H–I). qPCR (H) and gel electrophoresis (I) confirmed the amplification of the c-Jun-binding sites of VPS33B after ChIP using an antibody against c-Jun. IgG antibody was used as the negative control. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01. (J). EMSA identified c-Jun binding to the VPS33B promoter in A549 and H1975 cells. Labeled wild-type probe was incubated without (lane 1) or with (lane 4) cell nuclear proteins in the absence or presence of unlabeled probe (lanes 2–3). Unlabeled wild-type probe (lane 2) and mutant c-Jun probe (lane 3) were used to compete with c-Jun binding, each at 100-fold excess. Supershift assay (lane 5) was performed using an anti-c-Jun antibody. (K). Luciferase reporter assay demonstrated the luciferase activities of the wild-type and Mut VPS33B promoter in A549 and H1975 cells transfected with c-Jun plasmid. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01.
NESG1 knockdown reverses VPS33B-mediated migration and invasion inhibition of LUAD cells
To investigate whether NESG1 mediates VPS33B-induced suppression, siRNAs were used to reduce NESG1 expression (Fig. 5A) in VPS33B-overexpressed LUAD cells. NESG1 knockdown markedly restored the migration and invasion ability of VPS33B-overexpressing LUAD cells, as shown by Transwell (Fig. 5B) and Boyden (Fig. 5C) assays. We further observed that the VPS33B-modulated signals were reversed after silencing NESG1 in VPS33B-overexpressing LUAD cells. These reversed signals included the upregulation of EGFR-induced Ras/ERK signal, c-Myc and EMT-related protein factors (such as N-cadherin, vimentin, Snail, and c-Jun), and the downregulation of p53 and E-cadherin (Fig. 5D). In addition, reduced p53 mRNA levels were also observed (Fig. 5E). Finally, the ChIP assays indicated that the ability of c-Myc to bind the p53 promoter was enhanced in VPS33B-overexpressing LUAD cells upon NESG1 knockdown (Fig. 5F). There data showed that knocking down NESG1 reversed VPS33B-induced migration and invasion inhibition for LUAD cells.
Figure 5.
Knockdown of NESG1 reverses VPS33B-mediated inhibition of LUAD. (A) qPCR confirmed the effectiveness of NESG1 silencing in VPS33B-overexpressing A549 and H1975 cells. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01. (B–C) Transwell (B) and Boyden (C) assays demonstrated changes in the migration and invasion ability of VPS33B-overexpressing A549 and H1975 cells after transfection with NESG1 siRNA. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01. (D) Expression changes for NESG1, EGFR, Ras/ERK/p-ERK, c-Jun, c-Myc, p53, Snail, E-cadherin and Vimentin were detected by Western blot in VPS33B-overexpressing A549 and H1975 cells after transfection with NESG1 siRNA. β-Actin was used as a loading control. (E) qPCR found that knockdown of NESG1 by siNESG1 reduced p53 expression in VPS33B-overexpressing LUAD cells. Student's t-test, Mean ± SD, *P < 0.05,**P < 0.01. (F) NESG1 siRNA suppression increased the binding of c-Myc with the p53 promoter in VPS33B-overexpressing A549 and H1975 cells, as found by ChIP and qPCR. Student's t-test, Mean ± s.d., *P < 0.05, **P < 0.01.
Nicotine downregulates VPS33B via PI3K/AKT/c-Jun signaling
To determine whether nicotine modulates VPS33B and its mediated signaling, LUAD cells were incubated in nicotine-containing media. We observed that the levels of VPS33B mRNA were downregulated in LUAD cells treated with nicotine for 72 h at different concentrations (0.1, 1, 10, and 100 μmol/L) and over different lengths of time (48, 60, 72, 100, 132, and 144 h when treated with 10 μmol/L). The results showed that VPS33B mRNA was markedly reduced in LUAD cells at nicotine concentrations up to 10 μmol/L and when the treatment time was increased to 72 h in LUAD cells with a nicotine treatment concentration of 10 μmol/L (Fig. 6A). Moreover, PI3K/AKT/c-Jun signaling was reduced, whereas VPS33B protein expression (Fig. 6B) and mRNA expression (Fig. 6C) were elevated in the nicotine-treated LUAD cells transfected with the PI3K-specific inhibitor Ly294002 compared to the nicotine-treated cells, which was similar to the control cells. The interaction of c-Jun with the VPS33B promoter was significantly decreased when the PI3K-specific inhibitor Ly294002 was transfected into the nicotine-treated LUAD cells, but this interaction was similar to control cells (Fig. 6D and E). These results demonstrated that nicotine downregulates VPS33B expression via suppressing PI3K/AKT/c-Jun signaling.
