Abstract
Background
In insects, the chemosensory system is crucial in guiding their behaviors for survival. Plagiodera versicolora (Coleoptera: Chrysomelidae), is a worldwide leaf-eating forest pest in salicaceous trees. There is little known about the chemosensory genes in P. versicolora. Here, we conducted a transcriptome analysis of larvae heads in P. versicolora.
Results
In this study, 29 odorant binding proteins (OBPs), 6 chemosensory proteins (CSPs), 14 odorant receptors (ORs), 13 gustatory receptors (GRs), 8 ionotropic receptors (IRs) and 4 sensory neuron membrane proteins (SNMPs) were identified by transcriptome analysis. Compared to the previous antennae and foreleg transcriptome data in adults, 12 OBPs, 2 CSPs, 5 ORs, 4 IRs, and 7 GRs were newly identified in the larvae. Phylogenetic analyses were conducted and found a new candidate CO2 receptor (PverGR18) and a new sugar receptor (PverGR23) in the tree of GRs. Subsequently, the dynamic expression profiles of various genes were analyzed by quantitative real-time PCR. The results showed that PverOBP31, OBP34, OBP35, OBP38, and OBP40 were highly expressed in larvae, PverOBP33 and OBP37 were highly expressed in pupae, and PverCSP13 was highly expressed in eggs, respectively.
Conclusions
We identified a total of 74 putative chemosensory genes based on a transcriptome analysis of larvae heads in P. versicolora. This work provides new information for functional studies on the chemoreception mechanism in P. versicolora.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-09079-2.
Keywords: Plagiodera versicolora, transcriptome, odorant binding proteins, chemosensory proteins
Background
Insect chemosensory system mainly includes the olfactory system and gustatory system, which are involved in complex behaviors, such as feeding, mating, ovipositing, and avoiding dangers and enemies [1–4]. The major chemosensory proteins include odorant-binding proteins (OBPs), chemosensory proteins (CSPs), odorant receptors (ORs), gustatory receptors (GRs), ionotropic receptors (IRs), and sensory membrane proteins (SNMPs) [5–11]. In the proposed process of odorant and taste detection, molecules are first bound and transported by OBPs or/and CSPs within the sensillum lymph, then detected by ORs, GRs or/and IRs expressed at the membrane of olfactory sensory neurons. After that, electrical signals are transmitted to the central nervous system through nerve axons inducing an action potential [12–15]. Other protein families, such as SNMPs, are also critical for pheromone detection [16, 17].
The first insect OBP was a pheromone-binding protein that was discovered from an antenna extract of the giant moth Antheraea polyphemus [5]. The structure of OBPs is further stabilized by three interlocked disulfide bridges between conserved cysteines [18–20]. The classical OBP fold possesses a core of six α-helices, a member of the C-plus class of OBPs, which has a longer sequence than classical OBPs, and C-minus OBPs have a shorter conserved cysteine [21–24].
The first identified member of the CSP family, called p10, was first identified in the American cockroach Periplaneta americana [25]. Later, a similar protein, OS-D (or A10) proteins, were identified by subtractive hybridization in the antennae of Drosophila melanogaster [18]. OBPs and CSPs, belong to a class of small water-soluble proteins and are impregnated in the sensilla lymph [26, 27]. CSPs, instead, seem to form a more homogeneous group of proteins, some with only four instead of six α-helical domains [28]. A large number of OBPs and CSPs have been identified in different insect species, particularly in recent years, due to genome projects and transcriptome sequencing [29–33].
ORs and GRs are ligand-gated cation channels, and encode seven transmembrane domain proteins with an intracellular N terminus and an extracellular C terminus [34, 35]. Insect ORs are composed of two related heptahelical subunits: a divergent conventional ligand-binding OR subunit that confers odor specificity [36, 37] and a highly conserved co-receptor (Orco) subunit [38]. Unlike ORs, GRs can function independently as monomers, such as the sugar receptor [39, 40] or form obligatory heteromers of two receptors as the CO2 receptor [41]. Insect IRs act as ligand-based ion channels that are related to ionotropic glutamate receptors [42]. Similar to ORs, the antennal IRs form heteromeric complexes with one or more co-receptors (IR8a, IR25a or IR76b) to perform their functions [43, 44].
The willow leaf beetle Plagiodera versicolora (Coleoptera: Chrysomelidae), is a leaf-eating forest pest, which mainly damages salicaceous trees, such as willows (Salix) and poplars (Populus) [45, 46]. In our previous study, we obtained 111 chemosensory genes from antennae [47] and forelegs transcriptomes [48], including 41 ORs, 28 OBPs, 11 CSPs, 17 GRs, 10 IRs, and 4 SNMPs. Antennae and maxillae are the paired principal chemosensory organs located on the head of the larvae [49]. Considering that the head is the tissue where most olfactory and gustatory sensilla are located, we identified more chemosensory genes from the larvae head transcriptomes of P. versicolora. Phylogenetic analysis was conducted among these chemosensory genes and other insect species. The quantitative real-time PCR was utilized in our study to screen the expression patterns. This work could provide a basis to facilitate functional clarification of these chemosensory genes at the molecular level.
