Highlights
-
•
TMT-based proteomics was used to study and compare the muscle protein profiles of Pacific abalones between northern and southern China.
-
•
729 differential abundance proteins were identified in different regions.
-
•
Fatty acid synthase and other 3 proteins were identified as candidate biomarkers for identification of northern and southern abalone.
Keywords: Proteomics, Haliotis discus hannai, Protein biomarker, TMT
Abstract
Due to latitude, the growth cycle of abalone in southern China is significantly lower than that in the northern regions. Therefore, it often occurs merchants use southern abalone to disguise as northern abalone. This study aims to explore the differences in the muscle proteome of Pacific abalone (Haliotis discus hannai) in different regions. A total of 1,569 proteins were detected and 729 proteins were identified as differential abundance proteins (DAPs) in Haliotis discus hannai cultured in Northern (Liaoning Province) and Southern (Fujian Province) China. Bioinformatics analysis revealed and Western blot verified that fatty acid synthase, troponin I, calpain small subunit 1, and myosin light chain 6 are candidate biomarkers for abalone cultured in different regions. This study provides a deeper understanding of how to distinguish which region abalone is harvested from to improve abalone quality controls, and prevent food fraud.
1. Introduction
Pacific abalone (Haliotis discus hannai) is considered a delicacy with high nutritional value in many Asian countries, such as China, Japan, and South Korea (Shi et al., 2020). Abalone has been consumed in China for more than 2,000 years; thus, unsurprisingly, China is both the world's largest producer and consumer of abalones. The demand for abalones in China and other regions has created a huge market for abalone farming. Abalone from Dalian Province, in particular, are recognized as the best quality, having dominated the market for abalone farming in China for a long time. However, with the industrial application of hybrid technology in recent years, the abalone aquaculture industry has expanded southward. In 2017, Fujian Province produced about 123,400 tons of abalone, accounting for 83.1% of China's total abalone production. Due to the difficulty in distinguishing between north and south abalone, Dalian abalone is often counterfeited by cheap southern abalone, causing regional production to drop from 70% of the national annual production in 2004 to only 1.6% in 2017.
Proteomics refers to the characterization of the proteome, which can dynamically analyze the composition and content of proteins in samples of different origins, substances, or growth stages, including the expression, function, and modification of proteins at any stage (Aslam et al., 2017). It has been widely used in food inspection to reveal the mechanisms and factors that affect food quality and safety (Men et al., 2020). For example, researchers have used proteomics to characterize milk samples and their products (Agregán et al., 2021), and identify proteins related to quality traits of frozen mud shrimp (Solenocera melantho) (Shi et al., 2018). A study of pork proteomic changes associated with quality confirmed that tenderness (Warner Bratzler shear) was associated with latissimus dorsi (LD) muscle 6 protein (Lametsch et al., 2003). Moreover, post-mortem proteolysis of pork longissimus muscle was reported to correlate with pork meat quality traits (Hwang et al., 2005). It is well-known that the geographical distribution of food affects its quality and nutritional value, and it has become one of the indicators for product authenticity identification and quality testing, particularly for some of the more expensive ingredients. While researchers have explored the effect of the ripening chamber's geographical location on dry-cured Iberian ham's key odorants (Segura-Borrego et al., 2022) and geographic variations on the proteome of sea cucumber (Feng et al., 2020), similar studies on abalones are still limited. Among the few existing studies, Di et al. used 2-DE combined with matrix-assisted laser desorption/ionization time-of-flight-time-of-flight mass spectrometry (MALDI-TOF-TOF/MS) to analyze the differences in the expression of the proteome from three different origins of Haliotis diversicolor foot muscle to elucidate their molecular differentiation (Di et al., 2016).
In this study, a tandem mass tag (TMT)-based proteomic strategy was employed to differentiate cultured Pacific abalones of different geographic origins. Through bioinformatics analysis, FASN, TNNI1, CAPNS1 and MYL6 were found as potential biomarkers to differentiate farmed abalone between northern and southern China, and the results were preliminarily verified using the Western blot method. This study provides a deeper understanding of how to improve abalone breeding, maintain adequate quality controls, and prevent abalone fraud.
