Abstract
Background:
Several studies have revealed that chromatin modifications lead to activation or repression of multiple genes including oncogenes and tumor suppressor genes. Inactivation mutation in EZH2 gene would result in activation of oncogenes. The aim of this case-control study was to identify mutations in the EZH2 gene, to study their prevalence among Jordanian patients with colorectal adenoma and to determine how these mutations could be related to colorectal cancer (CRC) progression.
Methods:
EZH2 gene sequencing was done by Sanger method for 100 DNA samples, extracted from blood of 50 patients, and 50 controls. Sequencing results were analyzed by Chromaspro and mutational effects were predicted by Mutation Taster bioinformatics tool.
Results:
Four variants were identified in Jordanian patients with adenoma; Two novel variantsc.1941T>A and c.2201G>C and two reported variants, g.73038C>T and g.75508A>C. g.73038C>T is the most common germline variant identified in this study. A significant association between the presence of c.2201G>C mutation and colorectal adenoma was found (p value < 0.05).
Conclusion:
The present study identified several variants in EZH2 gene among Jordanians with colorectal adenoma.
Key Words: EZH2, colorectal polyps, mutation, DNA methylation
Introduction
According to the most recent data extracted from Jordan’s Cancer Registry (JCR), colorectal cancer (CRC) is the most common cancer among men and the second most common cancer among women (Khader et al., 2018). JCR data also indicates that CRC is the second most common cause of cancer related death. Global estimates predict a 4% increase in cancer incidence in underdeveloped countries, including Jordan, for the next 10 years (Jemal et al., 2010). The above estimates indicate that CRC is and will remain a significant cause of patient morbidity and mortality in Jordan. It is thus of significance to understand the environmental and/or genetic factors that increase cancer risk in Jordan and worldwide.
Accumulating evidence over the last few decades demonstrates that carcinogenesis is a multistep process where multiple consecutive genetic alterations accumulate over time eventually leading to the cancer phenotype (Winawer, 1999). Perhaps the best example of the above hypothesis is displayed in our current understanding of the pathogenesis of CRC. Indeed, in CRC it is currently established that most tumors start from precursor pre-neoplastic lesions histologically known as polyps. Polyps are clumps of epithelial cells that result from the overgrowth of tissues along the lining of the colon or rectum (Zhou et al., 2014). The vast majority of polyps are benign; however, some can transform into cancer and acquire the ability to invade neighboring tissues and eventually metastasize to distant organs (Triantafillidis et al., 2009). There are multiple types of colorectal polyps; the most common are adenomatous polyps which carry the highest transformation potential. Other types of polyps include inflammatory, serrated and hamartomata’s polyps (Padmanabhan et al., 2018).
Given the above discussion, it is evident that the change of colon tissue into polyps and its later transformation into cancer provides a unique opportunity to study the multiple steps that accompany malignant transformation at the molecular level. One of the earliest molecular events that precede polyposis involves inactivating mutations of the tumor suppressor gene Adenamatous Polyposis Coli (APC) (Baldin et al., 2015). In the course of the transformation of polyps into CRC, mutations in APC are followed by additional mutations in oncogenes or tumor suppressor genes like K-Ras or P53 (Goldstein, 2006). There are multiple factors that trigger the above mutations and the eventual transformation of polyps into cancer; these factors could be environmental like obesity, smoking and inflammatory bowel disease; or genetic like chromosomal and microsatellite instability, CpG island hypo or hypermethylation, and altered histone modification (Bedeir and Krasinskas, 2011). However, this remains an active area of investigation especially since the identification of novel genetic alterations and/or predisposing environmental factors is believed to aid in identifying individuals at a higher risk of transformation who would benefit the most from closer surveillance and/or life style modifications (Ernst et al., 2010; Tatton-Brown et al., 2011).
EZH2 is a gene that encodes for the catalytic subunit of the polycomb repressive complex 2 (PRC2). This complex is a histone H3 lysine 27 (H3K27) methyltransferase. The activity of this complex places chromatin in a transcriptionally inactive conformation eventually leading to reduced pluripotency and cellular senescence. Given the above role of EZH2 in mediating gene silencing and transcriptional repression, it is not surprising that mutations in EZH2 were reported in 5% to 10% of patients with Myelodysplastic Syndrome (MDS) and 20% of the patients with Chronic Myelomonocytic Leukemia (CMML)(Bhatia et al., 2018; Wang et al., 2018). Intriguingly, the majority of these mutations are inactivating frameshift, nonsense or missense mutations. Moreover, several studies reported that mutations in EZH2 are associated with a worse prognosis in patients with MDS independent of other more well-established prognostic factors (Morin et al., 2010).