Methods
Insect rearing and tissue collection
The larvae and adults of P. versicolora were collected from Sha Lake Park in Wuhan, Hubei Province, and maintained at the laboratory following conditions at 28 ± 1 °C, 70% ± 5% relative humidity, and a 12 h: 12 h light: dark (L: D) photoperiod. Larvae and adults were fed with a fresh leaf of willows. A total of 150 third instar larvae heads were collected and the tissues were immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
RNA extraction, cDNA Library Construction and Sequencing
The total RNA of the third instar larvae heads was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instruction. RNA integrity was verified by gel electrophoresis. RNA concentration was measured on a Nanodrop ND-2000 spectrophotometer (NanoDrop products, Wilmington, DE, USA). Three biological replicates were used in the study. A total RNA of about 2 μg of each sample was used to construct the cDNA libraries. The libraries were sequenced using the Illumina Novaseq 6000 and performed at Shanghai Majorbio Bio-pharm Technology Co., Ltd. following the detailed protocol.
Transcriptome assembly and annotation
Datasets of clean reads were obtained by removing adaptor reads, ploy-N and low-quality reads, from the raw data and were further assembled into transcripts using Trinity v2.5.1 with default parameters [50]. Functional annotation of the longest transcript sequence (unigenes) was performed against six databases, non-redundant database (Nr), Pfam, Clusters of Orthologous Groups (COG), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO), respectively.
Chemosensory Gene Identification and phylogenetic tree construction
To identify putative chemosensory genes (ORs, GRs, IRs, OBPs, CSPs, and SNMPs), we search the results of non-redundant protein (Nr) annotation from our transcriptome dataset. Annotations of all retrieved sequences were confirmed by blastx on the National Center for Biotechnology Information (NCBI). The Open reading frames (ORFs) of all putative chemosensory genes were predicted by ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The transmembrane domains of ORs, IRs and GRs were predicted by TMHMM2.0 (https://services.healthtech.dtu.dk/service.php? TMHMM-2.0). Putative N-terminal signal peptides of OBPs and CSPs were predicted using SignalP 5.0 (https://services.healthtech.dtu.dk/service.php? SignalP-5.0).
Phylogenetic analyses were performed based on amino acid sequences from candidate chemosensory genes in P. versicolora and other insects. For the OBPs and CSPs tree sets, we selected the protein sequences including four Coleoptera from different families (Table S1), Colaphellus bowringi [51], Monochamus alternatus [52], Dastarcus helophoroides [52] and Ophraella communa [53]. For the GRs tree, the protein sequences from two Coleoptera (Chrysomelidae) species (C. bowringi and O. communa), the sequences that have been functionally identified, including three Lepidoptera (Plutella xylostella [54], Helicoverpa armigera [51, 55, 56] and Bombyx mori [35, 39, 57]) and four Diptera (D. melanogaster [40, 58–60], Anopheles gambiae [61], Aedes aegypti [41] and Anopheles coluzzii [62]) were used. ClustalW (gap opening penalty: 10, gap extension penalty: 0.2) was employed for amino acid sequence alignment and the neighbor-joining method was constructed using MEGA6, the gap/missing data treatment was set as partial deletion with a 95% site coverage cutoff [63, 64]. Branch support was assessed with 1000 bootstrap replicates. Phylogenetic trees were visualized and edited by FigTree 1.4.3(http://tree.bio.ed.ac.uk/software/figtree/).
Expression analysis by quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed to verify the expression patterns of candidate chemosensory genes by using CFX Connect Real-Time System (Bio-Rad, CA, USA). Different stages, including eggs, first instar larvae, second instar larvae heads, third instar larvae heads and pupae, were collected. Total RNA was isolated using the methods described above and reverse transcribed into cDNA using HiScript® III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China). The qRT-PCR was conducted in a 20 μL reaction system, containing 10 μL ChamQ Universal SYBR qPCR Master Mix (2×), 0.4 μL each of the forward and reverse primers (10 μM), 1 μL cDNA template, and 8.2 μL of nuclease-free water. The reaction conditions were as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Gene expression profiles were analyzed using the 2 −ΔΔCT method [65]. The means and variances of the treatments were analyzed in a one-way ANOVA using SPSS 26.0 software. Each reaction for each tissue was performed in three biological replicates. The RPS18 gene (accession number: OM885975) was used as the reference gene for normalizing the expression of various samples [66]. And RPS18 gene has been proven stable in different development stages, female and male adults, different tissues, different temperatures, and pathogen treatment [67]. The gene-specific primers were designed using Beacon Designer 8.0 (amplicon length: 100 ± 20 bp; Tm: 60 ± 2 °C; GC%: 45–55%) (PRIMER Biosoft International, CA, USA), and were listed in Supplementary Table S2.
Results
Transcriptome sequencing, assembly and analysis
To identify the chemosensory genes of P. versicolora, the transcriptome sequencing of third instar larvae heads was sequenced with three independent biological replicates. Approximately 75.14 million, 68.24 million and 77.56 million raw reads, and a total of 74.50 million, 67.73 million and 76.97 million clean reads were generated. Clean reads were assembled into 28,268 unigenes with an average length of 1128 bp, and an N50 of 2500 bp. Length distribution analysis showed that 45.35% of all unigenes were longer than 500 bp in size (Fig. S1).
After function annotations, a total of 10,030 unigenes (35.74%) and 13,310 unigenes (47.43%) were successfully annotated through GO and Nr datasets, respectively. In the GO analysis, the unigenes were assigned to three main functions: biological process (13,448), cellular component (16,640), and molecular function (12,036). Within three classes, the subcategories “binding” (5321), “cell part” (4857) and “cellular process” (4475) contained the majority of the unigenes, respectively (Fig. S2).
