Abstract
The prognostic value of the traditional pathologic parameters that form part of the American Joint Committee on Cancer staging system and genetic classifications using monosomy chromosome 3 and structural alterations in chromosome 8 are well established and are part of the diagnostic workup of uveal melanoma (UM). However, it has not been fully clarified whether nuclear protein expression of the tumor suppressor gene BAP1 (nBAP1) by immunohistochemistry alone is as powerful a predictor of overall survival (OS) and/or disease-specific survival (DSS) as chromosome analysis. The protein expression of nBAP1 was evaluated in a retrospective cohort study of 308 consecutive patients treated by primary enucleation between January 1974 and December 2022. We correlated clinical, pathologic, and cytogenetic characteristics to identify the best prognostic indicators for OS and DSS. Loss of nBAP1 was detected in 144/308 (47%) of patients. Loss of nBAP1 expression was significantly associated with poor survival. In patients with disomy chromosome 3, nBAP1 negative is significantly associated with poorer OS but not DSS. We observed that older age (>63 years), presence of metastasis, and nBAP1 negative remained independent prognostic factors in multivariate analysis. nBAP1 protein expression proved to be a more reliable prognostic indicator for OS than the American Joint Committee on Cancer staging, M3 status, or The Cancer Genome Atlas classification in this cohort. This study provides support for accurate prognostication of UM patients in routine histology laboratories by immunohistochemistry for nBAP1 alone.
Key Words: BAP1 protein, immunohistochemistry, prognostic factor, uveal melanoma
Uveal melanoma (UM) is the most common intraocular cancer in adults, encompassing 3% to 5% of all melanomas. UM can arise from melanocytes of the choroid (85% to 90%), iris (3% to 5%), and ciliary body (5% to 8%). Genetic mutations in GNAQ and GNA11 genes are frequently observed, and prognosis is based on chromosomal abnormalities, including monosomy chr 3, amplification of chr 8q, loss of chr 8p, 6q, 1p loss, and the mutation status of BAP1, SF3B1, and EIF1AX.1 Despite the advances in local disease control, ~50% of UM patients will ultimately develop metastasis with a median overall survival (OS) of 1.07 years.2,3 In the great majority of cases, the liver is the first site of metastatic disease. Most patients with primary UM will be enrolled in surveillance programs of regular hepatic imaging. The treatment for metastatic UM, when diagnosed, is, however, highly unsatisfactory, and the disease is, with rare and anecdotal exceptions, invariably fatal. The median survival is <1 year.4 For patients who express HLA A2.01, tebentafusp (an immune-mobilizing monoclonal T-cell receptor against cancer) produces a modest improvement in OS.5 Immune checkpoint inhibitors are much less active than in cutaneous melanoma. Liver-directed therapies have been shown to enhance hepatic disease control but have little impact on OS.6
There are several recognized prognostic factors. Tumor size and depth are related to metastatic risk and, together with other pathologic parameters, are routinely assessed in anatomic pathology reports in the TNM American Joint Committee on Cancer (AJCC) classification.7,8 There has been considerable interest in biomarkers. It has long been recognized that monosomy of chromosome 3 (M3) imparts a worse prognosis in UM.9 Monosomy or loss of chromosome 3, as well as gain or loss in either or both arms of chromosome 8, are associated with disease spread and death from metastasis.10,11 The Cancer Genome Atlas (TCGA) project has provided valuable insights into the genetic makeup of UM, helping to categorize it into 4 main groups that differ in prognosis. The TCGA system classifies tumors based on the disomy or not of chromosomes 3 and 8. It has been postulated by some studies that when genetic testing is available, the TCGA classification is a more accurate way to identify patients at high risk of metastasis who might benefit from adjuvant therapy than the classic staging of tumors using the TNM classification of the AJCC system.12 Gelmi et al13 showed that the combination of both TCGA and AJCC enhances prognostic precision.