2. Materials and methods
2.1. Sample preparation
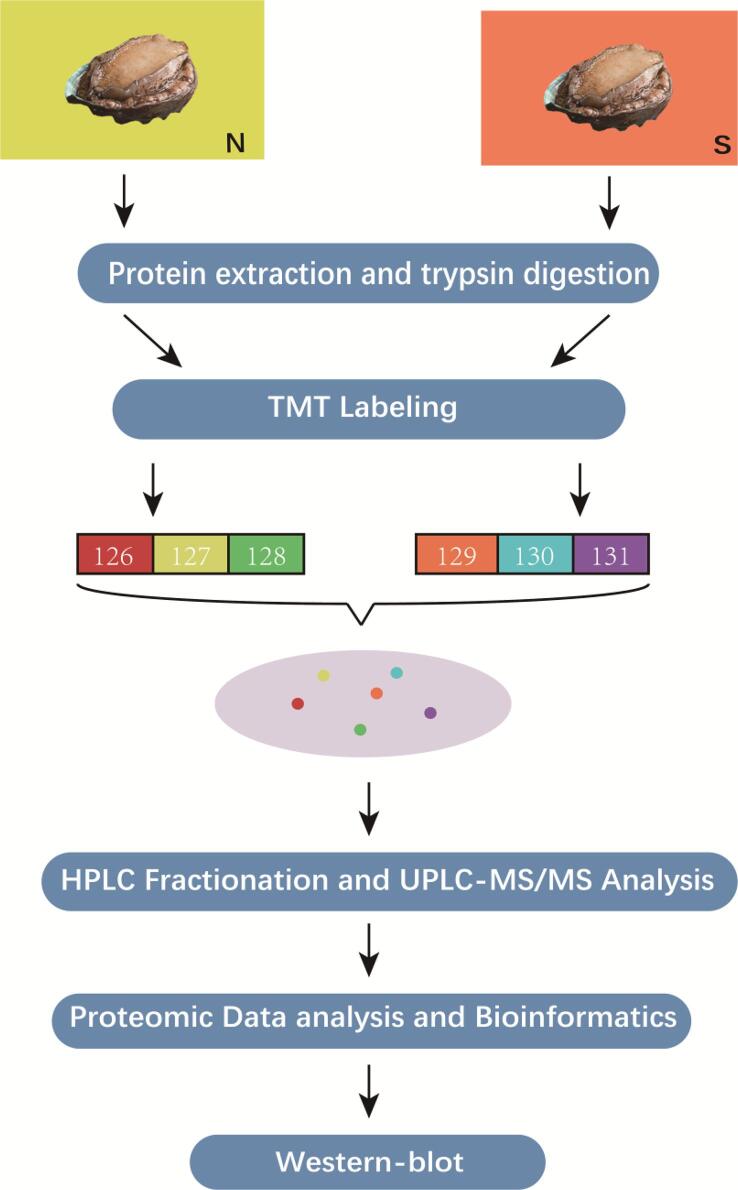
Pacific abalones (Haliotis discus hannai) (48.14 ± 2.47 g) were purchased from aquaculture companies in Fujian and Dalian, respectively. Samples were randomly divided into 2 groups: Dalian Abalone (N) and Fujian Abalone (S).
2.2. Measurement of quality traits
The method for measuring muscle mass has been described in previous studies by measuring CIE yellowing (b*), lightness (L*) and redness (a*) using a WSL-2 automatic colorimeter (Lovibond, Germany). The pH of abalone foot musculature was analyzed using a pH meter (Model Accumet) and texture profile analysis (TPA), including chewability, hardness, and elasticity, was performed using a TA.XT Plus texture analyzer (Stable Micro System, UK).
2.3. Protein extraction
A total of 3 biological replicates were used in each group (Fig. 1). Immediately after snap-frozen in liquid nitrogen, samples were added with lysis buffer (8 M urea, 1% protease inhibitors) for sonication. Cell debris was removed by centrifugation at 12,000 g for 10 min at 4 °C, and the supernatant was transferred to a new centrifuge tube and the protein concentration was determined using the BCA kit (Men et al., 2020).
Fig. 1.
Experimental design and workflow of this experiment.
2.4. Trypsin digestion and TMT labelling
The protein was taken out for equal lysis, an appropriate amount of standard protein was added, and the lysis buffer was adjusted to an equal volume. The protein lysate was mixed with a final concentration of 20% TCA, centrifuged at 4,500 g for 5 min at 4 °C, and the supernatant was discarded. After washing the pellet with acetone, TEAB was added to a final concentration of 200 mM, and after sonication, 1:50 trypsin was added overnight for enzymatic digestion. Dithiothreitol (DTT) was added to a final concentration of 5 mM, incubated at 56 °C for 30 min, then iodoacetamide (IAA) was added to a final concentration of 11 mM and incubated at room temperature for 15 min in the dark. According to the kit manufacturer's protocol (126 N, 127C, and 128 N for Group N and 129C, 130 N, and 131C for Group S; Thermo Fisher Scientific, USA), they were then incubated at 37 °C for 3 h and dried in speed-vac.
2.5. HPLC fractionation and LC-MS/MS analysis
The TMT-labeled mixture was separated by high performance reversed-phase high performance liquid chromatography using an Agilent 300Extend C18 column (C18, 5 μm, 4.6 × 250 mm). Mobile Phase A was 0.1% formic acid and 2% acetonitrile in water; mobile phase B was 0.1% formic acid and 90% acetonitrile in water. Smooth gradient: 0–26 min, 6%∼25% B; 26–34 min, 25%∼35% B; 34–37 min, 35%∼80% B; 37–40 min, 80% B, flow rate maintained at 500 nL/min. The 60 components were separated in 60 min using an 8–32% acetonitrile gradient. Peptides were separated by UHPLC, ionized by injecting an NSI ion source, and analyzed by Q Exactive™ HF-X mass spectrometry. The scan range is 400–1600 m/z with a mass resolution of 120,000. The secondary mass spectrometer scan range was set to a threshold of 100 m/z, and the secondary scan resolution was set to 15,000 (Yang et al., 2022).
2.6. Database search
Maxquant search engine was used for database searches, full trypsin specificity was required and tolerance was set to 4 missing cleavages. Tandem mass spectra were searched against the UniProt database. The mass tolerance for precursor ions was set at 20 ppm in the first round of search, and the mass tolerance was set at 0.02 Da for fragment. For protein identification, data was filtered with a false discovery rate (FDR) of < 1% and at least one matched unique peptide (Shi et al., 2018).
2.7. Bioinformatics analysis
The Proteome Annotated Gene Selection (GO) was obtained from the UniProt GOA database (https://www.ebi.ac.uk/GOA/). Functional descriptions of protein domains can be found in the InterPro Domain Database (https://www.ebi.ac.uk/interpro/). Wolfpsort predicts subcellular localization and soft cello predicts subcellular localization in prokaryotic species. Information was analyzed from UniProtKB/Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ithology (GO) to analyze the functional enrichment of the identified proteins. Heatmap was produced using R package heatmap.