Identification of candidate OBPs
In this study, a total of 29 predicted PverOBP transcripts were identified from third instar larvae heads transcriptomes, 12 PverOBPs were new compared to antennae and forelegs. The complete ORF was detected in 22 PverOBPs, with lengths ranging from 131 to 241 amino acids. Among them, 22 transcripts with the complete sequence have a signal peptide sequence (Table S3). Sequence analysis showed that 6 PverOBPs belonged to the Classical OBP subclass with the typical six conserved cysteine residues. Fifteen PverOBPs belonged to the minus-C OBP subgroup with 4 conserved cysteine residues (Fig. 1). A phylogenetic analysis showed that PverOBP4 and OBP12 were on the same branch as MaltOBP3 and OBP10, which functions were identified in M. alternatus, respectively (Fig. 2).
Fig. 1.
Multiple amino acid alignments of the predicted Classic OBPs and Minus-C OBPs. Conserved cysteine residues are marked with a red box (C2 and C5), black (C3, C4 and C6) and pink (C1) background. The number of 61 and 107 amino acids were deleted at the begin of OBP14 and end of OBP38, respectively
Fig. 2.
Phylogenetic analysis of the OBPs (odorant-binding proteins) from four insect species: P. versicolora (Pver), C. bowringi (Cbow), M. alternatus (Malt), O. communa (Ocom). The P. versicolora genes are shown in blue. The values at the branch nodes represent bootstrap values based on 1000 replicates
Identification of candidate CSPs
Six transcripts encoding CSPs were identified from the P. versicolora transcriptomes and PverCSP12 and CSP13 were newly compared to antennae and forelegs transcriptomes. All but one transcript (PverCSP13) included full-length ORFs, with lengths from 122 to 137 amino acids and a predicted signal peptide. Five CSP proteins have four highly conserved cysteine residues, which are characteristic of typical insect CSPs (Fig. 3). Phylogenetic analyses of CSPs from P. versicolora and other four Coleoptera species showed that PverCSP9 was clustered with MaltCSP5 in M. alternatus that functional characteristics have been performed (Fig. 4).
Fig. 3.
Multiple amino alignments of the predicted CSPs. Conserved cysteine (C1-C4) residues are marked with a blue box
Fig. 4.
Phylogenetic analysis of the CSPs (chemosensory proteins) from four insect species: P. versicolora (Pver), C. bowringi (Cbow), M. alternatus (Malt), O. communa (Ocom) and D. helophoroides (Dhel). The P. versicolora genes are shown in blue. The values at the branch nodes represent bootstrap values based on 1000 replicates
Identification of candidate GRs
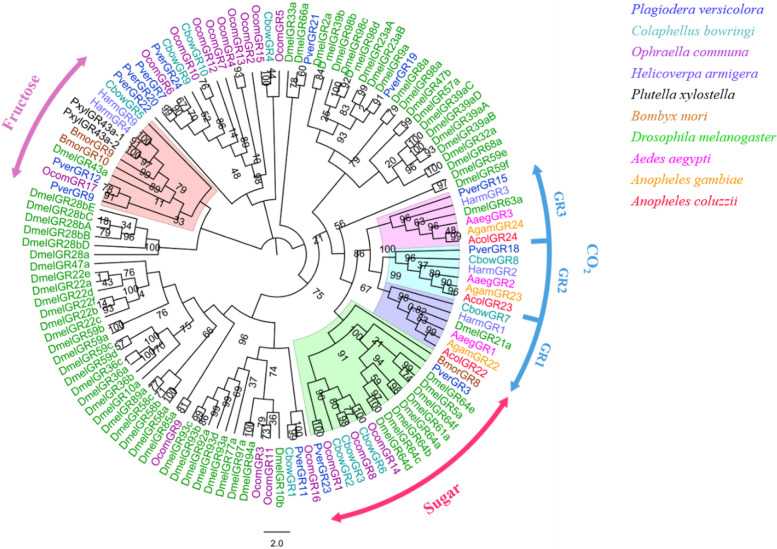
Thirteen PverGR transcripts were obtained from P. versicolora third instar larvae heads and 7 PverGRs were newly identified. Among 13 PverGRs, only PverGR15 has a full-length ORF, with 7 transmembrane domains (TMDs), which are characteristic of typical insect GRs. Based on the phylogenetic tree analysis that PverGR15 and GR18 (newly identified) were clustered in carbon dioxide (CO2) receptors, PverGR3 and GR23 (newly identified) were clustered in sugar receptors (except fructose). In addition, PverGR9 and GR12 were clustered with the reported fructose receptors (Fig. 5).
Fig. 5.
Phylogenetic analysis of the GRs (gustatory receptors) from ten insect species: P. versicolora (Pver), C. bowringi (Cbow), O. communa (Ocom), A. gambiae (Agam), A. aegypti (Aage), A. coluzzii (Acol), D. melanogaster (Dmel), H. armigera (Harm), P. xylostella (Pxyl) and B. mori (Bmor). The P. versicolora genes are shown in blue. The values at the branch nodes represent bootstrap values based on 1000 replicates
Identification of candidate ORs and IRs
From the assembled transcripts, 14 of them were annotated encoding ORs, and 5 PverORs were newly identified from the transcriptomes of third instar larvae heads. The Orco gene was also obtained in the transcriptomes (Table S4). None of these OR unigenes have full-length ORFs, encoding lengths from 45 to 289 amino acids. We identified 8 transcripts encoding candidate IRs in the P. versicolora transcriptome and 4 PverIRs were newly identified, compared to antennae and forelegs transcriptomes. These IRs were confirmed belong to IR family by the blast and phylogenetic analysis (Fig. S3). Of these, 3 PverIRs contained a putative full-length ORF, with three to four TMDs.
