Abstract
Background
Asthma exacerbations are highly prevalent in children, but only a few studies have examined the biologic mechanisms underlying exacerbations in this population.
Objective
High-resolution metabolomics analyses were performed to understand the differences in metabolites in children with exacerbating asthma who were hospitalized in a pediatric intensive care unit for status asthmaticus. We hypothesized that compared with a similar population of stable outpatients with asthma, children with exacerbating asthma would have differing metabolite abundance patterns with distinct clustering profiles.
Methods
A total of 98 children aged 6 through 17 years with exacerbating asthma (n = 69) and stable asthma (n = 29) underwent clinical characterization procedures and submitted plasma samples for metabolomic analyses. High-confidence metabolites were retained and utilized for pathway enrichment analyses to identify the most relevant metabolic pathways that discriminated between groups.
Results
In all, 118 and 131 high-confidence metabolites were identified in positive and negative ionization mode, respectively. A total of 103 unique metabolites differed significantly between children with exacerbating asthma and children with stable asthma. In all, 8 significantly enriched pathways that were largely associated with alterations in arginine, phenylalanine, and glycine metabolism were identified. However, other metabolites and pathways of interest were also identified.
Conclusion
Metabolomic analyses identified multiple perturbed metabolites and pathways that discriminated children with exacerbating asthma who were hospitalized for status asthmaticus. These results highlight the complex biology of inflammation in children with exacerbating asthma and argue for additional studies of the metabolic determinants of asthma exacerbations in children because many of the identified metabolites of interest may be amenable to targeted interventions.
Key words: Arginine, asthma control, asthma exacerbation, asthma in children, glycine, inflammation, metabolomics, phenylalanine, status asthmaticus
Asthma exacerbations are highly prevalent, affecting nearly half of all children with current asthma in the United States who are younger than 18 years,1 and they are a substantial driver of pediatric asthma–related health care costs.2 Although asthma exacerbations are primarily managed at home or in outpatient settings, 8.8 of every 100,000 children in the US population younger than 18 years (and approximately 1 of every 20 children with asthma) progress to status asthmaticus and require hospitalization.1 Hospitalization rates are even higher in socially vulnerable populations of children.3 Yet despite the substantial personal and societal impact of hospitalization for asthma, effective management of these children in the hospital setting remains a substantial knowledge gap.4,5 This is particularly concerning because hospital admission for asthma is a primary predictor of future hospital readmission6 and significantly increases the risk of asthma-related death.7
Although advances have been made in understanding the biologic bases of asthma in stable outpatient populations, there is a paucity of studies that have examined biologic mechanisms underlying asthma exacerbations in children. Indeed, much of what is known about the inflammatory underpinnings of asthma exacerbations stems from work in mice8, 9, 10 and work in stable populations of children, in which predetermined biomarkers are associated with exacerbation risk in either a prospective or retrospective manner.5,11 Only a few studies have examined inflammatory mechanisms in patients during acute exacerbations.12, 13, 14 Even fewer studies have examined inflammatory mechanisms in hospitalized children with status asthmaticus at the time of admission. As a result, hospital treatment of asthma exacerbations remains largely symptomatic7 and is not currently directed toward specific inflammatory pathways.
To address this important knowledge gap, we performed metabolomics analyses using HPLC with high-resolution, accurate mass spectrometry to understand the differences in metabolite patterns in children with exacerbating asthma who were hospitalized in a pediatric intensive care unit (PICU) for status asthmaticus. We hypothesized that compared with a similar population of stable outpatients with asthma, children with exacerbating asthma would have differing metabolite patterns with distinct clustering profiles.
Methods
Participants
Children aged 6 through 17 years with physician-diagnosed asthma evaluated at Children’s Healthcare of Atlanta were eligible for inclusion. Two groups of children were enrolled. Children with exacerbating asthma were enrolled within 48 hours of admission from a 36-bed PICU after receipt of any of the following interventions in the emergency department: (1) a third continuous nebulized albuterol treatment; (2) noninvasive respiratory support delivered by high-flow nasal cannula, bilevel positive airway pressure, or invasive mechanical ventilation; (3) an 80%:20% helium-oxygen mixture; or 4) at least a 50% fraction of inspired oxygen by Venturi mask or positive pressure ventilation strategies to maintain oxygen saturations of 92% or higher. The children with stable asthma were outpatient children participating in research-only clinic visits. Stable children were enrolled from an asthma specialty clinic if they had historical evidence of at least 12% reversibility in their FEV1 value relative to baseline after bronchodilator administration and if they had at least 1 asthma exacerbation necessitating treatment with systemic corticosteroids in the preceding year.15 In the event that an outpatient participant had a recent exacerbation treated with systemic corticosteroids, the research visit was postponed until at least 2 weeks after the last dose of systemic corticosteroids. Children were excluded from either group if they had a history of hematopoietic stem cell or solid organ transplant, an oncologic diagnosis, sickle cell anemia, a rheumatologic diagnosis, pulmonary aspiration, gastroesophageal reflux requiring acid suppression medication, bronchiectasis, congenital airway anomalies, a history of premature birth before 35-weeks' gestation, or a personal history of smoking or vaping. Permission to proceed with this study was granted by the Emory University and Children’s Healthcare of Atlanta institutional review boards. Informed written consent was obtained from legal guardians. Verbal assent was obtained from children aged 6 to 10 years, and written assent was obtained from children and adolescents aged 11 to 17 years. All study procedures were performed in accordance with the relevant guidelines and regulations in the Declaration of Helsinki.
Participant characterization procedures
Participants and their caregivers completed demographic and medical history questionnaires, the 8-item Pediatric Asthma Impact Scale (PAIS),16 and the Asthma Control Test (ACT)17 or Childhood ACT (C-ACT).18 The PAIS is measured with a T-score, with higher scores reflecting worse impact.16 ACT scores lower than 20 reflect uncontrolled asthma.19 Severe asthma was defined by a European Respiratory Society–American Thoracic Society guideline as asthma requiring treatment with high-dose inhaled corticosteroids plus a second controller to prevent it from becoming uncontrolled or asthma that remains uncontrolled despite this therapy.20 Total serum IgE level, aeroallergen sensitization as determined by specific IgE testing, and lung function data (while the patient's asthma was stable) were abstracted from the electronic medical record of hospitalized patients within 12 months of PICU admission. Aeroallergen sensitization was assessed in outpatients by specific IgE testing with 8 extracts: tree mix, grass mix, weed mix, mold mix (Alternaria alternata, Aspergillus fumagatis, and Cladosporium herbarum), dog dander, cat dander, Blatella germanica, and dust mite mix (Dermatophagoides farinae and Dermatophagoides pteronyssinus) (Greer Laboratories, Lenoir, NC). Spirometry (KoKo PDS, Ferraris, Louisville, Colo) was performed in outpatients according to technical standards21 from the best of 3 forced vital capacity (FVC) maneuvers. All spirometry data from the hospitalized patients and outpatients, including FVC, FEV1 value, and forced expiratory flow at 25% to 75% of FVC, were interpreted according to Global Lung Function Initiative prediction equations22 and expressed as percentages of predicted values. All participating children also submitted blood samples for plasma cytokines and plasma metabolomics. If hospitalized children were missing serum IgE values, these values were obtained with a commercially available ELISA kit (Invitrogen, Waltham, Mass). Plasma IFN-γ, IL-4, IL-5, and IL-13 levels were quantified with a commercial assay according to the manufacturers’ instructions (Millipore Sigma Milliplex MAP Human High Sensitivity T-Cell Panel, Burlington, Mass). A subset of participants also submitted blood samples for quantification of blood eosinophils (Children’s Healthcare of Atlanta, Atlanta, Ga).