2.8. Western blot analysis
Detailed protocol for western blot analysis can be found in a previous study (Wang et al., 2019). The antibodies of FASN and MYL6 were purchased from Thermo Fisher (USA), and the antibodies of TNNI1 and CAPNS1 were purchased from abcam (UK). GAPDH was used as a positive control in this study.
3. Results
3.1. Quality traits of abalone muscle
Muscle pH, color, and texture are the main indicators of abalone quality. The muscle mass characteristics of the studied two groups of abalone (N = north, S = south) are shown in Table 1. Compared with the N group, the hardness and chewing values of the S group increased while the pH, L*, a*, b* and elasticity decreased. The L* and b* scores of S muscle were significantly lower relative to N, and there was a significant difference between N and S muscle (P < 0.05).
Table 1.
Quality traits of abalone muscle in north (N) and south (S) groups.
| Quality parameters | S | N |
|---|---|---|
| pH | 6.56 ± 0.05a | 6.59 ± 0.02a |
| Lightness (L*) | 78.56 ± 0.99a | 81.40 ± 1.01c |
| Redness (a*) | −0.05 ± 0.20a | 1.10 ± 1.11a |
| Yellowness (b*) | 6.62 ± 0.82a | 8.91 ± 0.88c |
| Hardness (g) | 7.33 ± 1.81a | 6.34 ± 1.93a |
| Elasticity (mm) | 0.78 ± 0.90a | 0.80 ± 0.09a |
| Chewiness (mJ) | 2.72 ± 1.13a | 2.16 ± 0.60a |
Data are reported as means ± SD (n = 6). Different superscripts in the same row.
indicate significant difference (P < 0.05).
3.2. Identification of proteins from quantitative proteomics analysis
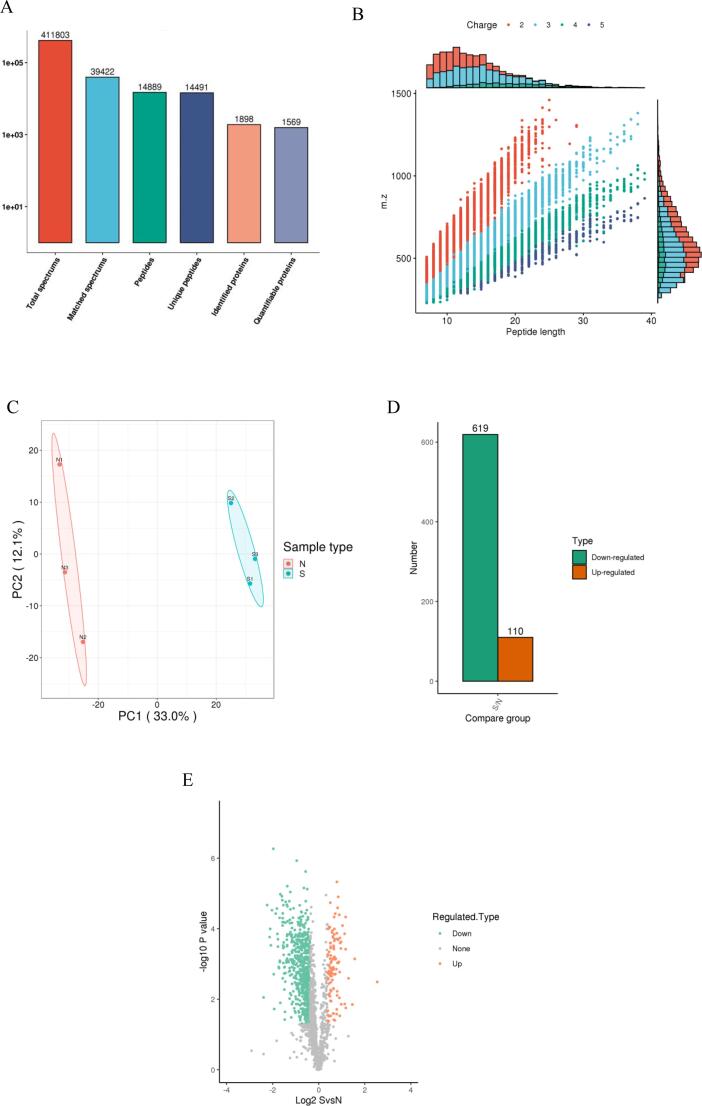
The total numbers of spectra, matching spectra, peptides, unique peptides, identified proteins, and quantitative proteins were 411,803, 39,422, 14,889, 14,491, 1,898 and 1,569, respectively (Fig. 2A). The coverage of the protein sequence identified according to the peptide length and the number of charges is presented in Fig. 2B and Supplementary table S1. Among all proteins identified, most correspond to more than two polypeptides (Fig. S1A). In addition, those proteins with high sequence coverage were identified. Most proteins had<30% sequence coverage: 47.91% had below 20% sequence coverage, and 25.37% had below 10% sequence coverage (Fig. S1B).
Fig. 2.
Results of proteome analysis. (A) Basic information for protein identification. (B) Length distribution of identified peptides. (C) Score plot of principal component analysis (PCA) of Dalian abalone (N) and Fujian abalone (S). (D) Differential protein statistics chart. (E) Differential protein volcano map.