BAP1 (BRCA1-associated protein) is a tumor suppressor gene involved in DNA damage repair, cell cycle control, chromatin modification, programmed cell death, and the immune response.14 Harbour et al15 found that the co-occurrence of M3 together with inactivating mutations of BAP1 on the remaining copy of Chr 3 is associated with the development of metastases. The germline BAP1 tumor predisposition syndrome (BAP1-TDS) is associated with increased occurrence of UM, cutaneous melanoma, renal cell carcinoma, and mesothelioma. UM is the most common and earliest presenting malignancy.16,17 van Essen et al18 speculated that the inactivation of BAP1 gene may be a main driving force for metastasis, and BAP1 levels influence survival. Koopmans et al19 have shown a strong correlation between BAP1 protein expression and genetic sequencing data. They showed that both variables predict poor survival in a study of 74 cases of UM and highlight BAP1 immunohistochemistry (IHC) as a good surrogate for BAP1 mutation status.
The aim of this study is to assess BAP1 protein expression as a prognostic indicator in a large series of UM tumors treated by primary enucleation and to identify if BAP1 protein expression can separate subgroups with different survival outcome in the group of tumors. We further aimed to investigate whether BAP1 protein expression alone is sufficient to assess the risk clinically associated with genetic changes in chromosomes 3 and 8/TCGA classification and AJCC as prognostic indicators to allow histology laboratories to provide a rapid and low-cost way of providing valuable clinical information to guide clinical follow-up and entry onto therapeutic drug trials.
MATERIALS AND METHODS
Study Design
This retrospective study of 308 patients with a confirmed UM diagnosis with long-term follow-up was conducted in accordance with the Declaration of Helsinki, and Institutional Review Board approval was obtained from The Royal Victoria Eye and Ear Hospital, Dublin, Ireland. Patient histology samples were accessed by the National Ophthalmic Pathology Laboratory, Royal Victoria Eye and Ear Hospital, between 1974 and 2022. Clinical, histopathological, and molecular data were reviewed using charts and patient information systems. Exclusion criteria included insufficient clinical data or tissue or pretreatment with radiation therapy.
The clinicopathological features evaluated include patient age, sex, greatest tumor diameter (T1 to T4) of TNM AJCC subcategory20 tumor cell type using Callender classification.21 Cytogenetic data obtained consisted of FISH study results for Chr 3 and 8. These results were used to categorize the cases based on the TCGA classification: class A: melanoma with disomy chromosome 3 and 8; class B: disomy of chromosome 3 and chromosome 8q gain; class C: monosomy of chromosome 3 and chromosome 8q gain and class D: monosomy of chromosome 3 and multiple 8 q gains. We noted that this TCGA classification does not include a category for those cases exhibiting monosomy 3 and normal 8q status. For this study, we categorized those cases as class E. BAP1 IHC results were either obtained from historically stained BAP1 IHC slides or from UM tissue microarrays (TMA).
TMA Creation and BAP1 Immunohistochemical Scoring
Eighty-three whole sections and 225 histologically confirmed UM specimens were used to construct UM-TMA using the Manual Tissue Arrayer from Beecher Instruments (Beecher Instruments). Triplicate 0.6 mm cores from each tumor were distributed across the TMA alongside non-tumor tonsil reference samples. The minimum requirement for scoring samples was one assessable core. The Ventana Benchmark ultra fully automated staining system (Ventana Medical systems Inc., Tucson) with an alkaline phosphatase red detection kit was used. The tests were performed in a clinical laboratory accredited under EU standard (ISO 15189). Sections were deparaffinized and heated using heat-induced epitope retrieval for 64 minutes at 97°C; the sections were then incubated for 32 minutes at 37°C with the primary BAP1 antibody (sc-28383, Santa Cruz Biotechnology) at a concentration of 1:50. Target amplification was performed, followed by incubation with hematoxylin 11 (Harris hematoxylin, GCC diagnostics, Flints UK catalog no. S0500) counterstain for 8 minutes, an additional counterstain was performed with blueing reagent (Ventana Medical Systems Inc.).