Metabolomics
Detailed methods are presented in the Online Repository (available at www.jaci-global.org). Briefly, mass spectrometry was performed on plasma samples with a Vanquish Horizon Binary ultrahigh-performance liquid chromatography system coupled with a Q Exactive High-Field Hybrid Orbitrap mass spectrometer (ThermoFisher). Data were analyzed by using Compound Discoverer 3.3 (ThermoFisher). Metabolite retention times were aligned across runs, and sample extracts combined as a global pooled sample were used as quality controls to correct instrumental response draft and evaluate measurement reproducibility (relative SD < 30%). Metabolites were considered high-confidence annotations and retained for analysis if they included accurate mass (5 ppm) match and either (1) retention time (±5%) matching to previously analyzed reference standards, and/or (2) matching to mass spectrometry2 spectra in mzCloud (mzcloud.org) with at least 1 product ion match.
Metabolite validation analyses
Validation of selected metabolites was performed in an independent sample of 215 outpatient children with asthma from metropolitan Atlanta, Georgia, who were considered at high risk for exacerbation. Details of the sample, participant characterization, and the metabolomics methodology were published previously.23 Metabolites were compared in children with acute uncontrolled asthma versus in children with stable asthma. Acute uncontrolled asthma was defined by either an Asthma Control Questionnaire 6-item score higher than 1.524 or Global Initiative for Asthma criteria for uncontrolled asthma,7 which include at least 3 of the following: (1) daytime asthma symptoms more than twice per week, (2) any night awakening due to asthma, (3) short-acting bronchodilator use for symptoms more than twice per week, and (4) any activity limitation due to asthma.7 Metabolites were also compared in children with severe versus nonsevere asthma.
Exploratory analyses
Exploratory analyses focused on associations between significant metabolites and PAIS scores, lung function measures (FEV1 value and FEV1/FVC ratio), and asthma severity.
Statistical analyses
Features between groups were compared by performing chi-squared analyses and t tests using IBM SPSS Statistics Software (version 28, IBM, Armonk, NY). Data that were not normally distributed were logarithmically transformed before statistical analyses. Metabolomics data were analyzed with RStudio software.25 Metabolites were log10 transformed, median normalized, and range scaled to minimize outliers and produce more normal distributions. Univariate analyses were performed on each ionization mode separately.26 Metabolite comparisons between exacerbating and stable asthma groups were made by ANOVA, adjusting for race as a binary variable (Black versus non-Black race). Results were adjusted for multiple comparisons by using the Benjamini-Hochberg procedure, with a false discovery rate of 10%.27 Metabolites were considered significantly different only if they displayed an unadjusted P value less than .05 and adjusted q value less than 0.1. Partial least squares discriminant analysis (PLS-DA) was then performed by using MetaboAnalyst 5.028 on all significantly different metabolites between groups. The predictive ability of the PLS-DA model was tested by using 10-fold cross-validation.29 Pathway enrichment analysis, which considers the connectedness of metabolites within a pathway, was conducted by using MetaboAnalyst 5.0 with utilization of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, a hypergeometric test, and relative betweenness centrality.30 For validation analyses, mass-to-charge features for preselected metabolites were z score–normalized and compared with the Welch t test values without assumption of equal SDs. For exploratory analyses, associations were assessed with Pearson correlation analyses and Welch t tests. Validation and exploratory analyses utilized a significance level of.05 without adjustment for multiple comparisons.
Results
A total of 98 children were enrolled (69 with exacerbating asthma and 29 with stable asthma) (Table I). The groups did not differ by age, sex, weight percentile, family history of asthma, daily asthma controller medications, exposures, or asthma-related health care utilization. However, there were slightly more self-reported Black children in the exacerbating group. Although asthma control in the prior 4 weeks (as reflected by ACT or C-ACT scores) did not differ between the groups, asthma impact in the previous week (as reflected by PAIS scores) was significantly worse in the exacerbating children. Although not specifically a requirement for inclusion, more than 93% of children in both groups had aeroallergen sensitization confirmed by specific IgE testing. Both groups also had similar elevations in serum IgE level. Blood eosinophil counts during hospitalization were elevated (ie, >150 cells/μL) in the exacerbating children but were significantly lower than those of the stable children. Lung function values for FVC; FEV1; forced expiratory flow at 25% to 75% of FVC (obtained while the patient was stable before or after hospitalization in the case of exacerbating patients); and plasma IFNγ, IL-4, IL-5, and IL-13 concentrations were also not different between groups (Table I). Although respiratory viral panels were not performed in all exacerbating children, in the subset in which they performed (n = 33), a respiratory virus was identified in 61% of children (n = 20), with enterovirus and/or rhinovirus present in 95% of the positive samples (n = 19).
Table I.