Figure 6.
Nicotine negatively modulated VPS33B in LUAD. (A) qPCR showed that mRNA levels of VPS33B were downregulated in A549 and H1975 cells treated with different concentrations of nicotine (0.1, 1, 10, or 100 μmol/L) for 72 h and for different times (24, 48, 72, 100, 132 and 144 h) at 10 μmol/L nicotine. (B) Changes in PI3K/AKT/p-PI3K/p-AKT, c-Jun and VPS33B expression were detected by Western blot in nicotine-treated A549 and H1975 cells after transfection with the PI3K inhibitor Ly294002. β-Actin was used as a loading control. (C) VPS33B was upregulated in nicotine-treated A549 and H1975 cells after transfection with the PI3K inhibitor Ly294002. Student's t-test, mean ± s.d, *P < 0.05, **P < 0.01. (D) c-Jun binding to the VPS33B promoter was examined by qPCR in nicotine-treated A549 and H1975 cells after transfection with Ly294002. Student's t-test, mean ± s.d, **P < 0.01. (E) Gel electrophoresis confirmed that c-Jun-binding sites in VPS33B were enhanced after treatment with nicotine by using ChIP with an anti-c-Jun antibody. IgG antibody was used as the negative control.
Discussion
In a previous study, Wang et al used a VPS33B-knockout mouse model to report that VPS33B acted in a tumor suppressor role during the carcinogenesis of hepatocellular carcinoma. Subsequently, we found that VPS33B suppresses NPC growth, which further supports VPS33B as being a potential tumor suppressor. In the current study, we explored the role of VPS33B in LUAD. Immunochemical staining indicated that VPS33B protein in LUAD tissues was significantly downregulated relative to that in lung bronchial epithelium tissues. Reduced VPS33B protein expression also indicated a poor prognosis for LUAD patients. These data suggest that VPS33B protein has a potential suppressive role in LUAD.
Previous studies have indicated VPS33B as a tumor suppressor involved with hepatocarcinogenesis and NPC growth. However, whether VPS33B suppresses the tumor metastases of LUAD cells is still undetermined. In the current study, we observed that VPS33B acts as a antitumor-metastasis factor to reduce cell migration, invasion and metastasis in vitro and in vivo. Furthermore, the overexpression of VPS33B also markedly reduced NPC cell resistance to DDP chemoresistance in vitro and in vivo. These findings suggest that VPS33B protein is a potential suppressor of tumor metastasis in LUAD.
EGFR influences the initiation and promotion of tumor pathogenesis.4,35, 36, 37 Most non-small-cell lung cancers express epidermal growth factor receptor and its natural ligand. The distribution and expression of EGFR in lung cancer has been well established. In 2002,38 Piyathilake et al detected the expression of EGFR in tumor tissues and normal lung tissues of 60 lung cancer patients by immunohistochemistry. The expression of EGFR in lung cancer tissues was significantly higher than that in normal lung tissues. The expression in pre-lesions and lung cancer tissues is progressively elevated. The activation of EGFR-induced Ras/ERK and its downstream EMT signals are key elements that promote metastasis and resistance to chemotherapy.39, 40, 41 In this study, we examined the changes in EGFR/Ras/ERK signaling and its downstream c-Myc, p53, c-Jun and EMT signaling in VPS33B-overexpressed LUAD cells. Notably, the overexpression of VPS33B reduced EGFR-induced Ras/ERK signaling, which further suppressed its downstream c-Myc and EMT pathways, including the upregulated expression of E-cadherin and the downregulated expression of Snail, N-cadherin, and Vimentin in LUAD cells. Furthermore, p53 was elevated in VPS33B-overexpressed LUAD cells. Finally, the expression of the oncogenic transcription factor c-Jun was also reduced in VPS33B-overexpressing LUAD cells. Taken together, the aforementioned findings suggest that VPS33B is a suppressor of tumor metastasis and downregulates EGFR/Ras/ERK signaling and its downstream c-Myc, c-Jun, and EMT signaling and induces p53 expression.