3.3. Sample repeatability inspection
The mass spectrum of abalone muscle tissue was further processed by baseline correction, peak extraction, and peak normalization, and the comparison matrix of each mass spectrum was obtained. Principal Component Analysis (PCA) was applied to distinguish abalones produced in different regions and to detect biomarkers contributing to their separation. As shown in Fig. 2C, abalone muscle tissues from different sources were well-separated in PCA score plot, indicating that there are significant different.
3.4. Analysis of differential abundance proteins
Differential abundance protein (DAP) is defined as fold change (FC) greater than 1.3 or FC < 0.77 with p-value < 0.05. A total of 729 DAPs were identified by S/N comparison, of which 110 were up-regulated and 619 were down-regulated (Fig. 2D). All DAPs are listed in Supplementary table S2 and shown as a heatmap in Fig. S2. The volcano plots for DAPs are presented in Fig. 2E.
3.5. Functional classification and enrichment analysis of DAPs
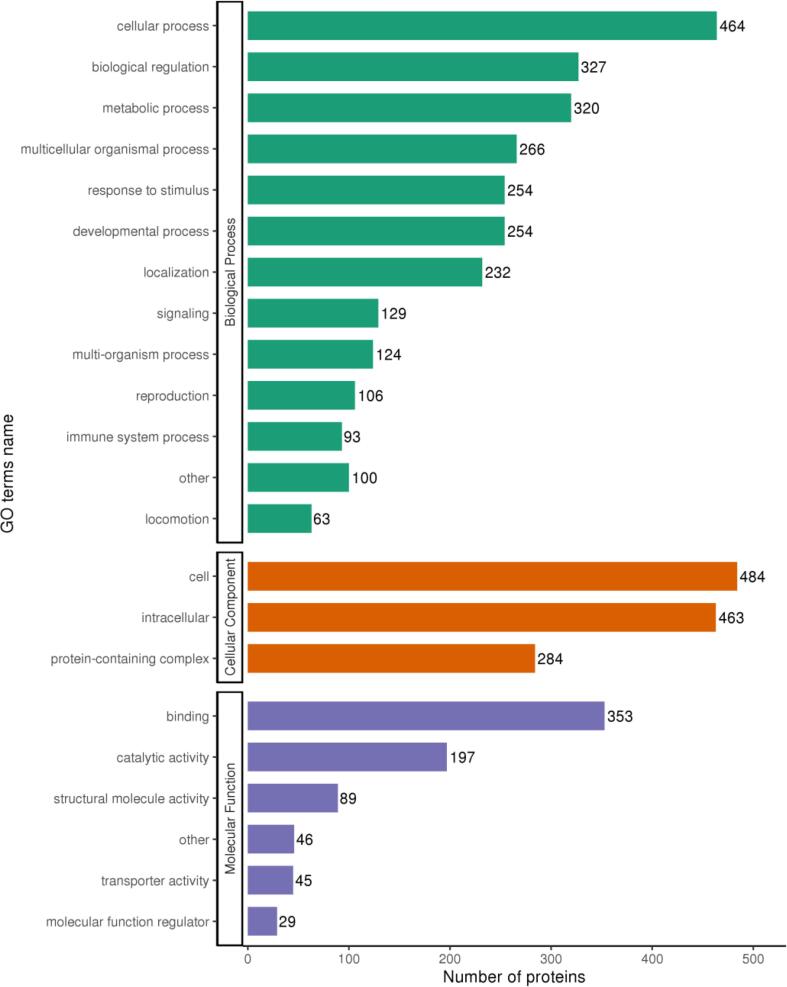
GO annotations were used to describe the distinct properties of 22 differentially expressed proteins grouped into three broad categories: biological process (BP), cellular component (CC), and molecular function (MF), as shown in Fig. 3. Analysis of biological processes revealed that these proteins were mainly involved in cellular processes and metabolic regulation, and the analysis of cellular components showed that these proteins were mainly distributed in the intercellular space. In addition, molecular function analysis showed that these proteins were mainly related to binding, catalytic activity, and structural molecular activities. Increased DAP regulation in the S/N group was significantly enriched in 20 KEGG pathways, with the highest accumulation found in cardiomyopathy (Fig. S3A); down-regulated DAPs had significantly enriched ribosomal signaling pathways (Fig. S3B), all of which were enriched in KEGG. The signaling and binding proteins are listed in Supplementary table S3.
Fig. 3.
Gene ontology (GO) categorize of the DAPs in the Dalian abalone (N) and Fujian abalone (S).