Features of the participants
| Feature | With exacerbating asthma (n = 69) | With stable asthma (n = 29) | P value |
|---|---|---|---|
| Age (y), median (25th, 75th percentile) | 11 (9, 14) | 10 (8, 14) | .328 |
| Males, no. (%) | 42 (60.9) | 14 (48.3) | .250 |
| Self-reported race, no. (%) | .020 | ||
| White | 4 (5.8) | 8 (27.6) | |
| Black | 58 (84.1) | 19 (65.5) | |
| Asian or American Indian | 2 (2.9) | 2 (6.9) | |
| ≥1 Race | 3 (4.3) | 0 | |
| Declined to answer | 2 (2.9) | 0 | |
| Weight percentile, median (25th, 75th percentile) | 76 (54, 99) | 77 (60, 99) | .500 |
| Family history, no. (%) | |||
| Mother with asthma | 23 (33.3) | 10 (35.7) | .823 |
| Father with asthma | 20 (29.0) | 8 (28.6) | .967 |
| Daily asthma controller medications, no. (%) | |||
| Montelukast | 28 (40.6) | 15 (51.7) | .310 |
| Inhaled corticosteroid | 39 (56.5) | 22 (75.9) | .071 |
| Long-acting β-agonist | 23 (33.3) | 10 (34.5) | .912 |
| Exposures, no. (%) | |||
| Smokers in the home | 23 (33.3) | 6 (20.7) | .211 |
| Dog or cat in the home | 22 (31.9) | 9 (31.0) | .934 |
| Previous PICU admission for asthma (ever in lifetime), no. (%) | 26 (37.7) | 16 (55.2) | .110 |
| Intubation for asthma (ever), no. (%) | 5 (7.2) | 5 (17.2) | .136 |
| Unscheduled visits for asthma symptoms (in past 12 mo), median (25th, 75th percentile) | 2 (1, 3) | 2 (1, 4) | .466 |
| Severe asthma, median (25th, 75th percentile)∗ | 38 (55.1) | 11 (37.9) | .121 |
| ACT or C-ACT score, median (25th, 75th percentile) | 14 (11, 16) | 18 (14, 21) | .181 |
| ACT or C-ACT score of <20, no. (%) | 49 (71.0) | 18 (62.1) | .116 |
| PAIS score, median (25th, 75th percentile) | 58 (54, 66) | 53 (46, 57) | .010 |
| Aeroallergen sensitization, no. (%)† | 53 (98.1) | 27 (93.1) | .240 |
| Serum IgE (kU/L), median (25th, 75th percentile) | 443 (202, 697) | 346 (167, 765) | .359 |
| Blood eosinophils (per μL), median (25th, 75th percentile)‡ | 217 (59, 551) | 396 (217, 551) | .001 |
| Lung function (% predicted), median (25th, 75th percentile)§ | |||
| FVC | 95 (83, 114) | 99 (88, 108) | .284 |
| FEV1 | 87 (72, 104) | 89 (79, 99) | .180 |
| FEV1/FVC | 91 (77, 94) | 91 (84, 99) | .401 |
| FEF25-75 | 70 (42, 80) | 73 (59, 86) | .165 |
| Plasma cytokines (pg/mL), median (25th, 75th percentile) | |||
| IFN-γ | 10.1 (5.3, 16.4) | 9.0 (5.8, 12.3) | .286 |
| IL-4 | 20.1 (8.3, 39.8) | 17.6 (10.1, 65.9) | .935 |
| IL-5 | 3.1 (2.3, 4.0) | 2.0 (1.0, 4.5) | .230 |
| IL-13 | 1.7 (1.1, 0.8) | 1.6 (0.8, 9.1) | .974 |
C-ACT, Childhood ACT; FEF25-75, forced expiratory flow at 25% to 75% of forced vital capacity.
Defined as the requirement for high-dose inhaled corticosteroids plus an additional controller medication for asthma control.
Exacerbating asthma (n = 54).
Exacerbating asthma (n = 35).
Exacerbating asthma (n = 43).
Metabolomic analyses
In all, 118 and 131 high-confidence metabolites were identified in positive and negative ionization mode, respectively. A total of 103 unique metabolites differed significantly between the exacerbating and stable asthma groups. These metabolites, along with the log2 fold change and q value, are listed in Tables E1 and E2 (in the Online Repository at www.jaci-global.org). PLS-DA, which is a “supervised” version of principal components analysis, was performed to visualize the variability and assess the discriminatory and predictive ability of metabolite profiles for the predetermined patient groups (Fig 1, A and B). Component 1 and component 2 explained 22.1% to 24.5% and 6.3% to 7% of the variance in positive and negative modes, respectively. The exacerbating children had higher within-group variability than did the stable children, as indicated by the size of the ellipse over the point spread (Fig 1, A and B). Classification performance of the PLS-DA model was evaluated for accuracy, goodness of fit (R2), and predictive ability (Q2) for the top 5 components for positive and negative modes. With use of 10-fold cross-validation, the model with 3 components best classified the positive mode model, with an accuracy of 0.97, an R2 value of 0.89, and a Q2 value of 0.79 (see Fig E1, A in the Online Repository at www.jaci-global.org). The model with 2 components best classified the negative mode model, with an accuracy of 0.99, an R2 value of 0.85, and a Q2 value of 0.78 (see Fig E1, B), confirming that the models were predictive and not overfitted.
Fig 1.
A and B, PLS-DA of metabolites from positive mode (A) and negative mode (B) that discriminated between exacerbating asthma (red circles) and stable asthma (green circles) in children. The number in parentheses reflects the variance explained by each component.
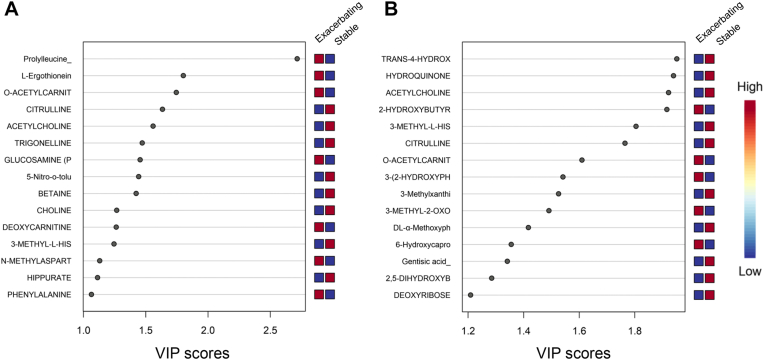
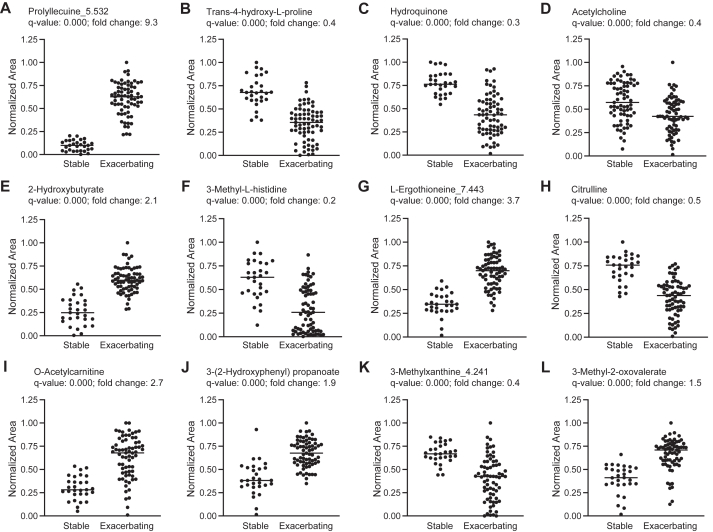
Variable importance of projection (VIP) scores were then generated to estimate the importance of each metabolite in the projection used in the PLS-DA model. Metabolites with a VIP score close to or higher than 1 were considered important in the model. The metabolites that had the highest predictive power in classifying exacerbating and stable asthma were then visualized by the plot of the first component of normalized metabolites by group for positive and negative modes. This approach yielded 15 predictive metabolites in positive mode and 15 metabolites in negative mode (Fig 2, A and B). The plots of the normalized concentrations of the top 12 metabolites shown in the VIP score plot, regardless of ionization mode, are shown by group in Fig 3, A-L. These 12 metabolites included prolylleucine, trans-4-hydroxy-L-proline, hydroquinone, acetylcholine, 2-hydroxybutyrate, 3-methyl-L-histidine, L-ergothioneine, citrulline, O-acetylcarnitine, 3-(2-hydroxyphenyl) propanoate, 3-methylxanthine, and 3-methyl-2-oxovalerate.