Specific expression profiles of candidate genes
The qRT-PCR was used to measure the specific expression levels of candidate chemosensory genes during the eggs, larvae and pupae stage (Fig. 6). The PCR efficiencies of primers ranged from 95–105% with high correlation coefficient (R2) values (0.991–0.999). PverOBP31, OBP34, OBP35, OBP38 and OBP40 were significantly higher expressed in 1st to 3rd instar larvae, while PverOBP33 and OBP37 were significantly higher expressed in pupae. PverCSP12 and CSP13 have significantly higher expression in 3rd instar larvae and eggs, respectively. PverGR22 has significantly higher expression in eggs, 3rd instar larvae and pupae, while PverGR24 has no significant difference in the tested tissues. PverOR42 was significantly higher expressed in pupae, moderately in 2nd and 3rd instar larvae, while the expression level of PverOR41 was no significant difference among eggs, larvae and pupae. Both PverIR12 and IR14 were no significant difference among eggs, larvae and pupae.
Fig. 6.
Expression levels of chemosensory genes in different stages performed by qRT-PCR. E, eggs; 1st, first instar larvae; 2nd, second instar larvae heads; 3rd, third instar larvae heads; P, pupae. Error bars indicate standard error of three biological replicates
Discussion
Compared to Dipterans and Lepidopterans, the molecular basis of chemoreception in Coleopterans is relatively poorly understood. In the current study, we sequenced and analyzed the transcriptome of the third instar larvae heads from P. versicolora. In total, 74 chemosensory genes were identified, including 29 OBPs, 6 CSPs, 14 ORs, 13 GRs, 8 IRs and 4 SNMPs. Compared to our previous antennae and forelegs transcriptome data in adults, 12 OBPs, 2 CSPs, 5 ORs, 4 IRs and 7 GRs were newly identified (Fig. S4). Moreover, the expression profile of 16 chemosensory genes in different tissues was validated by qRT-PCR, which would facilitate the exploration of the function of these genes. Systematic research on chemosensory genes in larvae may provide valuable information on understanding the molecular mechanisms of insect chemosensory systems.
The phylogenetic tree of OBPs and CSPs of P. versicolora was constructed using various Coleoptera species of C. bowringi, M. alternatus and O. communa. PverOBP4, PverOBP12 and PverCSP9 were clustered with MaltOBP3 (bound the beetle- or host-plant-related compounds) [68], MaltOBP10 (bound the volatiles from pine bark) [69] and MaltCSP5 (bound the odor molecules of pine volatiles) [70], respectively, suggesting that PverOBPs and PverCSPs may have functions (detect the host-plant-related volatiles) similar to these of OBPs and CSPs. Notably, PverOBP26 (identified in forelegs transcriptome) and five PverOBPs (PverOBP29, 30, 32, 35 and 39) formed a cluster with CbowOBP24, indicating that these genes of the clade may have suffered gene duplication.
The phylogenetic tree of GRs of P. versicolora was constructed using different species, including Coleoptera, Lepidoptera and Diptera. Insect GRs usually have three CO2 receptors (GR1, GR2 and GR3) family [61, 62, 71], and a new CO2 receptor (PverGR18), belonging to GR2 family, was identified from our larvae transcriptome. Considering three CO2 receptors have been identified, the function of these putative CO2 receptors genes could elucidate in the future. BmGr10 plays an important role in the myo-inositol recognition required for B. mori larval feeding behavior [57]. PverGR9 and GR12 were homologues to BmGr10, indicating three PverGRs were expressed in larval heads may guide several chemosensory behaviors. OR41 and OR52 were obtained from larval antennae and maxillae transcriptome in H. armigera, and responded to a four-component blend that strongly attracted larvae [72]. It meant that some ORs were identified in larval heads play role in chemosensory-guided behaviors. However, no PverORs were homologues to HarmOR41/OR52 (Fig. S5), as the function of 5 PverORs need to identify in the future.
OBPs are commonly postulated to play an important role in the perception of environmental volatiles, in both adult and larval stages [73, 74]. PverOBP31 and OBP34 have a highly expression level in the third instars larval heads, and PverOBP35 showed increased expression levels from the first to third instars. Usually, higher instar larvae of agriculture and forestry pests are crucial for crop and tree feeding [45, 75]. The studies have indicated that PverOBP31, PverOBP34 and PverOBP35 may participate in larval food searching.
Some pieces of evidence inciting that a relatively simple system, OBPs, abundantly expressed in the larval antennae, also involves the detection of pheromones in larvae [76, 77]. In vitro expression and ligand-binding of SexiOBP13 (highly expressed in the larval heads, and showed increased expression levels from the first- to third-instars), the result showed that it displayed a high binding affinity to Z9, E12–14:Ac, the major sex pheromone component of S. exigua, while low affinities to the tested host plant volatiles [78]. In general, PBPs have significantly higher expression in the antennae of adults. This report on S. exigua provides a new idea for identifying the function of PBPs in P. versicolora that may have a significantly higher expression level (PverOBP31, 34, 35, 38 and 40) in larvae. In addition, these five PverOBPs may detection of any other compounds, such as host or non-host volatiles.