C-Myc is a key oncogenic factor and had been reported to suppress p53 expression in tumors.42, 43, 44 However, its detailed molecular basis has not been reported. In this study, bioinformatics analysis predicted that c-Myc could bind to the p53 promoter. Further, we confirmed that c-Myc did indeed bind to the p53 promoter and suppressed its expression at the transcription level. Finally, EGFR transfection led to decreased p53 and upregulated the EGFR/Ras/ERK/c-Myc pathway in VPS33B-overexpressing LUAD cells. These findings demonstrate that VPS33B suppresses EGFR/Ras/ERK/c-Myc signaling and thus induces p53 expression. P53 is a classical tumor suppressor and has been reported to bind to Snail protein, thereby suppressing EMT signaling.45 Thus, VPS33B suppresses EMT signaling by reducing the EGFR/Ras/ERK/c-Myc-induced transcriptional suppression of p53.
Protein–protein interactions are vital when exploring cellular signals.46, 47, 48 In prior studies, we cloned and revised the NESG1 coding sequence. Further, we identified this protein as a tumor suppressor in NPC and NSCLC.22,23 In the current study, we used the yeast two-hybrid approach to screen interaction proteins of NESG1. Interestingly, VPS33B was found to be a potential interacting protein of NESG1. We then validated the interaction of VPS33B and NESG1 and co-located the complex in the cytoplasm and vesicles using Co-IP and laser confocal fluorescence microscopy in LUAD cells. In addition, we found that VPS33B and NESG1 exhibit mutually stimulated expression by reducing Ras/ERK signaling to downregulate c-Jun expression. C-Jun, as an oncogenic transcription factor, was identified to bind directly to the VPS33B and NESG1 promoters and thus suppressed the expression of both genes. Knockdown of NESG1 in VPS33B-overexpressing LUAD cells markedly reversed the VPS33B-modulated signaling, including upregulating the expression levels of EGFR/Ras/ERK/c-Myc, EMT signals and the oncogenic transcription factor c-Jun, and decreasing p53 expression. These findings demonstrated that NESG1 interacts with VPS33B and participates in VPS33B-mediated LUAD metastasis suppression.
Cigarette smoke is considered a high-risk factor for various cancers, including lung cancer, gastric cancer, and pancreatic cancer.49 Nicotine is the most important component of smoking. After cigarette smoke inhalation, nicotine is rapidly absorbed into the lungs, resulting in a relatively high nicotine concentration in the volume of blood leaving the heart. A previous study showed that blood or plasma nicotine concentrations that were sampled in the afternoon in smokers generally ranged from 10 to 50 ng/mL−1 (0.06–0.31 μM) for long-term smoking populations.50 Stable low nicotine concentrations continue to damage the bronchial epithelium and lead to the loss of expression of some genes that maintain the normal function of bronchial epithelial cells, which eventually induces lung cancer pathogenesis. In prior immunohistochemistry assays, we confirmed that VPS33B protein is highly expressed in human bronchial epithelial cells. We speculated that long-term smoking would cause the respiratory tract accumulation of harmful substances found in tobacco, including nicotine, which would destroy bronchial epithelial cells and inhibit VPS33B expression. In the current study, due to the lack of a smoking-induced lung cancer model, we observed the effect of nicotine on the expression of VPS33B at the cellular level. Different from the long-term impact of low nicotine concentrations in humans, we used higher nicotine concentrations to treat LUAD cells to quickly observe the impacts. The mRNA and protein levels of VPS33B were downregulated in nicotine-treated LUAD cells by stimulating PI3K/AKT/c-Jun-mediated transcription suppression. These findings demonstrate that a reduction of VPS33B is involved in the smoking-induced pathogenesis of LUAD. This suggests that patients who are regular smokers with low VPS33B expression levels should quit smoking to possibly improve their survival prognosis.
Conclusion
Together, these findings indicate that downregulated VPS33B protein is an unfavorable factor in LUAD and that VPS33B is suppressed by nicotine. VPS33B interacts with NESG1 and modulates EGFR/Ras/ERK/c-Myc/p53 and its downstream EMT signals, which thus reduces cell migration, invasion, metastasis and chemoresistance to DDP in LUAD (Fig. 7). Our study is the first to provide insights into the significance of VPS33B as an antitumor-metastasis factor in LUAD.
Figure 7.
Working model of VPS33B. Molecular mechanism involving VPS33B,NESG1,EGFR/Ras/ERK/c-Myc/p53 pathway and its downstream EMT signals in LUAD.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgements
This study was supported by national nature science fund of China (grant number 81572247), Nature science fund of Guangdong Province (grant numbers 2017A030313702, 2015A030311005), Shenzhen health system science foundation (grant number SZBC2018019) and the Supporting plan for Special Talents in Guangdong Province (grant number 2016TQ03R466), Guangzhou science and technology plan (grant number 201804010023).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2019.12.009.