Given that (a) changes in gene methylation could increase the risk of polyposis and/or increase the risk of transformation of polyps into CRC and (b) the fact that EZH2 plays a well-established role in mediating epigenetic changes including changes in methylation (Fitzgerald et al., 2015), we hypothesized that germ line mutations in the coding region of EZH2 are associated with an increased risk of polyposis including progression into CRC. This hypothesis was tested through mutational analysis of EZH2 gene in patients with a confirmed diagnosis of colorectal adenoma that was followed up with a mandatory biopsy after one year for transformation into CRC.
Materials and Methods
Subjects description, sample collection and extraction of genomic DNA
Subjects recruitment took place at the Endoscopy unit of the Gastroenterology clinic of King Abdullah University Hospital (KAUH), a tertiary hospital affiliated with Jordan University of Science and Technology (JUST), between January 2016 and February 2017. Prior to subject recruitment, the study was approved by the appropriate Institutional Review Board (IRB) of KAUH and JUST. Subjects recruited to participate in the study were patients scheduled to have an endoscopy for suspicion of colorectal cancer. A biopsy was procured of polyp-like lesions. Histopathological examination of the lesions by a board-certified pathologist confirmed the presence of colorectal adenoma. Subjects that had tubular, villous or tubulovillous adenoma were asked to participate in the study. Other inclusion criteria were the presence of 10 or more polyps as indicated by the endoscopic examination, the absence of previous history of prior surgical removal of any polyp or polyp like lesion and the absence of prior diagnosis of irritable bowel syndrome or Crohn’s disease. The objectives of the study where then verbally explained to the subjects including the presence of a mandatory follow up endoscopy following one year of the diagnosis. Subjects who were willing to participate in the study were then asked to sign an informed consent. Relevant clinical and anthropometric data was then collected from the patients’ records by a clinical research coordinator who entered data into a data-collection sheet customized for the study. Subjects who performed an endoscopy that showed the absence of any polyp or polyp-like lesion were recruited as controls. Whole blood samples (5 mls in volume), were collected in EDTA tubes (AFCO, Amman, Jordan) from all the study participants (cases and controls) and transferred on ice to Princess Haya Biotechnology Center located at KAUH. Blood samples were then directly used for the extraction of genomic DNA.
Genomic DNA was isolated from whole blood samples using Qiagen ® genomic DNA purification kit (QIAGEN, USA) according to the instructions of the manufacturer. The quality of the purified DNA was assessed by running the DNA in a 1% agarose gel stained with ethidium bromide, and visualizing the DNA under UV light. Genomic DNA was considered of sufficient quality if it appeared under UV light as a single high molecular weight band (>10 Kb). DNA concentration was then determined using a Nano-Drop 2000 (Thermo Scientific, USA). DNA samples were stored at -80°C, until further processing.
Mutational analysis of EZH2 gene
Mutational analysis was performed on six exons of EZH2 gene. A schematic representation of the anatomy of EZH2 gene is shown in Figure 1. First, a DNA fragment that contains the entire coding region in addition to the exon-intron boundary of each respective exon was amplified by Polymerase Chain Reaction (PCR). The primers were designed using the Primer3 plus software (available at http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The sequence of all primers used for amplifying DNA fragments of each respective exon are listed in Table 1. PCR was carried out in a final reaction volume of 25µls containing 12.5µls of a commercially available master mix (iNtRON, Burlington, USA) that contains (0.05U/µl Taq DNA polymerase, reaction buffer, 4mM MgCl2, 0.4mM of each dNTP). The final reaction-mix also contained 2 µls of each of the forward and reverse primers, 7.5 µl of nuclease free water and 1 µl of the genomic DNA template. Five microliters from each PCR product were then subjected to gel electrophoresis in a 2% agarose gel stained with ethidium bromide. The electrophoresis was run at 150 volts for 30 minutes and the resulting DNA bands were visualized under UV light using a Doc™XR system.
Figure 1.
Schematic Representation of DesignAed EZH2 Primers. EZH2 5' untranslated region, 6 exons and exons-intron boundaries were amplified using 6 primer sets
Table 1.