Two special PverOBPs (OBP33 and 37) were highly expressed in pupae which were also consistent with the results obtained in other species. For example, five OBPs in Procecidochares utilis, OBP44a in Bactrocera dorsalis, and three OBPs (OBP15, 17, and 25) in Galeruca daurica were found to be abundantly expressed in the pupal stage, respectively [79–81]. In addition, PverOR42 was highly expressed in pupae, the same as three ORs in P. utilis and eight ORs in Chlorops oryzae [79, 82]. These results suggest that these OBPs and ORs in P. versicolora may be related to the beginning of the development of chemosensory tissue during pupation. On the other side, maybe it is also likely that these ORs and OBPs are a consequence of development.
Different expression profiles during development indicate that these CSP genes might have different functions. PverCSP12 has a high expression level in the third instar larva stage, which might suggest a role of these proteins in food searching and feeding. Particularly interestingly, PverCSP13 was highly expressed in the eggs. It has been demonstrated that CSP5 in the eggs of honeybees is required for the correct development of the embryo [83, 84]. It suggested that PverCSP13 may involve in the development of the eggs.
Supplementary Information
Additional file 1: Table S1. The classification and number among different species used in OBPs, CSPs and GRs in the phylogenetic trees. OBPs and CSPs in non-coleoptera species were not used in the phylogenetic analyses. The number of GR in non-coleoptera species of annotated proteins in those genomes were marked in green, in the transcriptome data was marked in bule. The italic numbers were used to structure the GR tree. Table S2. Primers for qRT-PCR of chemosensory genes in P. versicolora. Table S3. The Blastx match of P. versicolora candidate CSP and OBP genes. The new genes compared to previous reports were marked with bule color. Table S4. The Blastx match of P. versicolora candidate GR, IR, OR and SNMP genes. The new genes compared to previous reports were marked with bule color. Fig. S1. Distribution of unigene size of transcriptome assembly from third instar larvae head in P. versicolora. Fig. S2. Gene ontology (GO) classification of transcriptome unigenes from third instar larvae head in P. versicolora. Figure S3. Phylogenetic analysis of the IRs. The P. versicolora genes are shown in blue. The values at the branch nodes represent bootstrap values based on 1000 replicates. Figure S4. The Venn diagrams of chemosensory genes in P. versicolora from the transcriptome of antennae, forelegs and larvae. Figure S5. Phylogenetic analysis of the ORs. The P. versicolora genes are shown in blue. Pver, P. versicolora; Harm, Helicoverpa armigera; Cbow, Colaphellus bowringi. The values at the branch nodes represent bootstrap values based on 1000 replicates. File S1. The amino acid sequences of Plagiodera versicolora putative chemosensory genes.
Acknowledgements
We thank Le-Tian Xu (Hubei University, Wuhan, China) for help in the technical assistance.
Abbreviations
- OBP
Odorant binding protein
- CSP
Chemosensory protein
- OR
Odorant receptor
- GR
Gustatory receptor
- IR
Ionotropic receptor
- SNMP
Sensory neuron membrane protein
- Nr
Non-redundant database
- COG
Clusters of Orthologous Groups
- KEGG
Kyoto Encyclopedia of Genes and Genome
- GO
Gene Ontology
- NCBI
National Center for Biotechnology Information
- ORF
Open reading frame
- TMD
Transmembrane domain
Authors’ contributions
Performed the experiments: ZW, NT, JG. Conceived and designed the experiments: ZW, FJ, NT, YL, XL. Analyzed the data and wrote the manuscript: ML, XL. All authors read and approved the final manuscript.
Funding
This project is supported by the Hubei University National talent project (1070017364).
Availability of data and materials
The raw reads of the transcriptomes in this study have been submitted in the NCBI SRA database, under the accession number of SAMN29260310 (larvae head 1), SAMN29260311 (larvae head 2) and SAMN29260312 (larvae head 3).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
The original version of this article was revised: “The legends for Fig. 2 and Fig. 3 were reversed”. This issue has been corrected.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhe-Ran Wu and Jian-Ting Fan contributed equally to this work.
Change history
5/5/2023
A Correction to this paper has been published: 10.1186/s12864-023-09132-8
Contributor Information
Min Lu, Email: lumin@hubu.edu.cn.
Xiao-Long Liu, Email: bruceliu2021@outlook.com.