Contributor Information
Weiyi Fang, Email: fangweiyi1975@163.com.
Ping Xu, Email: ping-xu@hotmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zou Y., Wei J., Xia Y., Meng F., Yuan J., Zhong Z. Targeted chemotherapy for subcutaneous and orthotopic non-small cell lung tumors with cyclic RGD-functionalized and disulfide-crosslinked polymersomal doxorubicin. Signal Transduct Target Ther. 2018;3:e32. doi: 10.1038/s41392-018-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cafarotti S., Lococo F., Froesh P., Zappa F., Andre D. Target therapy in lung cancer. Adv Exp Med Biol. 2016;893:127–136. doi: 10.1007/978-3-319-24223-1_6. [DOI] [PubMed] [Google Scholar]
- 3.Luo W., Tian P., Wang Y. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. In J Cancer. 2018;143(7):1696–1705. doi: 10.1002/ijc.31542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim I.J., Quigley D., To M.D. Rewiring of human lung cell lineage and mitotic networks in lung adenocarcinomas. Nature communications. 2013;4:e1701. doi: 10.1038/ncomms2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohrherr J., Haber M., Breitenecker K. JAK-STAT inhibition impairs K-RAS-driven lung adenocarcinoma progression. Int J Cancer. 2019;145(12):3376–3388. doi: 10.1002/ijc.32624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin X., Zuo S., Luo R. HBX-induced miR-5188 impairs FOXO1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics. 2019;9(25):7583–7598. doi: 10.7150/thno.37717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng H., Tie Y., Fang Z. Jumonji domain-containing 6 (JMJD6) identified as a potential therapeutic target in ovarian cancer. Signal Transduct Target Ther. 2019;4:e24. doi: 10.1038/s41392-019-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Yin Z., Fan J., Zhang S., Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther. 2019;4:e47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Q., Song X., Liu Z. miRomics and proteomics reveal a miR-296-3p/PRKCA/FAK/Ras/c-Myc feedback loop modulated by HDGF/DDX5/β-catenin complex in lung adenocarcinoma. Clin Cancer Res. 2017;23(20):6336–6350. doi: 10.1158/1078-0432.CCR-16-2813. [DOI] [PubMed] [Google Scholar]
- 10.Xie M., Bu Y. SKA2/FAM33A: a novel gene implicated in cell cycle, tumorigenesis, and psychiatric disorders. Genes Dis. 2018;6(1):25–30. doi: 10.1016/j.gendis.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carim L., Sumoy L., Andreu N., Estivill X., Escarceller M. Cloning, mapping and expression analysis of VPS33B, the human orthologue of rat Vps33b. Cytogenet Cell Genet. 2000;89(1–2):92–95. doi: 10.1159/000015571. [DOI] [PubMed] [Google Scholar]
- 12.Gengyo-Ando K., Kage-Nakadai E., Yoshina S. Distinct roles of the two VPS33 proteins in the endolysosomal system in Caenorhabditis elegans. Traffic. 2016;17(11):1197–1213. doi: 10.1111/tra.12430. [DOI] [PubMed] [Google Scholar]
- 13.Wartosch L., Gunesdogan U., Graham S.C., Luzio J.P. Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic. 2015;16(7):727–742. doi: 10.1111/tra.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu G.D., Salazar G., Zlatic S.A. SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol biol cell. 2009;20(4):1223–1240. doi: 10.1091/mbc.E08-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., Zhang J. Arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome: from molecular genetics to clinical features. Ital J Pediatr. 2014;40:e77. doi: 10.1186/s13052-014-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang B., Zhang G., Ye S. Characterization of a novel integrin binding protein, VPS33B, which is important for platelet activation and in vivo thrombosis and hemostasis. Circ. 2015;132(24):2334–2344. doi: 10.1161/CIRCULATIONAHA.115.018361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosales A., Mhibik M., Gissen P., Segarra O., Redecillas S., Ariceta G. Severe renal Fanconi and management strategies in arthrogryposis-renal dysfunction-cholestasis syndrome: a case report. BMC nephrol. 2018;19(1):e144. doi: 10.1186/s12882-018-0926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C., Cheng Y., Zhang X. Vacuolar protein sorting 33B is a tumor suppressor in hepatocarcinogenesis. Hepatol. 2018;68(6):2239–2253. doi: 10.1002/hep.30077. [DOI] [PubMed] [Google Scholar]