List of Specific PCR primers for EZH2 Gene
| Exon | Forward sequence | Reverse sequence | Product size |
|---|---|---|---|
| 7 | TGGGTAGAGAAAATGAAAGATCAAAA | AGCACTCTCCAAGCTGCTTT | 947 |
| 9 | CAGCTTTTCATGCAACACCCA | CCTTCTGCAGGTGCCATTCA | 714 |
| 10 | TCGTAGGAGGGAGCAAAGGA | TTGAGGCTTCAGCACCACTC | 822 |
| 16 | CACCACCAAAAGGTTTTCTGT | ATCAGTGCCTTACCTCTCCAC | 350 |
| 18 | AGGCAAACCCTGAAGAACTG | TTCAAGTCCATCATCACAGGA | 250 |
| 20 | TGCACCCACTATCTTCAGCA | TGTTGAGCAGTCCTGAAAGC | 747 |
Prior to running sequencing reaction, the PCR products representing each respective exon were purified from impurities using an EZ spin PCR purification kit (iNtRON, Burlington, USA) according to the instructions of the manufacturer. The DNA was eluted in a final elution volume of 20 µls. The purified PCR products were then subjected to DNA sequencing using one primer and BigDye R-R reagent. The reaction mixture consisted of 4µls of the sequencing buffer, 4µls of nuclease free water, 1µl of R-R, 1µl of the sequencing primer (either forward or reverse primer) and 4µls of the purified PCR product.
Before loading into genetic analyzer, the excess of (Dye Deoxy™) terminator, primer dimers and other impurities were removed from the DNA sequencing reaction using Zymo Research clean up kit (Zymo Research, CA, USA). The cleaning procedure was performed according to the instructions of the manufacturer. Sequencing of cleaned cycle-sequenced products was carried out on an ABI prism 3130xL Genetic analyzer (Applied Bio systems, USA) at the Genomics Sequencing Laboratory of PHBC. Sequence analysis of each exon were ran using Chromas Pro software (http://technelysium.com.au/wp/chromas/).
Bioinformatics and Computational Analysis of Variants
Several bioinformatics tools were used in this study; novel variants of EZH2 gene discovered in this investigation were mapped and named according to an integrated genome viewer (http://software.broadinstitute.org/software/igv/). The guidelines of the human genome variation society were followed in the above process (http://www.hgvs.org/mutnomen/). The Mutation Taster software was used to predict if any genetic variant is likely to cause disease (http://www.mutationtaster.org/).
Statistical Analysis
Statistical analyses were performed using Statistical Package for Social Studies (JMP) software. Chi square was used to test for significant differences in genotype frequency of the genetic variants between patients with colorectal adenoma and their controls. A p-value less than 0.05 was used as a cut-off for significance.
Results
Characteristics of the study participants
The 50 recruited patients were 30 (60%) males and 20 (40%) females, Jordanian patients with colorectal adenoma. Also, the 50 recruited controls were 28 (56%) males and 22 (44%) females. Forty (80%) and 33 (66%) individuals from controls and patients, respectively, ages between 30 and 50 years old. 8 (16%) from controls and 12 (24%) from patients were between 51 and 70 years old. In addition, 2 (4%) controls and 5 (10%) patients were above 70. As shown in Table 2, the study subjects were age and sex matched. Various clinical parameters were compared between the study groups. For instance, histopathological classification, CRC progression and metastasis. The most prevalent histopathological type among study patients was tubular adenoma with 23(64%) subjects from the study group. Moreover, 11(22%) patients were with villous adenoma. Tubulovillous adenoma patients comprising 7(14%) of the study patients. For the progression of colorectal adenoma into colorectal cancer, 27(54%) from patients included in this study had recovered after polypectomy compared to 23(46%), who had progressed to colorectal cancer. However, no significant difference was found between CRC progression and sex among the study patients (p-value =0.642), Table 3.
Table 2.
General Characteristics of the Participants (N=100)
| Variable | Control N (%) | Patient N (%) | † P-value |
|---|---|---|---|
| Gender | 0.749 | ||
| Male | 28 (56%) | 30 (60%) | |
| Female | 22 (44%) | 20 (40%) | |
| Total | 50 | 50 | |
| Age groups | |||
| 30-50 | 40 (80%) | 33 (66%) | |
| 51-70year | 8 (16%) | 12 (24%) | 0.252 |
| >70 | 2 (4%) | 5 (10%) | |
† A p-value < 0.05 was considered significant
Table 3.