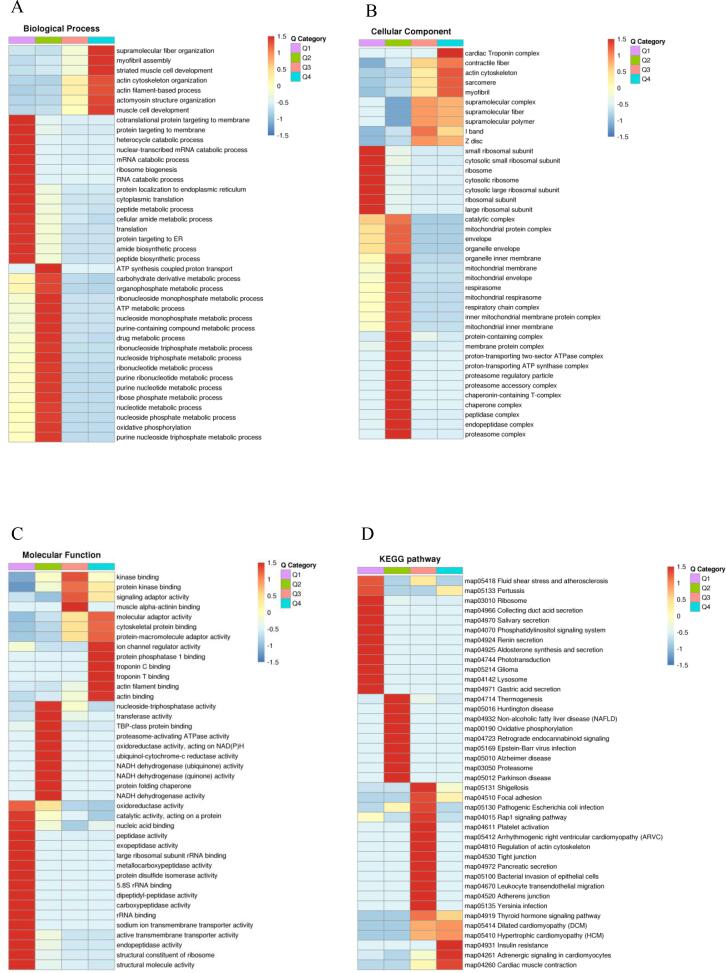
3.6. Cluster analysis of protein expression patterns
After GO classification and KEGG pathway enrichment analysis of DAPs, the correlation of DAP cluster analysis results for the control group were determined (see Fig. 4). According to the differential expression fold, the heatmap can be grouped into four horizontal clusters, named Q1 to Q4; the color blocks correspond to different Q groups representing the degree of enrichment, and the strong and weak enrichment are represented by red and blue, respectively. In the combined cluster, amide biosynthesis, peptide metabolism, myofibril assembly, etc. were strongly enriched in the BP category (Fig. 4A); ribosomes, mitochondria, and protein-containing complexes were strongly enriched in the CC category (Fig. 4B); transporter activity, ion channel modulator activity, and muscle α-actin activity were strongly enriched in the MF category (Fig. 4C). Regulation of ribosomes, oxidative phosphorylation, actin cytoskeleton, etc., all belonged to the category of strong enrichment of KEGG (Fig. 4D).
Fig. 4.
The functions in different Q groups are drawn into heatmaps through hierarchical clustering. (A) Biological Process. (B) Cellular Component. (C) Molecular Function. (D) KEGG cluster analysis.
3.7. Validation of the DAPs using Western blot
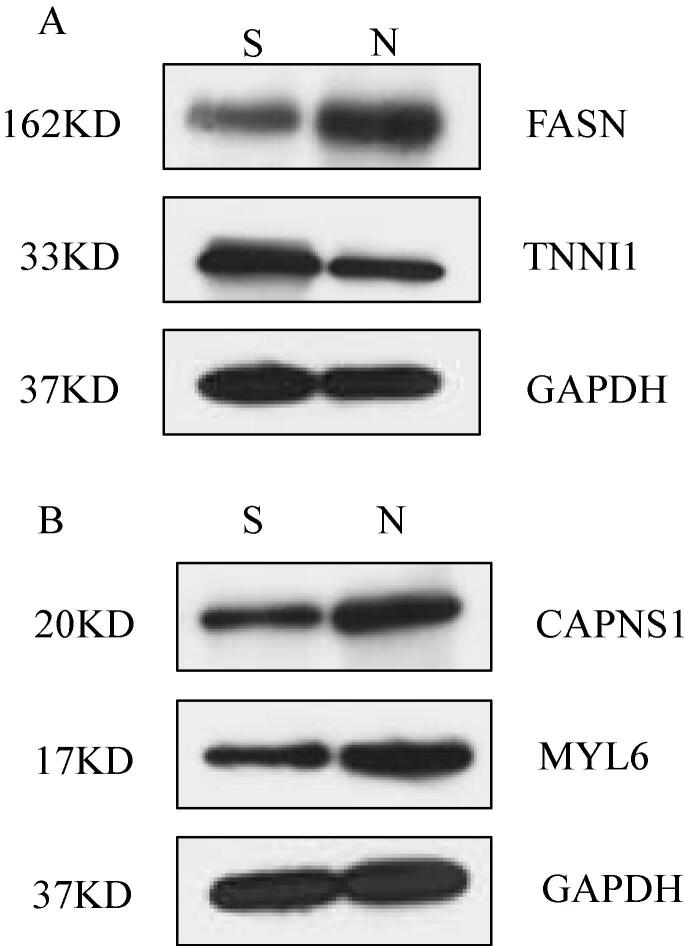
Four representative proteins were selected to further confirm their abundance using western blot, with one increased regulatory protein and three decreased proteins, as shown in Fig. 5. The expression level of TNNI1 (troponin I) in group S was higher than that in group N. The results of the western blot were consistent with the results of proteomics while the expression level of FASN (fatty acid synthase) was lower than that in group N (Fig. 5A). As shown in Fig. 5B, the expression levels of CAPNS1 (calpain small subunit 1) and MYL6 (myosin light chain 6) in group N were higher than those in group S.
Fig. 5.
Biomarkers used to characterize abalone were confirmed by Western blot analysis. (A) Expression of FASN and TNNI1: Fatty acid synthase and Troponin I subunit in the S and N groups. (B) Expression of calpain small subunit 1 and myosin light chain 6 in groups S and N, GAPDH as a positive control.