- 19.Liang Z., Liu Z., Cheng C. VPS33B interacts with NESG1 to modulate EGFR/PI3K/AKT/c-Myc/P53/miR-133a-3p signaling and induce 5-fluorouracil sensitivity in nasopharyngeal carcinoma. Cell Death Dis. 2019;10(4):e305. doi: 10.1038/s41419-019-1457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z., Li X., He X. Decreased expression of updated NESG1 in nasopharyngeal carcinoma: its potential role and preliminarily functional mechanism. Int J Cancer. 2011;128(11):2562–2571. doi: 10.1002/ijc.25595. [DOI] [PubMed] [Google Scholar]
- 21.Li Z., Yao K., Cao Y. Molecular cloning of a novel tissue-specific gene from human nasopharyngeal epithelium. Gene. 1999;237(1):235–240. doi: 10.1016/s0378-1119(99)00234-6. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z., Mai C., Yang H. Candidate tumour suppressor CCDC19 regulates miR-184 direct targeting of C-Myc thereby suppressing cell growth in non-small cell lung cancers. Journal of cellular and molecular medicine. 2014;18(8):1667–1679. doi: 10.1111/jcmm.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., Luo W., Zhou Y. Potential tumor suppressor NESG1 as an unfavorable prognosis factor in nasopharyngeal carcinoma. PloS one. 2011;6:e27887. doi: 10.1371/journal.pone.0027887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCartney D.L., Stevenson A.J., Hillary R.F. Epigenetic signatures of starting and stopping smoking. EBioMedicine. 2018;37:214–220. doi: 10.1016/j.ebiom.2018.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaiman D. Mother smoking leads to methylation anomalies on 'smoke' genes in the offspring: indelible traces of previous injuries. EBioMedicine. 2018;38:11–12. doi: 10.1016/j.ebiom.2018.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie Y., Huang C., Zhong S. Cigarette smoke extract (CSE) induces transient receptor potential ankyrin 1(TRPA1) expression via activation of HIF1alphain A549 cells. Free Radical Biol Med. 2016;99:498–507. doi: 10.1016/j.freeradbiomed.2016.07.028. [DOI] [PubMed] [Google Scholar]
- 27.Kesh K., Subramanian L., Ghosh N. Association of MMP7 -181A-->G promoter polymorphism with gastric cancer risk: influence of nicotine in differential allele-specific transcription via increased phosphorylation of cAMP-Response Element-Binding Protein (CREB) J Biol Chem. 2015;290(23):14391–14406. doi: 10.1074/jbc.M114.630129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Liu Z., Wang H., Tang Z. VPS33B negatively modulated by nicotine functions as a tumor suppressor in colorectal cancer. Int J Cancer. 2020;146(2):496–509. doi: 10.1002/ijc.32429. [DOI] [PubMed] [Google Scholar]
- 29.Nishioka T., Luo L.Y., Shen L. Nicotine increases the resistance of lung cancer cells to cisplatin through enhancing Bcl-2 stability. Br J Canc. 2014;110(7):1785–1792. doi: 10.1038/bjc.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann P.C., Sancho P., Canamero M. Nicotine promotes initiation and progression of KRAS-induced pancreatic cancer via Gata6-dependent dedifferentiation of acinar cells in mice. Gastroenterol. 2014;147(5):1119–1133. doi: 10.1053/j.gastro.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Lin H., Zhang X., Feng N. LncRNA LCPAT1 mediates smoking/particulate matter 2.5-induced cell autophagy and epithelial-mesenchymal transition in lung cancer cells via RCC2. Cell Physiol Biochem. 2018;47(3):1244–1258. doi: 10.1159/000490220. [DOI] [PubMed] [Google Scholar]
- 32.Pillai S., Trevino J., Rawal B. beta-arrestin-1 mediates nicotine-induced metastasis through E2F1 target genes that modulate epithelial-mesenchymal transition. Cancer Res. 2015;75(6):1009–1020. doi: 10.1158/0008-5472.CAN-14-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., Yu P., Zhu L., Zhao Q., Lu X., Bo S. Blockade of alpha7 nicotinic acetylcholine receptors inhibit nicotine-induced tumor growth and vimentin expression in non-small cell lung cancer through MEK/ERK signaling way. Oncol Rep. 2017;38(6):3309–3318. doi: 10.3892/or.2017.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boo H.J., Min H.Y., Jang H.J. The tobacco-specific carcinogen-operated calcium channel promotes lung tumorigenesis via IGF2 exocytosis in lung epithelial cells. Nat Commun. 