Fig 2.
A and B, VIP score plot of normalized metabolites by group in positive mode (A) and negative mode (B). Higher concentrations are in red and lower concentrations are in blue.
Fig 3.
Scatterplots of normalized concentrations of the top 12 metabolites explaining the variable importance of projection (VIP) of the first component in the PLS-DA. Data reflect either positive or negative mode for prolylleucine (A), trans-4-hydroxy-L-proline (B), hydroquinone (C), acetylcholine (D), 2-hydroxybutyrate (E), 3-methyl-L-histidine (F), L-ergothioneine (G), citrulline (H), O-acetylcarnitine (I), 3-(2-hydroxyphenyl) propanoate (J), 3-methylxanthine (K), and 3-methyl-2-oxovalerate (L).
Metabolite pathway analysis
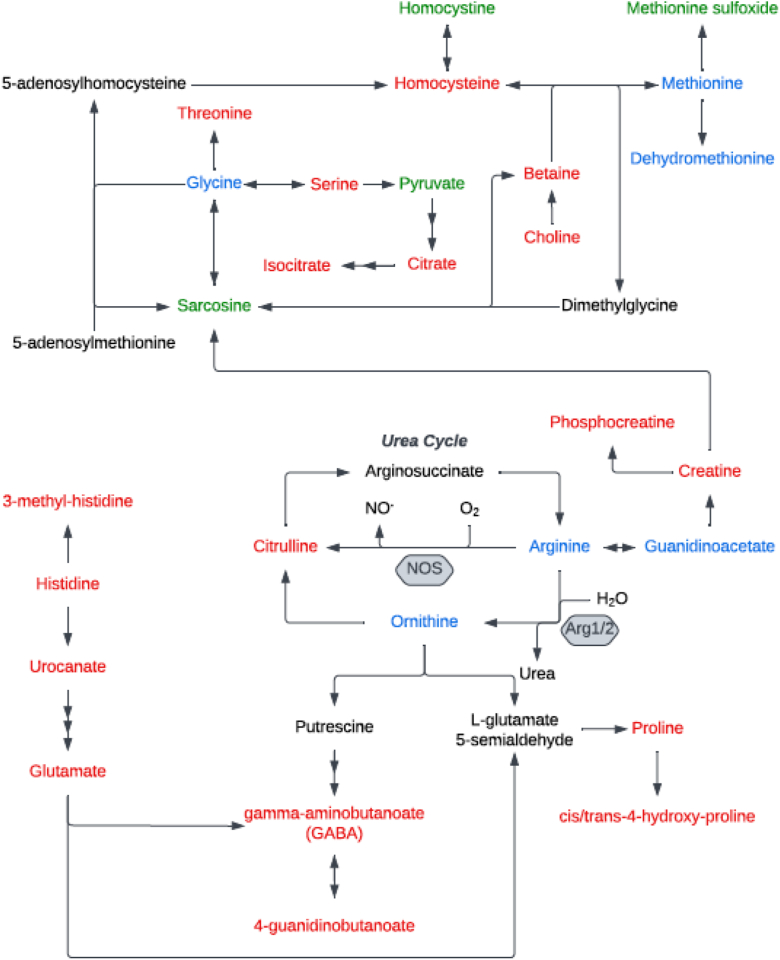
Pathway enrichment analysis was then performed by using the KEGG database to identify the most relevant metabolic pathways involved in discriminating children with exacerbating versus stable asthma. Of the 103 unique metabolites that differed between groups (see Tables E1 and E2), 83 had KEGG identification numbers and 8 significant metabolic pathways were identified (Fig 4). These significantly enriched pathways, in order of significance from highest to lowest, included (1) arginine and proline metabolism; (2) aminoacyl-tRNA synthesis (substrates that determine how the genetic code is interpreted as amino acids); (3) phenylalanine metabolism; (4) glycine, serine, and threonine metabolism; (5) valine, leucine, and isoleucine (branched-chain amino acids) biosynthesis; (6) histidine metabolism; (7) phenylalanine, tyrosine, and tryptophan biosynthesis; and (8) butanoate metabolism. The interconnectedness of the identified pathways is shown in Fig 5.
Fig 4.
Pathway enrichment analysis showing the differences between children with exacerbating versus stable asthma. Significant metabolic pathways with an impact of .05 or higher are shown. tRNA, Transfer RNA.
Fig 5.
Interconnectedness of significant pathways from pathway enrichment analysis. Metabolites with concentrations that were decreased in children with exacerbating asthma are shown in red, metabolites with concentrations that were increased are shown in green. Metabolites that were identified but did not differ in terms of concentration are shown in blue, and metabolites that were not identified by our methods are shown in black.
Metabolite validation analyses
To determine whether the identified metabolites might be generalizable to other samples of similar children, we performed validation analyses in an independent sample of 215 outpatient children with asthma from metropolitan Atlanta who were who were at high risk for exacerbation. The top 12 metabolites shown in the VIP score plot differentiating exacerbating from stable asthma in the present study were compared between children with acute uncontrolled asthma and children with stable asthma. Features of the validation sample are shown in Table E3 (in the Online Repository at www.jaci-global.org). The z score–normalized mass-to-charge ratios for 9 of these 12 metabolites, including prolylleucine, trans-4-hydroxy-L-proline, acetylcholine (as acetyl-β-methylcholine), 2-hydroxybutyrate, 3-methyl-L-histidine, citrulline, O-acetylcarnitine, 3-(2-hydroxyphenyl) propanoate, and 3-methyl-2-oxovalerate, were available for analysis. This validation analysis confirmed the following 5 metabolites in children with acute uncontrolled versus stable asthma: (1) higher 3-(2-hydroxyphenyl) propanoate concentration, (2) lower trans-4-hydroxy-L-proline concentration, (3) lower acetyl-β-methylcholine concentration, (4) lower 3-methyl-L-histidine concentration, and (5) lower citrulline concentration (Fig 6, A-E). Further analysis by asthma severity, irrespective of asthma control, also demonstrated higher 3-(2-hydroxyphenyl) propanoate, lower acetyl-β-methylcholine, lower 3-methyl-L-histidine, and lower citrulline concentrations in children with severe (n = 76) versus nonsevere (n = 139) asthma Fig E2 (in the Online Repository at www.jaci-global.org).