4. Discussion
Meat quality is a complex trait regulated by several proteins' coordinated activity. According to previous studies, the number, composition, and distribution of muscle fibers, intramuscular fat content, and fat composition play a decisive role in the color, tenderness, and flavor of meat (Listrat et al., 2016, Picard et al., 2012). A TMT-based quantitative proteomic method was used to compare two different types of H. discus hannai from Southern and Northern China in this study. Compared to the S group, the L* and b* of the muscles were significantly increased in the N group, which could be explained by the effect of myoglobin on the brightness of the flesh. This finding agrees with studies on meat color, showing that there are significant differences in L*, a*, and b* of lamb loin of different origins; in particular, the difference in L* and b* between different origins is greater, which may be due to the effect of myoglobin on the brightness of meat color (Calnan et al., 2016). In this study, a total of 729 differentially expressed proteins in southern and northern abalone, were detected, of which 110 were up-regulated and 619 were down-regulated. Statistical analysis revealed that these proteins are related to apoptosis, actin cytoskeleton, and thyroid hormone signaling, and are involved in various biological processes and signal transduction pathways (Huang et al., 2017).
Differential proteins upregulated in the KEGG pathway were significantly enriched in the map05414 dilated cardiomyopathy (DCM) and map05410 hypertrophic cardiomyopathy (HCM) pathways (Fig. S3A). DCM often involves mutations in genes responsible for the cytoskeleton and sarcomeres, and contractility is impaired in patients with DCM (Reichart et al., 2019). There are data suggesting that HCM alters sarcomere function, reducing the force or speed with which muscle cells contract (Ommen, 2011). The study showed that mean firmness and chewiness were higher in group S than group N and elasticity was lower in group S than group N, which is possibly related to the impaired contraction of DCM and HCM. The differential proteins downregulated by the KEGG pathway in the S/N group were significantly enriched in the map03010 (ribosome) pathway (Fig. S3B). Changes in proteins associated with ribosomal signaling pathways have been reported under cold stress in plants and animals (Fan et al., 2013, Zhou et al., 2014, Ji et al., 2020) and cold acclimation is known to trigger structural reprogramming of the ribosome proteome. Ribosome biosynthesis, the central mechanism driving the increased translation capacity of muscle cells, is important for muscle hypertrophy, implying that a ribosome-mediated reduction in the rate of protein synthesis affects muscle hypertrophy (Figueiredo et al., 2021, Wen et al., 2016).
In this study, four proteins, fatty acid synthase (FASN), troponin I (TNNI1), calpain small subunit 1 (CAPNS1), and myosin 6 light chain (MYL6), were selected as candidates for predicting the structure of Pacific abalones. The FASN gene is an important enzyme that catalyzes the biosynthesis of saturated fatty acids, which can improve meat quality and its quality characteristics by affecting fatty acid composition (Clop et al., 2003, Wood et al., 2004; Cameron & Enser, 1991). Studies have shown that FASN is mainly concentrated in down-regulated proteins, and some speculate that the quality traits of northern and southern abalone are related to fatty acid parameters. Troponin I (TnI) contractile protein is a component of striated myofilaments of the troponin complex, and its gene expression may affect the constituent fibers of the muscle, thereby affecting meat quality. The composition of the troponin I component in fast-twitch fibers of fish (Oreochromis genus) has been shown to have temperature-dependent contractile properties (Crockford et al., 1991). Following cold acclimation, the specific recruitment of TnI isoforms is important for maintaining optimal contractile function under different physiological conditions (Alderman et al., 2012). According to this study, the expression of TnI was lower in group N than in group S, which may be related to the water temperature (Yang et al., 2010). Calpain is a Ca2+-dependent intracellular cysteine protease that affects postmortem muscle proteolysis and meat tenderization (Melody et al., 2004). The CAPNS1 gene has been found to be associated with beef quality traits (Chung & Davis, 2011), which is consistent with our observations that northern abalone was significantly brighter than southern abalone, thus making CAPNS1 a potential biomarker associated with abalone color. The expression of CAPNS1 in northern abalone was higher than that in southern abalone and the average meat quality indicated a slight increase in tenderness than that seen in southern abalone, although the difference was not significant. One possible reason is that the experimental abalone is only two years old and the growth time is not long enough for the analysis. Myosin light chains are members of the calmodulin family that play key roles in the mechanoenzymatic function of myosin holoenzymes (Heissler & Sellers, 2014). Changes in the expression of MLC1 and skeletal muscle actin 6 lead to altered muscle stiffness when the animal is stressed (Li et al., 2022). and their levels correlate with meat tenderness (Anderson et al., 2012). The average hardness of northern abalone was lower than that of southern abalone while the elasticity was higher than that of southern abalone, suggesting that MYL6 may also circumlocutorily modulate the quality of abalone. The above four proteins were corroborated by Western blot and be recognized as potential biomarkers for abalone quality and geographic variation.