References
- 1.Bargen H, Saudhof K, Poehling H-M. Prey finding by larvae and adult females of Episyrphus balteatus. Entomologia Experimentalis et Applicata. 1998;87:245–254. doi: 10.1046/j.1570-7458.1998.00328.x. [DOI] [Google Scholar]
- 2.Field LM, Pickett JA, Wadhams LJ. Molecular studies in insect olfaction. Insect Mol Biol. 2000;9:545–551. doi: 10.1046/j.1365-2583.2000.00221.x. [DOI] [PubMed] [Google Scholar]
- 3.Verheggen FJ, Arnaud L, Bartram S, Gohy M, Haubruge E. Aphid and plant volatiles induce oviposition in an aphidophagous hoverfly. J Chem Ecol. 2008;34:301–307. doi: 10.1007/s10886-008-9434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 5.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 6.Wanner KW, Robertson HM. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol Biol. 2008;17:621–629. doi: 10.1111/j.1365-2583.2008.00836.x. [DOI] [PubMed] [Google Scholar]
- 7.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Ma H, Xie H, Xuan N, Guo X, Fan Z, et al. Biotype characterization, developmental profiling, insecticide response and binding property of Bemisia tabaci chemosensory proteins: role of CSP in insect defense. PLoS One. 2016;11:e0154706. doi: 10.1371/journal.pone.0154706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X-L, Sun S-J, Khuhro SA, Elzaki MEA, Yan Q, Dong S-L. Functional characterization of pheromone receptors in the moth Athetis dissimilis (Lepidoptera: Noctuidae) Pestic Biochem Physiol. 2019;158:69–76. doi: 10.1016/j.pestbp.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H-J, Xu W, Chen Q-M, Sun L-N, Anderson A, Xia Q-Y, et al. A phylogenomics approach to characterizing sensory neuron membrane proteins (SNMPs) in Lepidoptera. Insect Biochem Mol Biol. 2020;118:103313. doi: 10.1016/j.ibmb.2020.103313. [DOI] [PubMed] [Google Scholar]
- 11.Liu X-L, Zhang J, Yan Q, Miao C-L, Han W-K, Hou W, et al. The molecular basis of host selection in a crucifer-specialized moth. Curr Biol. 2020;30:4476–4482.e5. doi: 10.1016/j.cub.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb) 2009;103:208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- 13.Olivier V, Monsempes C, François M-C, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. 2011;20:189–199. doi: 10.1111/j.1365-2583.2010.01057.x. [DOI] [PubMed] [Google Scholar]
- 14.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 15.Pelosi P, Iovinella I, Felicioli A, Dani FR. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 2014;5:320. doi: 10.3389/fphys.2014.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci U S A. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cassau S, Krieger J. The role of SNMPs in insect olfaction. Cell Tissue Res. 2021;383:21–33. doi: 10.1007/s00441-020-03336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 19.Scaloni A, Monti M, Angeli S, Pelosi P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem Biophys Res Commun. 1999;266:386–391. doi: 10.1006/bbrc.1999.1791. [DOI] [PubMed] [Google Scholar]
- 20.Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999;464:85–90. doi: 10.1016/S0014-5793(99)01683-X. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J-J. Odorant-binding proteins in insects. Vitam Horm. 2010;83:241–272. doi: 10.1016/S0083-6729(10)83010-9. [DOI] [PubMed] [Google Scholar]
- 22.Lagarde A, Spinelli S, Qiao H, Tegoni M, Pelosi P, Cambillau C. Crystal structure of a novel type of odorant-binding protein from Anopheles gambiae, belonging to the C-plus class. Biochem J. 2011;437:423–430. doi: 10.1042/BJ20110522. [DOI] [PubMed] [Google Scholar]
- 23.Spinelli S, Lagarde A, Iovinella I, Legrand P, Tegoni M, Pelosi P, et al. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem Mol Biol. 2012;42:41–50. doi: 10.1016/j.ibmb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Schultze A, Schymura D, Forstner M, Krieger J. Expression pattern of a “Plus-C” class odorant binding protein in the antenna of the malaria vector Anopheles gambiae. Insect Mol Biol. 2012;21:187–195. doi: 10.1111/j.1365-2583.2011.01125.x. [DOI] [PubMed] [Google Scholar]
- 25.Nomura A, Kawasaki K, Kubo T, Natori S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach) Int J Dev Biol. 1992;36:391–398. [PubMed] [Google Scholar]
- 26.Mameli M, Tuccini A, Mazza M, Petacchi R, Pelosi P. Soluble proteins in chemosensory organs of phasmids. Insect Biochem Mol Biol. 1996;26:875–882. doi: 10.1016/S0965-1748(96)00055-0. [DOI] [PubMed] [Google Scholar]
- 27.Angeli S, Ceron F, Scaloni A, Monti M, Monteforti G, Minnocci A, et al. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur J Biochem. 1999;262:745–754. doi: 10.1046/j.1432-1327.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- 28.Pelosi P, Zhou J-J, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du L, Zhao X, Liang X, Gao X, Liu Y, Wang G. Identification of candidate chemosensory genes in Mythimna separata by transcriptomic analysis. BMC Genomics. 2018;19:518. doi: 10.1186/s12864-018-4898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Jiang G-F, Shu X-H, Wang Y-Q, Li M-J. Identification and expression profile analysis of chemosensory genes from the antennal transcriptome of bamboo locust (Ceracris kiangsu) Front Physiol. 2020;11:889. doi: 10.3389/fphys.2020.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rondoni G, Roman A, Meslin C, Montagné N, Conti E, Jacquin-Joly E. Antennal transcriptome analysis and identification of candidate chemosensory genes of the harlequin ladybird beetle, Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae) Insects. 2021;12:209. doi: 10.3390/insects12030209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G-C, Nuo S-M, Wang Z-Q, Yang A-J, Liu N-Y. Identification and expression profiling of chemosensory membrane protein genes in Achelura yunnanensis (Lepidoptera: Zygaenidae) Comp Biochem Physiol Part D Genomics Proteomics. 2021;40:100876. doi: 10.1016/j.cbd.2021.100876. [DOI] [PubMed] [Google Scholar]