Fig 6.
Scatterplots of normalized concentrations of 5 significant (of 9 in total) metabolites assessed in a validation sample of outpatient children with uncontrolled and stable asthma, including 3-(2-hydroxyphenyl) propanoate (A), trans-4-hydroxy-L-proline (B), acetyl-β-methylcholine (C), 3-methyl-L-histidine (D), and citrulline (E).
Exploratory analyses of potential clinical relevance
We then explored whether the 103 high-confidence metabolites that differed between exacerbating and stable asthma were associated with PAIS scores, lung function, and/or severe asthma. Of the 103 metabolites that differentiated exacerbating asthma, 31 were also associated with PAIS scores (see Table E4 in the Online Repository at www.jaci-global.org). In all, 7 metabolites were associated with FEV1 value and 8 metabolites were associated with FEV1/FVC ratio (see Table E5 in the Online Repository at www.jaci-global.org), but only 2 metabolites, acetoacetate and lactate, were common between FEV1 value and FEV1/FVC ratio (see Fig E3 in the Online Repository at www.jaci-global.org). Of the 103 high-confidence metabolites, 15 were also associated with severe asthma (see Table E6 in the Online Repository at www.jaci-global.org).
Discussion
In this study, we performed metabolomic analyses to understand differences in plasma metabolite patterns in children with exacerbating asthma who were hospitalized for status asthmaticus. Consistent with our hypothesis, we observed distinct metabolomic profiles in children with exacerbating versus stable asthma that were largely associated with alterations in arginine, phenylalanine, and glycine metabolism. Other pathways of interest and other high-confidence metabolites that could not be mapped to specific pathways but were also identified warrant further investigation. Interestingly, these identified metabolic alterations were not associated with systemic perturbations in classical type 1 (ie, IFN-γ) and type 2 (ie, IL-4, IL-5, and IL-13) cytokines. These results highlight the complex biology of inflammation in children with exacerbating asthma and argue for additional studies of the metabolic determinants of asthma exacerbations in children because many of the identified metabolites of interest may be amenable to targeted interventions.
Metabolomic studies in isolated plasma have previously been performed in children with asthma,31, 32, 33, 34 but to our knowledge, this is the first study to perform metabolomic analyses on children during an acute exacerbation. This is also one of very few studies to perform molecular characterization of hospitalized children with status asthmaticus. Whereas other pediatric metabolomic studies have primarily compared metabolites in children with asthma as a whole versus in healthy controls or other nonasthma populations,31, 32, 33, 34 this study utilized an asthma comparison group, which permitted study of the unique metabolites that are altered during an acute exacerbation necessitating hospitalization. Therefore, unlike other studies with healthy controls, our study did not identify eicosanoids (derived from oxidation of arachidonic acid or other polyunsaturated fatty acids),34 nicotinamide pathway metabolites (which regulate glycolysis, β-oxidation, and oxidative phosphorylation),32 or fatty acids (which drive inflammation through production of arachidonic acid, linoleic acid, eicosapentaenoic acid, and docosahexaenoic acid),31,33,34 even though our chromatographic technique was optimized for hydrophilic molecules. Nonetheless, our findings do have biologic plausibility. In our previous metabolomic analysis of children with severe asthma refractory to treatment with high-dose inhaled and systemic corticosteroids, children with severe asthma, compared with children with mild-to-moderate asthma, were differentiated by alterations in glycine, serine, and threonine metabolism, which are closely linked to arginine and proline metabolism and biosynthesis of valine, leucine, and isoleucine (branched-chain amino acids).35 We have also previously highlighted metabolomic perturbations in children with exacerbation-prone asthma (defined as those children having 3 or more exacerbations in the previous year) that were largely associated with arginine, lysine and methionine metabolism.23 In that study, comparison of the identified metabolite concentrations with reported values for healthy controls revealed that most of the metabolites were decreased in asthma, but significantly more so in children with exacerbation-prone asthma.23 Other studies have similarly identified disruptions in phenylalanine metabolism36 and histidine metabolism37 in children with severe asthma who failed to respond to systemic corticosteroids or omalizumab, respectively.
Arginine is a versatile amino acid that serves as a precursor for synthesis of nitric oxide, urea, creatine, polyamines, proline, glutamate, agmatine, and homoarginine. Metabolism of arginine is complex because multiple enzymes utilize arginine as a substrate. Furthermore, cells may contain multiple arginine pools, not all of which are accessible to arginine metabolic enzymes.38 Therefore, whether arginine metabolite concentrations are “increased” or “decreased” in children with asthma is difficult to assess and may be influenced by the chronicity of disease and current asthma control. For example, previous work in children with chronic, well-controlled asthma observed lower concentrations of arginine but significantly increased concentrations of ornithine and citrulline, which may represent a compensatory response.39 A separate study of children with exacerbating asthma presenting to an emergency department similarly noted decreased arginine concentrations and also lower citrulline concentrations compared with the concentrations in healthy controls.40 We also observed decreased plasma citrulline concentrations in otherwise stable children with exacerbation-prone asthma23 and children with obese asthma whose asthma was uncontrolled.41 However, other studies in young stable children with mild persistent asthma have shown no associations between arginine or its metabolites and features of asthma (such as allergic sensitization) or exacerbations over 48 weeks.42 Studies of older school-age children with asthma have also found no differences43 or increases44 in arginine metabolite concentrations compared with those in healthy controls.