Clinical Characteristics of the Patients (N=50)
| Variable | Male N (%) |
Female N (%) |
Total N (%) |
† P-value |
|---|---|---|---|---|
| Histopathology | ||||
| Tubular Adenoma | 20 (40%) | 12 (24%) | 32 (64%) | |
| Villous Adenoma | 6 (12% ) | 5 (10%) | 11 (22%) | 0.89 |
| Tubovillous Adenoma | 4 (8%) | 3 (6%) | 7 (14%) | |
| Total | 30 | 20 | 50 | |
| CRC progression | ||||
| Yes | 13 (26%) | 10 (20%) | 23 (46%) | |
| No | 17 (34%) | 10 (20%) | 27 (54%) | 0.642 |
| Total | 30 | 20 | 50 | |
| Metastasis | ||||
| Yes | 10 (20%) | 9 (18%) | 19 (38%) | |
| No | 20 (40%) | 11 (22%) | 31 (62%) | 0.405 |
| Total | 30 | 20 | 50 |
† A p-value < 0.05 was considered significant
Mutational analyses reveal two novel germline mutations among patients with colorectal adenoma
Four variants were identified in Jordanian patients with adenoma; two of them are novel. As shown in Table 4, c.1941T>A, g.73038C>T, g.75508A>c and c.2201G>C have been found among the study subjects. g.73038C>T is the most common germline variant identified in this study, found in 25(50%) subjects. As shown in Table 8, two variants were found to be disease predisposing, c.1941T>A and c.2201G>C. g.75508A>C and g.73038C>T were polymorphisms.
Table 4.
Genotype and Allele Frequency of Germline Mutations Found in Jordanian Patients with Colorectal Adenoma (N=50)
| Variant identified | Nucleotide (wt/vt)** |
Genotype frequency (wt/wt)* (wt/vt)** (vt/vt)*** |
Allele frequency (wt:vt)** N (%) |
|---|---|---|---|
| c.1941T>A | T:A | 34 (68%) 16(32%) 0(0%) | T: 84 (84%) A:16 (16%) |
| g.73038C>T | C:T | 25 (50%) 23(46%) 2(4%) | |
| g.75508A>C | A:C | 35 (70%) 11(22%) 4(8%) | A:81 (81%) C:19 (19%) |
| c.2201G>C | G>C | 39 (78%) 11(22%) 0(0%) | G:89 (89%) C:11 (11%) |
*, (Wild type/ wild type); **, (wild type/variant); ***, (variant type/ variant type)
Table 8.
Characteristic of Germline Mutations Found in Jordanian Patients with Colorectal Adenoma (N=50)
| Mutation Type and AA. Change | dbSNPID (rs) | Mutation Taster prediction (score) | |
|---|---|---|---|
| c.1941T>A | Non-sense (C647*) | Novel | Disease predisposing (6.0) |
| Polymorphism | |||
| g.73038C>T | - | rs2072407 | |
| g.75508A>C | - | rs10268879 | Polymorphism |
| c.2201G>C | Missense (S734T) | Novel | Disease predisposing (58) |
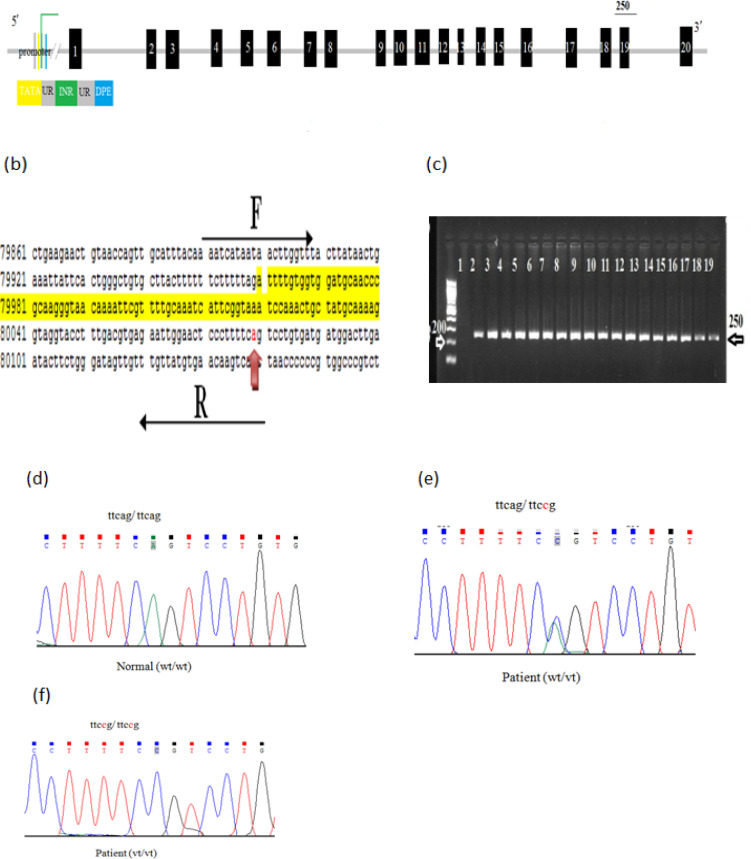
Exon 16 of EZH2 gene has been amplified (yielded a 350 bp PCR products). Moreover, two variants have been found in this exon; a novel variant c.1941T>A was found in 16 (32%) patients as a heterozygous pattern which is non-sense mutation instead of substituting one amino acid for another, however, the altered DNA sequence prematurely signals the cell to stop building a protein which may be abrogates methyltransferase activity as the Mutation Taster software predicted (Figure 3).