- 33.He M, Ma Y-F, Guo H, Liu X-Z, Long G-J, Wang Q, et al. Genome-wide identification and expression pattern analysis of novel chemosensory genes in the German cockroach Blattella germanica. Genomics. 2022;114:110310. doi: 10.1016/j.ygeno.2022.110310. [DOI] [PubMed] [Google Scholar]
- 34.Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H-J, Anderson AR, Trowell SC, Luo A-R, Xiang Z-H, Xia Q-Y. Topological and functional characterization of an insect gustatory receptor. PLoS One. 2011;6:e24111. doi: 10.1371/journal.pone.0024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Butterwick JA, Del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, et al. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560:447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato K, Tanaka K, Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc Natl Acad Sci U S A. 2011;108:11680–11685. doi: 10.1073/pnas.1019622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto T, Slone J, Song X, Amrein H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151:1113–1125. doi: 10.1016/j.cell.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erdelyan CNG, Mahood TH, Bader TSY, Whyard S. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol Biol. 2012;21:119–127. doi: 10.1111/j.1365-2583.2011.01120.x. [DOI] [PubMed] [Google Scholar]
- 42.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Utsumi S, Ando Y, Ohgushi T. Evolution of feeding preference in a leaf beetle: the importance of phenotypic plasticity of a host plant. Ecol Lett. 2009;12:920–929. doi: 10.1111/j.1461-0248.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Xu S, Sun L, Zhang Y, Luo J, Bock R, et al. Synergistic action of the gut microbiota in environmental RNA interference in a leaf beetle. Microbiome. 2021;9:98. doi: 10.1186/s40168-021-01066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Tong N, Wu Z, Li Y, Ma M, Liu P, et al. Identification of chemosensory genes based on the antennal transcriptomic analysis of Plagiodera versicolora. Insects. 2021;13:36. doi: 10.3390/insects13010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Z, Tong N, Li Y, Guo J, Lu M, Liu X. Foreleg transcriptomic analysis of the chemosensory gene families in Plagiodera versicolora (Coleoptera: Chrysomelidae) Insects. 2022;13:763. doi: 10.3390/insects13090763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itagaki H, Hildebrand JG. Olfactory interneurons in the brain of the larval sphinx moth Manduca sexta. J Comp Physiol A. 1990;167:309–320. doi: 10.1007/BF00192566. [DOI] [PubMed] [Google Scholar]
- 50.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X-M, Zhu X-Y, Wang Z-Q, Wang Y, He P, Chen G, et al. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genomics. 2015;16:1028. doi: 10.1186/s12864-015-2236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Li D-Z, Min S-F, Mi F, Zhou S-S, Wang M-Q. Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp Biochem Physiol Part D Genomics Proteomics. 2014;11:1–8. doi: 10.1016/j.cbd.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Ma C, Zhao C, Cui S, Zhang Y, Chen G, Chen H, et al. Identification of candidate chemosensory genes of Ophraella communa LeSage (Coleoptera: Chrysomelidae) based on antennal transcriptome analysis. Sci Rep. 2019;9:15551. doi: 10.1038/s41598-019-52149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X-L, Sun S-J, Hou W, Zhang J, Yan Q, Dong S-L. Functional characterization of two spliced variants of fructose gustatory receptor in the diamondback moth, Plutella xylostella. Pestic Biochem Physiol. 2020;164:7–13. doi: 10.1016/j.pestbp.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Xu W, Zhang H-J, Anderson A. A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J Chem Ecol. 2012;38:1513–1520. doi: 10.1007/s10886-012-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang X-J, Ning C, Guo H, Jia Y-Y, Huang L-Q, Qu M-J, et al. A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect Biochem Mol Biol. 2015;60:39–46. doi: 10.1016/j.ibmb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Kikuta S, Endo H, Tomita N, Takada T, Morita C, Asaoka K, et al. Characterization of a ligand-gated cation channel based on an inositol receptor in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2016;74:12–20. doi: 10.1016/j.ibmb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 58.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 59.Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci U S A. 2007;104:14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu T, Qiu YT, Wang G, Kwon JY, Rutzler M, Kwon H-W, et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu F, Ye Z, Baker A, Sun H, Zwiebel LJ. Gene editing reveals obligate and modulatory components of the CO2 receptor complex in the malaria vector mosquito, Anopheles coluzzii. Insect Biochem Mol Biol. 2020;127:103470. doi: 10.1016/j.ibmb.2020.103470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 64.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Ma M, Tu C, Luo J, Lu M, Zhang S, Xu L. Metabolic and immunological effects of gut microbiota in leaf beetles at the local and systemic levels. Integr Zool. 2021;16:313–323. doi: 10.1111/1749-4877.12528. [DOI] [PubMed] [Google Scholar]
- 67.Tu C, Xu P, Han R, Luo J, Xu L. Defining suitable reference genes for qRT-PCR in Plagiodera versicolora (Coleoptera: Chrysomelidae) under different biotic or abiotic conditions. Agronomy. 2022;12:1192. doi: 10.3390/agronomy12051192. [DOI] [Google Scholar]
- 68.Gao X, Wang M-Q. A cDNA library from the antenna of Monochamus alternatus Hope and binding properties of odorant-binding proteins. J Appl Entomol. 2015;139:229–236. doi: 10.1111/jen.12136. [DOI] [Google Scholar]