The aromatic amino acid phenylalanine is considered inflammatory, with higher concentrations of phenylalanine reported in adults with coronary artery disease45,46 and infants at risk for cardiovascular disease with more frequent infections during the first year of life.47 Previous studies have shown that phenylalanine concentrations are higher in children with acute exacerbations of asthma than in healthy controls,48 in obese children with uncontrolled versus controlled asthma,41 and in children with severe asthma who failed to respond to systemic corticosteroid therapy.36 Likewise, imbalances in the glycine, serine, and threonine metabolism pathway are also considered inflammatory and may promote sustained oxidative stress. Serine is a precursor of cysteine, which is itself a precursor of glutathione. Disturbances in serine biosynthesis may account for alterations in the cysteine and glutathione redox balance that we have previously reported in children with severe refractory asthma.49,50 The present study did not detect cysteine or glutathione disulfide, prohibiting evaluation of their respective redox balances. However, we did identify methionine and 2 of its oxidation products (methionine sulfoxide and dehydromethionine), with methionine being a precursor of cysteine and glutathione. Methionine sulfoxide concentration was significantly higher in children with exacerbating asthma and may be associated with neutrophil-mediated lung damage, similar to what is seen in children with cystic fibrosis.51
Serine is also a critical reagent for the synthesis of sphingolipids, including sphingomyelins, which have a phosphocholine head group. We found that both serine and choline concentrations were decreased in children with exacerbating asthma, whereas the concentrations of 2 sphingomyelins were increased. Sphingolipids play key roles in regulation and function of immune cells52 and may drive lung inflammation.53 Although the precise roles of sphingolipids in asthma is unclear, decreased sphingolipid synthesis due to increased expression of the ORMDL3 (orosomucoid-like 3) gene, which inhibits serine palmitoyl transferase,54 has been implicated in increased susceptibility to wheezing and early-onset asthma.55, 56, 57, 58, 59 However, increased sphingolipid levels may drive asthma progression because higher levels of ceramides (in particular long-chain ceramides) and sphingosine-1-phosphate have been noted in adult patients with asthma,60, 61, 62, 63 especially in patients whose asthma is uncontrolled.62
Strengths of the study include characterization of the included patients, who were essentially all sensitized with mild airflow limitation and poor asthma control in the 4 weeks before enrollment, which permits generalization to a very large group of children with asthma encountered in both primary care and specialty settings. Another strength is the high-confidence annotations in our metabolites. Nonetheless, this study does have limitations. First, given the acute nature of children with exacerbating asthma, the findings in this group might result from hypoxia, infection, acidosis, and other factors rather than from asthma itself. Although our groups were clinically similar, some confounding by indication is also expected because the medications and interventions used for management of acute asthma differ. Therefore, the observations observed here might be an epiphenomenon and therefore require more validation. Second, pathway enrichment analyses were not performed on all metabolites because many of these could not be mapped to the KEGG database. These metabolites may be of clinical importance. For example, the dipeptide prolylleucine was the most significant single metabolite identified, yet it is not included in the KEGG metabolic pathways. Indeed, the dipeptide isoleucine/leucine-proline (isomeric with prolylleucine, and indistinguishable by our current method) has been negatively associated with FEV1 value and positively associated with lung damage and bronchiectasis in children with cystic fibrosis, potentially through the proteases such as neutrophil elastase.64,65 Furthermore, viral panels were not clinically indicated and were not performed in all exacerbating children. Therefore, we are unable to make inferences related to exacerbation triggers. However, in the subset of hospitalized children in whom viral panels were performed, a respiratory virus was identified in 61% of cases, with enterovirus and/or rhinovirus present in 95% of the positive samples, which is consistent with the results of other pediatric studies.66 We were also potentially limited by the cross-sectional analyses and cannot comment on the temporal stability of the metabolome in either group. Future studies of the same patient before and after hospital discharge would also be informative.
In summary, targeted metabolomic analyses permitted discrimination of children with exacerbating versus stable asthma, largely owing to alterations in arginine, phenylalanine, and glycine metabolism. Although independent validation studies are warranted, this work highlights the biologic complexity of asthma exacerbations in children but also offers some insight into individual metabolites and pathways that might be amenable to future intervention.
Key messages.
-
•
Children with exacerbating asthma have significant perturbations in their plasma metabolites that are largely associated with alterations in arginine, phenylalanine, and glycine metabolism.
-
•
The identified metabolic alterations are not associated with systemic perturbations in classical type 2 inflammation and highlight the complex biology of inflammation in children with exacerbating asthma.
-
•
Additional studies of the metabolic determinants of asthma exacerbations in children are warranted because many of the identified metabolites of interest may be amenable to targeted interventions.
Disclosure Statement
Supported by National Institutes of Health grants R01 NR018666, K24 NR018866, T32 HL116271, and UL 1TR002378.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Supplementary data
References
- 1.Most recent national asthma data. US Centers for Disease Control and Prevention. Atlanta, Ga. http://cdc.gov/asthma/most_recent_national_asthma_data.htm Available at.
- 2.Sullivan P.W., Ghushchyan V., Navaratnam P., Friedman H.S., Kavati A., Ortiz B., et al. The national burden of poorly controlled asthma, school absence and parental work loss among school-aged children in the United States. J Asthma. 2018;55:659–667. doi: 10.1080/02770903.2017.1350972. [DOI] [PubMed] [Google Scholar]
- 3.Grunwell J.R., Opolka C., Mason C., Fitzpatrick A.M. Geospatial analysis of social determinants of health identifies neighborhood hot spots associated with pediatric intensive care use for life-threatening asthma. J Allergy Clin Immunol Pract. 2022;10:981–991.e1. doi: 10.1016/j.jaip.2021.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papadopoulos N.G., Custovic A., Cabana M.D., Dell S.D., Deschildre A., Hedlin G., et al. Pediatric asthma: an unmet need for more effective, focused treatments. Pediatr Allergy Immunol. 2019;30:7–16. doi: 10.1111/pai.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray C.S., Jackson D.J., Teague W.G. Prevention and outpatient treatment of asthma exacerbations in children. J Allergy Clin Immunol Pract. 2021;9:2567–2576. doi: 10.1016/j.jaip.2021.03.035. [DOI] [PubMed] [Google Scholar]