2016;7:e12961. doi: 10.1038/ncomms12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song X., Shao Y., Jiang T. Radiotherapy upregulates programmed death ligand-1 through the pathways downstream of epidermal growth factor receptor in glioma. EBioMedicine. 2018;28:105–113. doi: 10.1016/j.ebiom.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia Y., Li X., Jiang T. EGFR-targeted therapy alters the tumor microenvironment in EGFR-driven lung tumors: implications for combination therapies. Int J Cancer. 2019;145(5):1432–1444. doi: 10.1002/ijc.32191. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y.N., Tsai M.F., Wu S.G. Acquired resistance to EGFR tyrosine kinase inhibitors is mediated by the reactivation of STC2/JUN/AXL signaling in lung cancer. Int J Cancer. 2019;145(6):1609–1624. doi: 10.1002/ijc.32487. [DOI] [PubMed] [Google Scholar]
- 38.Piyathilake C.J., Frost A.R., Manne U. Differential expression of growth factors in squamous cell carcinoma and precancerous lesions of the lung. Clin Cancer Res. 2002;8(3):734–744. [PubMed] [Google Scholar]
- 39.Pirazzoli V., Nebhan C., Song X. Acquired resistance of EGFR-mutant lung adenocarcinomas to afatinib plus cetuximab is associated with activation of mTORC1. Cell Rep. 2014;7(4):999–1008. doi: 10.1016/j.celrep.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezanejad Bardaji H., Asadi M.H., Yaghoobi M.M. Long noncoding RNA VIM-AS1 promotes colorectal cancer progression and metastasis by inducing EMT. Eur J Cell Biol. 2018;97(4):279–288. doi: 10.1016/j.ejcb.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Kfoury A., Le Corf K., El Sabeh R. MyD88 in DNA repair and cancer cell resistance to genotoxic drugs. J Natl Cancer Inst. 2013;105(13):937–946. doi: 10.1093/jnci/djt120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Liu X., Lin X. Chemical compound cinobufotalin potently induces FOXO1-stimulated cisplatin sensitivity by antagonizing its binding partner MYH9. Signal Transduct Target Ther. 2019;4:e48. doi: 10.1038/s41392-019-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng X., Liu Z., Liu X. miR-296-3p negatively regulated by nicotine stimulates cytoplasmic translocation of c-Myc via MK2 to suppress chemotherapy resistance. Mol Ther. 2018;26(4):1066–1081. doi: 10.1016/j.ymthe.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo P., Zhang C., Liao F. Transcriptional positive cofactor 4 promotes breast cancer proliferation and metastasis through c-Myc mediated Warburg effect. Cell Commun Signal. 2019;17(1):e36. doi: 10.1186/s12964-019-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S.H., Shen G.N., Jung Y.S. Antitumor effect of novel small chemical inhibitors of Snail-p53 binding in K-Ras-mutated cancer cells. Oncogene. 2010;29(32):4576–4587. doi: 10.1038/onc.2010.208. [DOI] [PubMed] [Google Scholar]
- 46.Zinatizadeh M.R., Momeni S.A., Zarandi K.P. The role and function of Ras-association domain family in cancer: a review. Genes Dis. 2019;6(4):378–384. doi: 10.1016/j.gendis.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mostafa S., Pakvasa M., Coalson E. The wonders of BMP9: from mesenchymal stem cell differentiation, angiogenesis, neurogenesis, tumorigenesis, and metabolism to regenerative medicine. Genes Dis. 2019;6(3):201–223. doi: 10.1016/j.gendis.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Jiang Q., Liu X. Cinobufotalin powerfully reversed EBV-miR-BART22-induced cisplatin resistance via stimulating MAP2K4 to antagonize non-muscle myosin heavy chain IIA/glycogen synthase 3β/β-catenin signaling pathway. EBioMedicine. 2019;48:386–404. doi: 10.1016/j.ebiom.2019.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z., Qi F., Wang Y. Cancer mortality attributable to cigarette smoking in 2005, 2010 and 2015 in Qingdao, China. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0204221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matta S.G., Balfour D.J., Benowitz N.L. Guidelines on nicotine dose selection for in vivo research. Psychopharmacol (Berl) 2007;190(3):269–319. doi: 10.1007/s00213-006-0441-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.