- 69.Li D-Z, Huang X-F, Yang R-N, Chen J-Y, Wang M-Q. Functional analysis of two odorant-binding proteins, MaltOBP9 and MaltOBP10, in Monochamus alternatus Hope. Front Physiol. 2020;11:317. doi: 10.3389/fphys.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ali S, Ahmed MZ, Li N, Ali S, a. I, Wang M-Q. Functional characteristics of chemosensory proteins in the sawyer beetle Monochamus alternatus Hope. Bull Entomol Res. 2019;109:34–42. doi: 10.1017/S0007485318000123. [DOI] [PubMed] [Google Scholar]
- 71.Xu W, Anderson A. Carbon dioxide receptor genes in cotton bollworm Helicoverpa armigera. Sci Nat. 2015;102:11. doi: 10.1007/s00114-015-1260-0. [DOI] [PubMed] [Google Scholar]
- 72.Di C, Ning C, Huang L-Q, Wang C-Z. Design of larval chemical attractants based on odorant response spectra of odorant receptors in the cotton bollworm. Insect Biochem Mol Biol. 2017;84:48–62. doi: 10.1016/j.ibmb.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 73.Zhang R, Wang B, Grossi G, Falabella P, Liu Y, Yan S, et al. Molecular basis of alarm pheromone detection in aphids. Curr Biol. 2017;27:55–61. doi: 10.1016/j.cub.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 74.Sun L, Li Y, Zhang Z, Guo H, Xiao Q, Wang Q, et al. Expression patterns and ligand binding characterization of Plus-C odorant-binding protein 14 from Adelphocoris lineolatus (Goeze) Comp Biochem Physiol B Biochem Mol Biol. 2019;227:75–82. doi: 10.1016/j.cbpb.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Jacquin-Joly E, Vogt RG, François MC, Nagnan-Le MP. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem Senses. 2001;26:833–844. doi: 10.1093/chemse/26.7.833. [DOI] [PubMed] [Google Scholar]
- 76.Zhu J, Ban L, Song L-M, Liu Y, Pelosi P, Wang G. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem Mol Biol. 2016;72:10–19. doi: 10.1016/j.ibmb.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 77.Han W-K, Yang Y-L, Si Y-X, Wei Z-Q, Liu S-R, Liu X-L, et al. Involvement of GOBP2 in the perception of a sex pheromone component in both larval and adult Spodoptera litura revealed using CRISPR/Cas9 mutagenesis. Insect Biochem Mol Biol. 2022;141:103719. doi: 10.1016/j.ibmb.2022.103719. [DOI] [PubMed] [Google Scholar]
- 78.Jin R, Liu NY, Liu Y, Dong SL. A larval specific OBP able to bind the major female sex pheromone component in Spodoptera exigua (Hübner) J Integr Agric. 2015;14:1356–1366. doi: 10.1016/S2095-3119(14)60849-2. [DOI] [Google Scholar]
- 79.Li L, Gao X, Gui H, Lan M, Zhu J, Xie Y, et al. Identification and preliminary characterization of chemosensory-related proteins in the gall fly, Procecidochares utilis by transcriptomic analysis. Comp Biochem Physiol Part D Genomics Proteomics. 2020;36:100724. doi: 10.1016/j.cbd.2020.100724. [DOI] [PubMed] [Google Scholar]
- 80.Li L, Zhou Y-T, Tan Y, Zhou X-R, Pang B-P. Identification of odorant-binding protein genes in Galeruca daurica (Coleoptera: Chrysomelidae) and analysis of their expression profiles. Bull Entomol Res. 2017;107:550–561. doi: 10.1017/S0007485317000402. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J, Luo D, Wu P, Li H, Zhang H, Zheng W. Identification and expression profiles of novel odorant binding proteins and functional analysis of OBP99a in Bactrocera dorsalis. Arch Insect Biochem Physiol. 2018;98:e21452. doi: 10.1002/arch.21452. [DOI] [PubMed] [Google Scholar]
- 82.Qiu L, Tao S, He H, Ding W, Li Y. Transcriptomics reveal the molecular underpinnings of chemosensory proteins in Chlorops oryzae. BMC Genomics. 2018;19:890. doi: 10.1186/s12864-018-5315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forêt S, Wanner KW, Maleszka R. Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol. 2007;37:19–28. doi: 10.1016/j.ibmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 84.Maleszka J, Forêt S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera) Dev Genes Evol. 2007;217:189–196. doi: 10.1007/s00427-006-0127-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The classification and number among different species used in OBPs, CSPs and GRs in the phylogenetic trees. OBPs and CSPs in non-coleoptera species were not used in the phylogenetic analyses. The number of GR in non-coleoptera species of annotated proteins in those genomes were marked in green, in the transcriptome data was marked in bule. The italic numbers were used to structure the GR tree. Table S2. Primers for qRT-PCR of chemosensory genes in P. versicolora. Table S3. The Blastx match of P. versicolora candidate CSP and OBP genes. The new genes compared to previous reports were marked with bule color. Table S4. The Blastx match of P. versicolora candidate GR, IR, OR and SNMP genes. The new genes compared to previous reports were marked with bule color. Fig. S1. Distribution of unigene size of transcriptome assembly from third instar larvae head in P. versicolora. Fig. S2. Gene ontology (GO) classification of transcriptome unigenes from third instar larvae head in P. versicolora. Figure S3. Phylogenetic analysis of the IRs. The P. versicolora genes are shown in blue. The values at the branch nodes represent bootstrap values based on 1000 replicates. Figure S4. The Venn diagrams of chemosensory genes in P. versicolora from the transcriptome of antennae, forelegs and larvae. Figure S5. Phylogenetic analysis of the ORs. The P. versicolora genes are shown in blue. Pver, P. versicolora; Harm, Helicoverpa armigera; Cbow, Colaphellus bowringi. The values at the branch nodes represent bootstrap values based on 1000 replicates. File S1. The amino acid sequences of Plagiodera versicolora putative chemosensory genes.
Data Availability Statement
The raw reads of the transcriptomes in this study have been submitted in the NCBI SRA database, under the accession number of SAMN29260310 (larvae head 1), SAMN29260311 (larvae head 2) and SAMN29260312 (larvae head 3).