- 6.Grunwell J.R., Travers C., Fitzpatrick A.M. Inflammatory and comorbid features of children admitted to a PICU for status asthmaticus. Pediatr Crit Care Med. 2018;19:e585–e594. doi: 10.1097/PCC.0000000000001695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Global strategy for asthma management and prevention. Updated 2022. Global Initiative for Asthma. http://www.ginasthma.org Available at:
- 8.Toussaint M., Jackson D.J., Swieboda D., Guedan A., Tsourouktsoglou T.D., Ching Y.M., et al. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat Med. 2017;23:681–691. doi: 10.1038/nm.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oikonomou N., Schuijs M.J., Chatzigiagkos A., Androulidaki A., Aidinis V., Hammad H., et al. Airway epithelial cell necroptosis contributes to asthma exacerbation in a mouse model of house dust mite-induced allergic inflammation. Mucosal Immunol. 2021;14:1160–1171. doi: 10.1038/s41385-021-00415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L., Netto K.G., Zhou L., Liu X., Wang M., Zhang G., et al. Single-cell transcriptomic analysis reveals the immune landscape of lung in steroid-resistant asthma exacerbation. Proc Natl Acad Sci U S A. 2021:118. doi: 10.1073/pnas.2005590118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.di Palmo E., Cantarelli E., Catelli A., Ricci G., Gallucci M., Miniaci A., et al. The predictive role of biomarkers and genetics in childhood asthma exacerbations. Int J Mol Sci. 2021;22:4651. doi: 10.3390/ijms22094651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wardzynska A., Pawelczyk M., Rywaniak J., Kurowski M., Makowska J.S., Kowalski M.L. Circulating MicroRNAs and T-cell cytokine expression are associated with the characteristics of asthma exacerbation. Allergy Asthma Immunol Res. 2020;12:125–136. doi: 10.4168/aair.2020.12.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams T.C., Jackson D.J., Maltby S., Walton R.P., Ching Y.M., Glanville N., et al. Rhinovirus-induced CCL17 and CCL22 in asthma exacerbations and differential regulation by STAT6. Am J Respir Cell Mol Biol. 2021;64:344–356. doi: 10.1165/rcmb.2020-0011OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khaenam P., Rinchai D., Altman M.C., Chiche L., Buddhisa S., Kewcharoenwong C., et al. A transcriptomic reporter assay employing neutrophils to measure immunogenic activity of septic patients' plasma. J Transl Med. 2014;12:65. doi: 10.1186/1479-5876-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhlbrigge A., Peden D., Apter A.J., Boushey H.A., Camargo C.A., Jr., Gern J., et al. Asthma outcomes: exacerbations. J Allergy Clin Immunol. 2012;129:S34–S48. doi: 10.1016/j.jaci.2011.12.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeatts K.B., Stucky B., Thissen D., Irwin D., Varni J.W., DeWitt E.M., et al. Construction of the Pediatric Asthma Impact Scale (PAIS) for the Patient-Reported Outcomes Measurement Information System (PROMIS) J Asthma. 2010;47:295–302. doi: 10.3109/02770900903426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathan R.A., Sorkness C.A., Kosinski M., Schatz M., Li J.T., Marcus P., et al. Development of the asthma control test: a survey for assessing asthma control. J Allergy Clin Immunol. 2004;113:59–65. doi: 10.1016/j.jaci.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Liu A.H., Zeiger R., Sorkness C., Mahr T., Ostrom N., Burgess S., et al. Development and cross-sectional validation of the Childhood Asthma Control Test. J Allergy Clin Immunol. 2007;119:817–825. doi: 10.1016/j.jaci.2006.12.662. [DOI] [PubMed] [Google Scholar]
- 19.Schatz M., Sorkness C.A., Li J.T., Marcus P., Murray J.J., Nathan R.A., et al. Asthma Control Test: reliability, validity, and responsiveness in patients not previously followed by asthma specialists. J Allergy Clin Immunol. 2006;117:549–556. doi: 10.1016/j.jaci.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Holguin F., Cardet J.C., Chung K.F., Diver S., Ferreira D.S., Fitzpatrick A., et al. Management of severe asthma: a European Respiratory Society/American Thoracic Society guideline. Eur Respir J. 2020;55 doi: 10.1183/13993003.00588-2019. [DOI] [PubMed] [Google Scholar]
- 21.Graham B.L., Steenbruggen I., Miller M.R., Barjaktarevic I.Z., Cooper B.G., Hall G.L., et al. Standardization of spirometry 2019 update. An official American Thoracic Society and European Respiratory Society technical statement. Am J Respir Crit Care Med. 2019;200:e70–e88. doi: 10.1164/rccm.201908-1590ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quanjer P.H., Stanojevic S., Cole T.J., Baur X., Hall G.L., Culver B.H., et al. Multi-ethnic reference values for spirometry for the 3-95-yr age range: the global lung function 2012 equations. Eur Respir J. 2012;40:1324–1343. doi: 10.1183/09031936.00080312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cottrill K.A., Stephenson S.T., Mohammad A.F., Kim S.O., McCarty N.A., Kamaleswaran R., et al. Exacerbation-prone pediatric asthma is associated with arginine, lysine, and methionine pathway alterations. J Allergy Clin Immunol. 2023;151:118–127. doi: 10.1016/j.jaci.2022.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juniper E.F., Bousquet J., Abetz L., Bateman E.D., Committee G. Identifying 'well-controlled' and 'not well-controlled' asthma using the Asthma Control Questionnaire. Respir Med. 2006;100:616–621. doi: 10.1016/j.rmed.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 25.RStudio: integrated development for R. RStudio Team; 2020. RStudio, PBC, Boston, MA. Available at: http://www.rstudio.com. Accessed September 12, 2022.
- 26.R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. https://www.R-project.org/ Available from.
- 27.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple hypothesis testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 28.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymanska E., Saccenti E., Smilde A.K., Westerhuis J.A. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics. 2012;8:3–16. doi: 10.1007/s11306-011-0330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crestani E., Harb H., Charbonnier L.M., Leirer J., Motsinger-Reif A., Rachid R., et al. Untargeted metabolomic profiling identifies disease-specific signatures in food allergy and asthma. J Allergy Clin Immunol. 2020;145:897–906. doi: 10.1016/j.jaci.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly R.S., Sordillo J.E., Lasky-Su J., Dahlin A., Perng W., Rifas-Shiman S.L., et al. Plasma metabolite profiles in children with current asthma. Clin Exp Allergy. 2018;48:1297–1304. doi: 10.1111/cea.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee-Sarwar K.A., Kelly R.S., Lasky-Su J., Zeiger R.S., O'Connor G.T., Sandel M.T., et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol. 2019;144:442–454. doi: 10.1016/j.jaci.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng P., Bian X., Zhai Y., Li C., Li N., Hao C., et al. Metabolomics reveals a correlation between hydroxyeicosatetraenoic acids and allergic asthma: evidence from three years' immunotherapy. Pediatr Allergy Immunol. 2021;32:1654–1662. doi: 10.1111/pai.13569. [DOI] [PubMed] [Google Scholar]