Figure 3.
The Mutational Analysis of Exon 16 of EZH2 Gene.(a) Schematic representation of EZH2 showing exon 16. (b) The reference sequence of exon 16 obtained from Gene Bank (NG_032043) and the highlighted sequence is for exon 16. The PCR was run using the forward (F) and reverse (R) primers (sequence above and below the block arrows). The expected PCR products are 350bp. The red arrow shows the position of g.73038c>t variation and the green arrow shows the position of c.1941T>A variation. (c) 2% gel electrophoresis for exon 16. The exon was successfully amplified and 350bp bands were visualized by 2% agarose gel. The size was compared to 100 ladder (the first lane) and negative control (lane1) was used. Lanes from 2-6 are representative patients samples and lanes from 7-18 are representative samples from controls. (d) Representative partial chromatogram for exon16 normal control showing wild type sequence (wt/wt), (e) heterozygous g.73038C>T (wt/vt), and (f) homozygous (vt/vt) of g.73038C>t, respectively. (g) Representative partial chromatogram for exon16 from patients illustrating wild type sequence (wt/wt), and (h) heterozygous c.1941T>A (wt/vt)
A SNP g.73038C>T was found in 25(50%) patients, 23(46%) of them are heterozygous and 2(4%) are homozygous (Table 8 and Figure 3). Table 8 shows that c.1941T>A is statistically associated with colorectal adenoma patients included in this study in comparison to controls (p-value 0.0051). Furthermore; we applied Sanger sequencing to identify mutations in exon 18 of EZH2 gene. Overall, we identify a reported heterozygous variant, g.75508A>C, in 11(22%) Jordanian patients with colorectal adenoma. As well, 4(8%) homozygous variants (Figure 4). Clearly, no significant association between g.75508A>C variant and colorectal adenoma patients compared to controls (p-value 0.16) (Table 6).
Figure 4.
The Mutational Analysis of Exon 18 of EZH2 Gene. (a) Schematic representation of EZH2 showing exon 18. (b) The reference sequence of exon 18 obtained from Gene Bank (NG_032043) and the highlighted sequence is for exon 18. The PCR was run using the forward (F) and reverse (R) primers (sequence above and below the block arrows). The expected PCR products are 250bp. The red arrow shows the position of g.75508A>C variation. (c) 2% gel electrophoresis for exon 18. The exon was successfully amplified and 250bp bands were visualized by 2% agarose gel. The size was compared to 100 ladders (the first lane) and negative control (lane1) was used. Lanes from 2-6 are representative patients samples and lanes from 7-19 are representative samples from controls. (d) Representative partial chromatogram for exon18 normal control showing wild type sequence (wt/wt). (e) Representative partial chromatogram for exon18 from patients illustrating heterozygous, g.75508A>C, variant (wt/vt). (f) Representative partial chromatogram for exon18 patient showing variant sequence (vt/vt)
Table 6.