5. Conclusion
The present study aimed at differentiating the farmed abalones originated from southern (S) and northern (N) China using TMT-proteomics. A total of 729 differential abundance proteins (DAPs) were identified in the S/N comparison, of which 110 were up-regulated and 619 were down-regulated. After screening using bioinformatics analysis and verification via Western blot, fatty acid synthase and three other proteins were selected as candidate biomarkers for abalone farming in different regions. These findings provide a deeper understanding of the impact of different geographic farming conditions on abalone quality, providing new insights into improving abalone farming, quality controls, and other measures to prevent abalone fraud. These findings highlight the potential application of proteomics to put an end to food fraud.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was funded by Dalian’s High-level Talent Innovation Support Program (2019RQ046) and Ministry of Education Key Laboratory Open Project (KF2022003). The authors thank Jingjie PTM BioLab (Hangzhou). Co. Inc for providing technical support for proteomics.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2022.100355.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agregán R., Echegaray N., López-Pedrouso M., Kharabsheh R., Franco D., Lorenzo J.M. Proteomic Advances in Milk and Dairy Products. Molecules (Basel, Switzerland) 2021;26(13):3832. doi: 10.3390/molecules26133832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderman S.L., Klaiman J.M., Deck C.A., Gillis T.E. Effect of cold acclimation on troponin I isoform expression in striated muscle of rainbow trout. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2012;303(2):R168–R176. doi: 10.1152/ajpregu.00127.2012. [DOI] [PubMed] [Google Scholar]
- Anderson M.J., Lonergan S.M., Huff-Lonergan E. Myosin light chain 1 release from myofibrillar fraction during postmortem aging is a potential indicator of proteolysis and tenderness of beef. Meat Science. 2012;90(2):345–351. doi: 10.1016/j.meatsci.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Aslam B., Basit M., Nisar M.A., Khurshid M., Rasool M.H. Proteomics: Technologies and Their Applications. Journal of Chromatographic Science. 2017;55(2):182–196. doi: 10.1093/chromsci/bmw167. [DOI] [PubMed] [Google Scholar]
- Calnan H., Jacob R.H., Pethick D.W., Gardner G.E. Production factors influence fresh lamb longissimus colour more than muscle traits such as myoglobin concentration and pH. Meat Science. 2016;119:41–50. doi: 10.1016/j.meatsci.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Cameron N.D., Enser M.B. Fatty acid composition of lipid in Longissimus dorsi muscle of Duroc and British Landrace pigs and its relationship with eating quality. Meat Science. 1991;29(4):295–307. doi: 10.1016/0309-1740(91)90009-F. [DOI] [PubMed] [Google Scholar]
- Chung H.Y., Davis M.E. Effects of calpain genotypes on meat tenderness and carcass traits of Angus bulls. Molecular Biology Reports. 2011;38(7):4575–4581. doi: 10.1007/s11033-010-0589-x. [DOI] [PubMed] [Google Scholar]
- Clop A., Ovilo C., Perez-Enciso M., Cercos A., Tomas A., Fernandez A., Coll A., Folch J.M., Barragan C., Diaz I., Oliver M.A., Varona L., Silio L., Sanchez A., Noguera J.L. Detection of QTL affecting fatty acid composition in the pig. Mammalian Genome: Official Journal of the International Mammalian Genome Society. 2003;14(9):650–656. doi: 10.1007/s00335-002-2210-7. [DOI] [PubMed] [Google Scholar]
- Crockford T., Wommack K.E., Johnston I.A., McAndrew B.J., Mutungi G., Johnson T.P. Inter- and intra-specific variation in myosin light chain and troponin I composition in fast muscle fibres from two species of fish (genus Oreochromis) which have different temperature-dependent contractile properties. Journal of Muscle Research and Cell Motility. 1991;12(5):439–446. doi: 10.1007/BF01738328. [DOI] [PubMed] [Google Scholar]
- Di G., Miao X., Ke C., Kong X., Li H., You W. Protein changes in abalone foot muscle from three geographical populations of Haliotis diversicolor based on proteomic approach. Ecology and Evolution. 2016;6(11):3645–3657. doi: 10.1002/ece3.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Wang A., Wu Y. Comparative proteomic identification of the hemocyte response to cold stress in white shrimp, Litopenaeus vannamei. Journal of Proteomics. 2013;80:196–206. doi: 10.1016/j.jprot.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Feng J., Zhang L., Xia X., Hu W., Zhou P. Effect of geographic variation on the proteome of sea cucumber (Stichopus japonicus) Food Research International (Ottawa, Ont.) 2020;136:109498. doi: 10.1016/j.foodres.2020.109498. [DOI] [PubMed] [Google Scholar]