- 35.Fitzpatrick A.M., Park Y., Brown L.A., Jones D.P. Children with severe asthma have unique oxidative stress-associated metabolomic profiles. J Allergy Clin Immunol. 2014;133 doi: 10.1016/j.jaci.2013.10.012. 258-61.e1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park Y.H., Shi Y.P., Liang B., Medriano C.A., Jeon Y.H., Torres E., et al. High-resolution metabolomics to discover potential parasite-specific biomarkers in a Plasmodium falciparum erythrocytic stage culture system. Malar J. 2015;14:122. doi: 10.1186/s12936-015-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carraro S., di Palmo E., Licari A., Barni S., Caldarelli V., De Castro G., et al. Metabolomics to identify omalizumab responders among children with severe asthma: a prospective study. Allergy. 2022;77:2852–2856. doi: 10.1111/all.15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris S.M., Jr. Arginine metabolism revisited. J Nutr. 2016;146:2579S–2586S. doi: 10.3945/jn.115.226621. [DOI] [PubMed] [Google Scholar]
- 39.Kraj L., Krawiec M., Koter M., Grabon W., Kraj G., Cholojczyk M., et al. Altered L-arginine metabolism in children with controlled asthma. Allergy Asthma Proc. 2014;35:80–83. doi: 10.2500/aap.2014.35.3777. [DOI] [PubMed] [Google Scholar]
- 40.Morris C.R., Poljakovic M., Lavrisha L., Machado L., Kuypers F.A., Morris S.M., Jr. Decreased arginine bioavailability and increased serum arginase activity in asthma. Am J Respir Crit Care Med. 2004;170:148–153. doi: 10.1164/rccm.200309-1304OC. [DOI] [PubMed] [Google Scholar]
- 41.Fitzpatrick A.M., Mutic A.D., Mohammad A.F., Stephenson S.T., Grunwell J.R. Obesity is associated with sustained symptomatology and unique inflammatory features in children with asthma. J Allergy Clin Immunol Pract. 2022;10:815–826.e2. doi: 10.1016/j.jaip.2021.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris C.R., Mauger D.T., Suh J.H., Phipatanakul W., Sheehan W.J., Moy J.N., et al. Glutathione and arginine levels: predictors for acetaminophen-associated asthma exacerbation? J Allergy Clin Immunol. 2018;142:308–311.e9. doi: 10.1016/j.jaci.2018.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanusch B., Sinningen K., Brinkmann F., Dillenhofer S., Frank M., Jockel K.H., et al. Characterization of the L-arginine/nitric oxide pathway and oxidative stress in pediatric patients with atopic diseases. Int J Mol Sci. 2022;23:2136. doi: 10.3390/ijms23042136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matysiak J., Klupczynska A., Packi K., Mackowiak-Jakubowska A., Breborowicz A., Pawlicka O., et al. Alterations in serum-free amino acid profiles in childhood asthma. Int J Environ Res Public Health. 2020;17:4758. doi: 10.3390/ijerph17134758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deidda M., Noto A., Cadeddu Dessalvi C., Andreini D., Andreotti F., Ferrannini E., et al. Metabolomic correlates of coronary atherosclerosis, cardiovascular risk, both or neither. Results of the 2 x 2 phenotypic CAPIRE study. Int J Cardiol. 2021;336:14–21. doi: 10.1016/j.ijcard.2021.05.033. [DOI] [PubMed] [Google Scholar]
- 46.Di Marino S., Viceconte N., Lembo A., Summa V., Tanzilli G., Raparelli V., et al. Early metabolic response to acute myocardial ischaemia in patients undergoing elective coronary angioplasty. Open Heart. 2018;5 doi: 10.1136/openhrt-2017-000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansell T., Saffery R., Burugupalli S., Ponsonby A.L., Tang M.L.K., O'Hely M., et al. Early life infection and proinflammatory, atherogenic metabolomic and lipidomic profiles in infancy: a population-based cohort study. Elife. 2022;11 doi: 10.7554/eLife.75170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J., Li X., Liu X., Wang X., Li J., Lin K., et al. Untargeted metabolomic study of acute exacerbation of pediatric asthma via HPLC-Q-Orbitrap-MS. J Pharm Biomed Anal. 2022;215 doi: 10.1016/j.jpba.2022.114737. [DOI] [PubMed] [Google Scholar]
- 49.Fitzpatrick A.M., Stephenson S.T., Hadley G.R., Burwell L., Penugonda M., Simon D.M., et al. Thiol redox disturbances in children with severe asthma are associated with posttranslational modification of the transcription factor nuclear factor (erythroid-derived 2)-like 2. J Allergy Clin Immunol. 2011;127:1604–1611. doi: 10.1016/j.jaci.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephenson S.T., Brown L.A., Helms M.N., Qu H., Brown S.D., Brown M.R., et al. Cysteine oxidation impairs systemic glucocorticoid responsiveness in children with difficult-to-treat asthma. J Allergy Clin Immunol. 2015;136:454–461.e9. doi: 10.1016/j.jaci.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandler J.D., Margaroli C., Horati H., Kilgore M.B., Veltman M., Liu H.K., et al. Myeloperoxidase oxidation of methionine associates with early cystic fibrosis lung disease. Eur Respir J. 2018;52 doi: 10.1183/13993003.01118-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luthers C.R., Dunn T.M., Snow A.L. ORMDL3 and asthma: linking sphingolipid regulation to altered T cell function. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.597945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maceyka M., Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caliskan M., Bochkov Y.A., Kreiner-Moller E., Bonnelykke K., Stein M.M., Du G., et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loss G.J., Depner M., Hose A.J., Genuneit J., Karvonen A.M., Hyvarinen A., et al. The early development of wheeze. environmental determinants and genetic susceptibility at 17q21. Am J Respir Crit Care Med. 2016;193:889–897. doi: 10.1164/rccm.201507-1493OC. [DOI] [PubMed] [Google Scholar]
- 57.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 58.Galanter J., Choudhry S., Eng C., Nazario S., Rodriguez-Santana J.R., Casal J., et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torgerson D.G., Ampleford E.J., Chiu G.Y., Gauderman W.J., Gignoux C.R., Graves P.E., et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinke S.N., Gallart-Ayala H., Gomez C., Checa A., Fauland A., Naz S., et al. Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J. 2017;49 doi: 10.1183/13993003.01740-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bahlas S., Damiati L.A., Al-Hazmi A.S., Pushparaj P.N. Decoding the role of sphingosine-1-phosphate in asthma and other respiratory system diseases using next generation knowledge discovery platforms coupled with luminex multiple analyte profiling technology. Front Cell Dev Biol. 2020;8:444. doi: 10.3389/fcell.2020.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S.H., Jung H.W., Kim M., Moon J.Y., Ban G.Y., Kim S.J., et al. Ceramide/sphingosine-1-phosphate imbalance is associated with distinct inflammatory phenotypes of uncontrolled asthma. Allergy. 2020;75:1991–2004. doi: 10.1111/all.14236. [DOI] [PubMed] [Google Scholar]
- 63.Choi Y., Kim M., Kim S.J., Yoo H.J., Kim S.H., Park H.S. Metabolic shift favoring C18:0 ceramide accumulation in obese asthma. Allergy. 2020;75:2858–2866. doi: 10.1111/all.14366. [DOI] [PubMed] [Google Scholar]
- 64.Esther C.R., Jr., Turkovic L., Rosenow T., Muhlebach M.S., Boucher R.C., Ranganathan S., et al. Metabolomic biomarkers predictive of early structural lung disease in cystic fibrosis. Eur Respir J. 2016;48:1612–1621. doi: 10.1183/13993003.00524-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinn R.A., Adem S., Mills R.H., Comstock W., DeRight Goldasich L., Humphrey G., et al. Neutrophilic proteolysis in the cystic fibrosis lung correlates with a pathogenic microbiome. Microbiome. 2019;7:23. doi: 10.1186/s40168-019-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hossain F.M.A., Choi J.Y., Uyangaa E., Park S.O., Eo S.K. The interplay between host immunity and respiratory viral infection in asthma exacerbation. Immune Netw. 2019;19:e31. doi: 10.4110/in.2019.19.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.