BAP1-stained UM-TMA slides were evaluated for nuclear (nBAP1) and cytoplasmic (cBAP1) BAP1 protein expression. Specimens were scored based on a binary system of positive or negative nuclear staining as the staining was either strongly positive in >50% of tumor cells or entirely negative. The localization of cytoplasmic rather than nuclear staining of tumor cells was recorded as cBAP1 positive (Supplementary Figure 1, Supplemental Digital Content 1, http://links.lww.com/PAS/B739). Testis was used as positive control. Normal cells such as intratumoral vessels, retina cells, and inflammatory cells, provided in-built positive controls.
Cytogenetic FISH Analysis
Fish analysis using the Vysis Cep3 probe (specific for the centromeric region of chromosome 3) was analyzed on 100 cells. FISH analysis using the Zytolight CEN8/MYC Dual Colour probe (specific for the centromeric region of chromosome 8 and MYC [8q24]) was examined in 100 cells. The methods were as previously described.22 These tests were performed in a clinically accredited laboratory.
Data and Statistical Analysis
Descriptive statistics were used to document the clinical and histopathological factors of patients with UM. Continuous variables such as age were dichotomized by their median. All nominal data were analyzed as binary variables. Comparisons between nBAP1 positive and negative groups were performed by using the 2-sided χ2 test for the detection of statistical significance. A P-value <0.05 was considered statistically significant.
OS was calculated from the date of diagnosis/enucleation to the date of last follow-up or date of death for any cause. Disease-specific survival (DSS) was calculated from the date of diagnosis/enucleation to the date of last follow-up or the date of UM cancer-specific death. The cause of death was established by medical records, primary care physician, or national death registry. The Kaplan-Meier survival estimator method was applied to calculate OS and DSS, and the log-rank test was used for the assessment of statistical significance. Univariate Cox proportional hazard (PH) analyses were used to determine independent prognostic predictors of OS and DSS. Variables with a significant association (P<0.05) in the OS univariate analysis were analyzed with multivariate Cox regression. Stepwise forward regression was used for developing the multivariate Cox PH model at a 5% threshold level to find prognostic covariates correlated with OS. Thereafter, the Cox PH assumption was tested. All analyses were carried out using Stata v17.
RESULTS
Patient Demographics and Clinical Characteristics
Three hundred eight patients from the National Ophthalmic Pathology Laboratory’s database were analyzed, and clinical features are described in Table 1. The group consisted of 182 males and 126 females with a median age of 63 (range 11 to 96 years). As per the AJCC staging system, tumor size of T1 (10 mm or less and 2.5 mm in thickness) was observed in 53 (17%) cases; T2 (>10 mm <16 mm) in 62 (20%) cases; T3 (>16 mm and or>10 mm in height, no extraocular extension) in 132 (43%) of cases; T4 (>16 mm or >10 mm in height or extraocular extension) in 61 (20%) of cases. Histologic examination revealed 124 (40%) of the tumors analyzed were of the spindle cell type, 71 (23%) were epithelioid, and 113 (37%) were mixed cell type.
TABLE 1.
Clinical and Histopathological Features of Uveal Melanoma Cohort
| Clinical feature | Number (n=308) (%) |
|---|---|
| Sex | |
| Male | 182 (59) |
| Female | 126 (41) |
| Age [y] | |
| Median [range] | 63 [11–96] (51) |
| <63 y | 156 (49) |
| >63 y | 152 |
| Cell type | |
| Mixed | 113 (37) |
| Spindle | 124 (40) |
| Epithelioid | 71 (23) |
| TNM classification | |
| pT1 | 53 (17) |
| pT2 | 62 (20) |
| pT3 | 132 (43) |
| pT4 | 61 (20) |
| Metastasis | |
| No | 184 (60) |
| Yes | 124 (40) |
| Chromosome 3 | |
| Disomy | 112 (56) |
| Monosomy | 88 (44) |
| Chromosome 8q | |
| Disomy | 90 (46) |
| Gain | 106 (54) |
| TCGA staging system | |
| D3/D8 | 73 (36) |
| D3/8qG | 42 (21) |
| M3/8qG | 32 (16) |
| M3/8q multiple | 31 (15) |
| M3/D8 | 25 (12) |
| Outcome | |
| No evidence of disease | 137 (45) |
| Alive with disease | 16 (5) |
| Died of disease | 110 (36) |
| Died of other causes | 45 (15) |
| Follow-up [mo] | |
| Median [range] | 56 [1.6-557] |
BAP1 IHC data were available for all 308 cases. Nuclear BAP1 expression (nBAP1) was detected in 164 (53%) of cases, whereas 144 (47%) were nBAP1 negative. Cytoplasmic BAP1 (cBAP1) was observed in 30/144 (21%) of nBAP1-negative cases. One hundred twenty-four (40%) cases developed metastases, of which 51 (41%) were nBAP1 positive and 73 (59%) were nBAP1 negative.