Genotype and Allele Frequencies of g.75508A>C in EZH2 Gene Exon18 among Study Subjects (N=100)
| Genotypes and alleles | Control N (%) |
Patients N (%) |
†P-value |
|---|---|---|---|
| A/A | 41 (82%) | 35 (70%) | 0.61 |
| A/C | 9 (18%) | 11 (22%) | |
| C/C | 0 (0%) | 4 (8%) | |
| Total | 50 | 50 | |
| Allele A | 91 (91%) | 81 (81%) | 0.12 |
| Allele C | 9 (9%) | 19 (19%) | |
| Total | 100 | 100 |
†A p-value < 0.05 was considered significant
Finally, we amplified exon 20 using the primer set listed in Table1. A novel missense variant, c.2201G>C, was successfully identified in Jordanians with colorectal adenoma, this variant results in the substitution of one amino acid for another in the protein sequence,Serine to Threonine (S734T) (Figure 5). A significant association between the presence of c.2201G>C mutation and colorectal adenoma was found in this study (p value < 0.05) (Table 7). NCBI genome databases were used for mutational search. The variant was named according to Human Genome Variation Society guidelines (HGVS) available at (http://www.hgvs.org/mutnomen/examplesdna.html#sub).
Figure 5.
The Mutational Analysis of Exon 20 of EZH2 Gene.(a) Schematic representation of EZH2 showing exon 20. (b) The reference sequence of exon 20 obtained from Gene Bank (NG_032043) and the highlighted sequence is for exon 20. The PCR was run using the forward (F) and reverse (R) primers (sequence above and below the block arrows). The expected PCR products are 747bp. The red arrow shows the position of c.2201GG>C mutation. (c) 2% gel electrophoresis for exon 20. The exon was successfully amplified and 747 bp bands were visualized by 2% agarose gel. The size was compared to 100 ladders (the first lane) and negative control (lane1) was used. Lanes from 2-6 are representative patients samples and lanes from 7-19 are representative samples from controls. (d) Representative partial chromatogram for exon20 normal control showing wild type sequence (wt/wt). (e) Representative partial chromatogram for exon20 from patients illustrating heterozygous, c.2201G>C, variant (wt/vt). C.2201G>C is noval mutation found in Jordanian patients with colorectal adenoma
Table 7.
Genotype and Allele Frequencies of c.2201G>C in EZH2 Gene exon20 among Study Subjects (N=100)
| Genotypes and alleles | Control N (%) |
Patients N (%) |
†P-value |
|---|---|---|---|
| G/G | 47 (94%) | 39 (78%) | 0.030* |
| G/C | 3 (6%) | 11 (22%) | |
| C/C | 0 (0%) | 0 (0%) | |
| Total | 50 | 50 | |
| Allele G | 97 (97) | 89 (89%) | 0.027** |
| Allele C | 3 (3%) | 11 (11%) | |
| Total | 100 | 100 |
†A p-value < 0.05 was considered significant; *Odds ratio, 4.42 and Relative Risk: 3.67; **Odds Ratio, 4 and Relative Risk 3.67.
The high prevalence of g.73038C>T in Jordanian patients with colorectal adenoma emphasized the importance of examining the association between this mutation and colorectal adenoma. Therefore, the g.73038C>T was found to be significantly associated with colorectal adenoma (p-value< 0.05). Moreover, g.73038C>T was also found to be significantly associated with CRC metastasis (p-value< 0.05) (Figure 2).
Figure 2.
The Association between g.73038C>T and Colorectal Adenoma Malignancies. Strong association between g.73038C>T and Colorectal Adenoma (p-value=0.00002).
Table 5.
Genotype and Allele Frequencies of c.1941T>A in EZH2 Gene Exon16 among Study Subjects (N=100)
| Genotypes and alleles | Control N (%) |
Patients N (%) |
†P-value |
|---|---|---|---|
| T/T | 46 (92%) | 34 (68%) | 0.0051* |
| T/A | 4 (8%) | 16 (32%) | |
| A/A | 0 (0%) | 0 (0%) | |
| Total | 50 | 50 | |
| Allele T | 96 (96%) | 84 (84%) | |
| Allele A | 4 (4%) | 16 (16%) | 0.004** |
| Total | 100 | 100 |
†A p-value < 0.05 was considered significant; *Odds ratio, 5.41 and Relative Risk 4; **Odds Ratio, 4.57 and Relative Risk 4.