- Figueiredo V.C., D'Souza R.F., Van Pelt D.W., Lawrence M.M., Zeng N., Markworth J.F., Poppitt S.D., Miller B.F., Mitchell C.J., McCarthy J.J., Dupont‐Versteegden E.E., Cameron‐Smith D. Ribosome biogenesis and degradation regulate translational capacity during muscle disuse and reloading. Journal of Cachexia, Sarcopenia and Muscle. 2021;12(1):130–143. doi: 10.1002/jcsm.v12.110.1002/jcsm.12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler S.M., Sellers J.R. Myosin light chains: Teaching old dogs new tricks. Bioarchitecture. 2014;4(6):169–188. doi: 10.1080/19490992.2015.1054092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., You W., Luo X., Ke C. ITRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai. International Journal of Molecular Sciences. 2017;18(11):E2237. doi: 10.3390/ijms18112237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.H., Park B.Y., Kim J.H., Cho S.H., Lee J.M. Assessment of postmortem proteolysis by gel-based proteome analysis and its relationship to meat quality traits in pig longissimus. Meat Science. 2005;69(1):79–91. doi: 10.1016/j.meatsci.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Ji C.Y., Kim H.S., Lee C.-J., Kim S.-E., Lee H.-U., Nam S.-S., Li Q., Ma D.-f., Kwak S.-S. Comparative transcriptome profiling of tuberous roots of two sweetpotato lines with contrasting low temperature tolerance during storage. Gene. 2020;727:144244. doi: 10.1016/j.gene.2019.144244. [DOI] [PubMed] [Google Scholar]
- Lametsch R., Karlsson A., Rosenvold K., Andersen H.J., Roepstorff P., Bendixen E. Postmortem proteome changes of porcine muscle related to tenderness. Journal of Agricultural and Food Chemistry. 2003;51(24):6992–6997. doi: 10.1021/jf034083p. [DOI] [PubMed] [Google Scholar]
- Li X., Li S., Shi G., Xiong G., Shi L., Kang J., Su J., Ding A., Li X., Qiao Y.u., Liao L.i., Wang L., Wu W. Quantitative proteomics insights into gel properties changes of myofibrillar protein from Procambarus clarkii under cold stress. Food Chemistry. 2022;372:130935. doi: 10.1016/j.foodchem.2021.130935. [DOI] [PubMed] [Google Scholar]
- Listrat A., Lebret B., Louveau I., Astruc T., Bonnet M., Lefaucheur L., Picard B., Bugeon J. How Muscle Structure and Composition Influence Meat and Flesh Quality. TheScientificWorldJournal. 2016;2016:1–14. doi: 10.1155/2016/3182746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melody J.L., Lonergan S.M., Rowe L.J., Huiatt T.W., Mayes M.S., Huff-Lonergan E. Early postmortem biochemical factors influence tenderness and water-holding capacity of three porcine muscles. Journal of Animal Science. 2004;82(4):1195–1205. doi: 10.2527/2004.8241195x. [DOI] [PubMed] [Google Scholar]
- Men L., Li Y., Wang X., Li R., Zhang T., Meng X., Liu S., Gong X., Gou M. Protein biomarkers associated with frozen Japanese puffer fish (Takifugu rubripes) quality traits. Food Chemistry. 2020;327:127002. doi: 10.1016/j.foodchem.2020.127002. [DOI] [PubMed] [Google Scholar]
- Ommen S.R. Hypertrophic cardiomyopathy. Current Problems in Cardiology. 2011;36(11):409–453. doi: 10.1016/j.cpcardiol.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Picard B., Lefèvre F., Lebret B. Meat and fish flesh quality improvement with proteomic applications. Animal Frontiers. 2012;2(4):18–25. doi: 10.2527/af.2012-0058. [DOI] [Google Scholar]
- Reichart D., Magnussen C., Zeller T., Blankenberg S. Dilated cardiomyopathy: From epidemiologic to genetic phenotypes: A translational review of current literature. Journal of Internal Medicine. 2019;286(4):362–372. doi: 10.1111/joim.v286.410.1111/joim.12944. [DOI] [PubMed] [Google Scholar]
- Segura-Borrego M.P., Ríos-Reina R., Galán-Soldevilla H., Forero F.J., Venegas M., Ruiz Pérez-Cacho P., Morales M.L., Callejón R.M. Influence of the ripening chamber’s geographical location on dry-cured Iberian ham’s key odorants. Food Research International (Ottawa, Ont.) 2022;153:110977. doi: 10.1016/j.foodres.2022.110977. [DOI] [PubMed] [Google Scholar]
- Shi J., Zhang L., Lei Y., Shen H., Yu X., Luo Y. Differential proteomic analysis to identify proteins associated with quality traits of frozen mud shrimp (Solenocera melantho) using an iTRAQ-based strategy. Food Chemistry. 2018;251:25–32. doi: 10.1016/j.foodchem.2018.01.046. [DOI] [PubMed] [Google Scholar]
- Shi L., Hao G., Chen J., Ma S., Weng W. Nutritional evaluation of Japanese abalone (Haliotis discus hannai Ino) muscle: Mineral content, amino acid profile and protein digestibility. Food Research International (Ottawa, Ont.) 2020;129:108876. doi: 10.1016/j.foodres.2019.108876. [DOI] [PubMed] [Google Scholar]
- Wang D., Gou M., Hou J., Pang Y., Li Q. The role of serpin protein on the natural immune defense against pathogen infection in Lampetra japonica. Fish & Shellfish Immunology. 2019;92:196–208. doi: 10.1016/j.fsi.2019.05.062. [DOI] [PubMed] [Google Scholar]
- Wen Y., Alimov A.P., McCarthy J.J. Ribosome Biogenesis is Necessary for Skeletal Muscle Hypertrophy. Exercise and Sport Sciences Reviews. 2016;44(3):110–115. doi: 10.1249/JES.0000000000000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.D., Richardson R.I., Nute G.R., Fisher A.V., Campo M.M., Kasapidou E., Sheard P.R., Enser M. Effects of fatty acids on meat quality: A review. Meat Science. 2004;66(1):21–32. doi: 10.1016/S0309-1740(03)00022-6. [DOI] [PubMed] [Google Scholar]
- Yang H., Xu Z.Y., Lei M.G., Li F.E., Deng C.Y., Xiong Y.Z., Zuo B. Association of 3 polymorphisms in porcine troponin I genes (TNNI1 and TNNI2) with meat quality traits. Journal of Applied Genetics. 2010;51(1):51–57. doi: 10.1007/BF03195710. [DOI] [PubMed] [Google Scholar]
- Yang L.-B., Guo G., Tian Z.-Q., Zhou L.-X., Zhu L.-J., Peng J., Sun C.-Q., Huang M.-J. TMT-based quantitative proteomic analysis of the effects of novel antimicrobial peptide AMP-17 against Candida albicans. Journal of Proteomics. 2022;250:104385. doi: 10.1016/j.jprot.2021.104385. [DOI] [PubMed] [Google Scholar]
- Zhou H., Zhou M., Yang Y., Li J., Zhu L., Jiang D.…Zhuang C. RNase Z(S1) processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nature Communications. 2014;5(1) doi: 10.1038/ncomms5884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.