Discussion
The aim of this study was to create a better understanding of EZH2 and colorectal carcinogenesis in colorectal adenoma Jordanian patients by performing a mutational analysis of six exons of the EZH2 gene using Sanger sequencing. Also, we focused on how these mutations could be related to CRC progression. The data reported here describe, for the first time in Jordan four variants; two reported variants, g.73038C>T, g.75508A>C, and two novel variants, c.1941T>A and c.2201G>C. The variant c.1941T>A was found in exon 16 in 16 colorectal adenoma patients and in 4 normal controls. It is considered as disease-predisposing changing the amino acid sequence (p.C647*) and affects the protein features that may result in non-functional protein. The c.2201G>C novel variant was found as heterozygous in 11 patients and in 3 normal controls. It’s a missense variant (p.S734T) predicted to change the amino acid sequence without changing the function of the protein. The variant c.1941T>A was identified in this study as a heterozygous variant in 16 colorectal adenoma patients. In addition, a reported variant, g.73038C>T (rs2072407), was also found in the intronic region in the study participants as a homozygous in 2 patients and as heterozygous in 23 patients.
Control of gene expression is exerted at a number of different levels, one of which is the accessibility of genes and their controlling elements to the transcriptional machinery. Accessibility is dictated broadly by the degree of chromatin compaction, which is influenced in part by polycomb group proteins. EZH2 is a small subunit of the polycomb repressive complex 2(PRC2) enzyme, which is genetic silencer that is vital to cell proliferation and differentiation. Specifically, PRC2 trimethyl H3K27 subunit. PRC2 has precursor subunits (EZH2, SUZ12 and EED with co-factors JARID2, AEBP2 and RbAp46/48). So, the EZH2 is one subunit of PRC2 complex that pressing catalytic activity that controls genes methylation. Our study was limited to the genotyping of EZH2 as part of a protein consisting of many other units that may affect the methylation process. So, further studies are needed to examine the effect of other PRC2 components in colorectal adenoma progression. Concomitantly, it has been reported that adenoma-carcinoma sequence includes sequential mutation and deletion of various genes. Thus, knowing when these mutations occur in relation to adenoma-carcinoma sequence may establish a new understanding of how to avoid their occurrence and thus may prevent tumor progression. Recent work has identified that allelic losses of chromosome 5q have been observed in colorectal adenoma patients without polyposis (Goelz et al. , 1985; Sanpaolo et al., 2016; Sasaki et al., 1989). Thus, allelic loss is an early event in colorectal tumor progression. Another study (Vogelstein et al., 1988) revealed that mutations in RAS were observed in intermediate and late adenoma. Whereas, allelic loss of chromosome 17p was observed in late adenoma carcinoma. These allelic losses induce other alterations, including tumor suppressor gene hypomethylation (Fearon and Vogelstein, 1990) which has been identified to occur early in colorectal tumorgenesis (Vogelstein et al., 1988). Further studies needed to determine if mutations reported in the present study were occurred in an early event in colorectal carcinogenesis. This can help in understanding that EZH2 mutations may be involved in genetic alterations which occur early in the progression from normal epithelial cells to adenoma or in the progression to carcinoma.
The therapeutic modulation of epigenetic marks is an area of great interest. Currently, there is no histone demethylation agents available for treatment or in clinical trials. However, several studies reported that therapies that primarily directed to DNA methylation or histone acetylation can result in modulation of EZH2 level and H3K27 methylation. As previously reported, at enzymatic level, 3-deazane planocin (DZNep) showed reduce EZH2 level and H3K27 trimethylation in vitro that subsequently lead to reducing cell proliferation in breast and prostate cancer (Tatton-Brown et al., 2011). Previous studies proved that the in vitro and In Vivo preclinical models of pharmacological inhibition of EZH2 can block proliferation and survival in a subset of cancer cell lines, according to its function as an oncogene. It has been recently reported that, tazemetostat, a potent and selective EZH2 inhibitor currently in phase II clinical trials, induces potent antiproliferative and antitumor effects in SCCOHT cell lines and xenografts deficient in both SMARCA2 and SMARCA4 genes (Chan-Penebre et al., 2017). Another study investigated the therapeutic target of EZH2 in synovial sarcoma in vitro. it has been proved that knockdown of EZH2 by shRNA or siRNA generated an inhibition of cell growth and migration of synovial cell lines (Shen et al., 2016). These data support that targeting EZH2 may be a promising therapeutic technique in the treatment of many tumors such as synovial sarcoma.
A notable limitation of the present study is the small number of samples due to the single institutional study design. Further studies including a larger number of samples are therefore required.
In conclusion, germline mutations in EZH2 gene are prevalent among colorectal adenoma patients in Jordan. Further functional studies are required to determine the impact of variants c.1941T>A, c.2201G>C and other variants on DNA methylation phenotype. If proved to be important then these variants may be used as biomarkers for early diagnosis of colorectal cancer in Jordanians. These variants potentially can be considered as early biomarkers for colorectal prognosis in Jordan and may open up new insights in colorectal adenoma treatment before progression to colorectal carcinoma. The association between EZH2 mutations and expression of tumor suppressor genes during colorectal carcinogenesis must be analyzed in vitro.
Author Contribution Statement
Amjad Mahasneh: Conceptualization; Investigation; Supervision; Project administration; Critical review of the manuscript. Faten Alnegresh: Data collection; Methodology; Investigation; Critical review of the manuscript. Mahmoud Alfagih; Investigation; Supervision, Critical review of the manuscript.
Funding Statement
This work was supported by the Deanship of Research at Jordan University of Science and Technology, Jordan (Grant number 12/2018).
Ethics approval
This study was approved by the IRB of King Abdullah University Hospital (KAUH), a tertiary hospital affiliated with Jordan University of Science and Technology (JUST)
Consent to participate
A written informed consent was obtained from each participant.
Conflicts of interest
The authors declare that they have no conflict of interest
References
- Baldin RKS, Anselmi RA, Azevedo M, et al. Interobserver variability in histological diagnosis of serrated colorectal polyps. J Coloproctology (Rio de Janeiro) 2015;35:193–7. [Google Scholar]
- Bedeir A, Krasinskas AM. Molecular diagnostics of colorectal cancer. Arch Pathol Lab Med. 2011;135:578–87. doi: 10.5858/2010-0613-RAIR.1. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Yadav A, Tiwari R, et al. Polycomb Group protein EZH2-mediated transcriptional repression of microRNA-338/-421 drives SPINK1-positive prostate cancer. bioRxiv. 2018;1 [Google Scholar]
- Chan-Penebre E, Armstrong K, Drew A, et al. Selective killing of SMARCA2-and SMARCA4-deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of EZH2: in vitro and in vivo preclinical models. Mol Cancer Ther. 2017;16:850–60. doi: 10.1158/1535-7163.MCT-16-0678. [DOI] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Fitzgerald T, Gerety S, Jones W, et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein NS. Serrated pathway and APC (conventional)-type colorectal polyps: molecular-morphologic correlations, genetic pathways, and implications for classification. Am J Clin Pathol. 2006;125:146–53. [PubMed] [Google Scholar]
- Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- Khader YS, Sharkas GF, Arkoub KH, et al. The epidemiology and trend of cancer in Jordan, 2000-2013. J Cancer Epidemiol. 2018;2018:2937067. doi: 10.1155/2018/2937067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanpaolo E, Miroballo M, Corbetta S, et al. EZH2 and ZFX oncogenes in malignant behaviour of parathyroid neoplasms. Endocr. 2016;54:55–9. doi: 10.1007/s12020-016-0892-y. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Okamoto M, Sato C, et al. Loss of constitutional heterozygosity in colorectal tumors from patients with familial polyposis coli and those with nonpolyposis colorectal carcinoma. Cancer Res. 1989;49:4402–6. [PubMed] [Google Scholar]
- Shen JK, Cote GM, Gao Y, et al. Targeting EZH2-mediated methylation of H3K27 inhibits proliferation and migration of Synovial Sarcoma in vitro. Sci Rep. 2016;6:1–10. doi: 10.1038/srep25239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Hanks S, Ruark E, et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafillidis JK, Nasioulas G, Kosmidis PA. Colorectal cancer and inflammatory bowel disease: epidemiology, risk factors, mechanisms of carcinogenesis and prevention strategies. Anticancer Res. 2009;29:2727–37. [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen SY, Colborne S, et al. Histone deacetylase inhibitors synergize with catalytic inhibitors of EZH2 to exhibit antitumor activity in small cell carcinoma of the ovary, hypercalcemic type. Mol Cancer Ther. 2018;17:2767–79. doi: 10.1158/1535-7163.MCT-18-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winawer SJ. Natural history of colorectal cancer. Am J Med. 1999;106:3–6. doi: 10.1016/s0002-9343(98)00338-6. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Du WD, Wu Q, et al. EZH2 genetic variants affect risk of gastric cancer in the Chinese Han population. Mol Carcinog. 2014;53:589–97. doi: 10.1002/mc.21871. [DOI] [PubMed] [Google Scholar]