FISH analysis available in 200 cases found chromosome 3 monosomy (M3) in 88 cases (44%) and 112 (56%) disomy (D3). Analysis of chromosome 8q revealed a gain (8qG) in 106 cases (54%) and disomy (D8) in 90 (46%) of 196 cases available.
In the TCGA classification system, 73/203 (36%) UM were classified as A (D3/D8), 42/203 (21%) UM as B (D3/8qG), 32/203 (16%) as C (M3/8qG), and 31/203 (15%) D (M3/8qG multiple). The TGCA classes do not include a category of M3/D8. In our series 25/203 (12%) patients were in this category.
The median follow-up time for patients was 56 months (range 1.6 to 557). Survival outcome was determined as 137 (45%) with no evidence of disease; 16 (5%) were alive with disease, 110 (36%) died of disease, and 45 (15%) died of other causes.
Association Between nBAP1 Expression and Clinicopathological Features in UM
We assessed the association of nBAP1-positive and nBAP1-negative expression with relevant clinicopathological features (Table 2). There was no significant difference in age, sex, TNM classification, and follow-up time between patients with nBAP1-positive and nBAP1-negative expression. Significant association was observed between spindle cell type (P=0.005), metastasis (P<0.001), chromosome 3 monosomy (P<0.001), chromosome 8q gain (P=0.004), and outcome (P<0.001).
TABLE 2.
Association of Clinicopathological Features Stratified by nBAP1-positive Compared With nBAP1-negative Expression
| nBAP1 positive (n=164) | nBAP1 negative (n=144) | P | |
|---|---|---|---|
| Median age, years [range] | 61 [11–88] | 66.5 [11–96] | 0.381 |
| Sex | |||
| Male (n=182) | 104 | 78 | 0.100 |
| Female (126) | 60 | 66 | |
| Cell type | |||
| Mixed | 53 | 60 | 0.005 |
| Spindle | 80 | 44 | |
| Epithelioid | 31 | 40 | |
| TNM classification | |||
| pT1 | 33 | 20 | 0.138 |
| pT2 | 38 | 24 | |
| pT3 | 65 | 67 | |
| pT4 | 28 | 33 | |
| Metastasis | |||
| No | 113 | 71 | <0.001 |
| Yes | 51 | 73 | |
| Chromosome 3 | |||
| Disomy | 83 | 29 | <0.001 |
| Monosomy | 27 | 61 | |
| Chromosome 8q | |||
| Disomy | 60 | 30 | 0.004 |
| Gain | 49 | 57 | |
| TCGA staging system | |||
| D3/D8 | 56 | 17 | <0.001 |
| D3/8qG | 29 | 13 | |
| M3/8qG | 11 | 21 | |
| M3/8q multiple | 8 | 23 | |
| M3/D8 | 8 | 17 | |
| Outcome | |||
| No evidence of disease | 90 | 47 | <0.001 |
| Alive with disease | 12 | 4 | |
| Died of disease | 42 | 68 | |
| Died of other causes | 20 | 25 | |
| Median follow-up [mo] | 64 [1.6-357] | 49 [2.5-557] | 0.385 |
Prognostic Role of nBAP1 Expression in UM
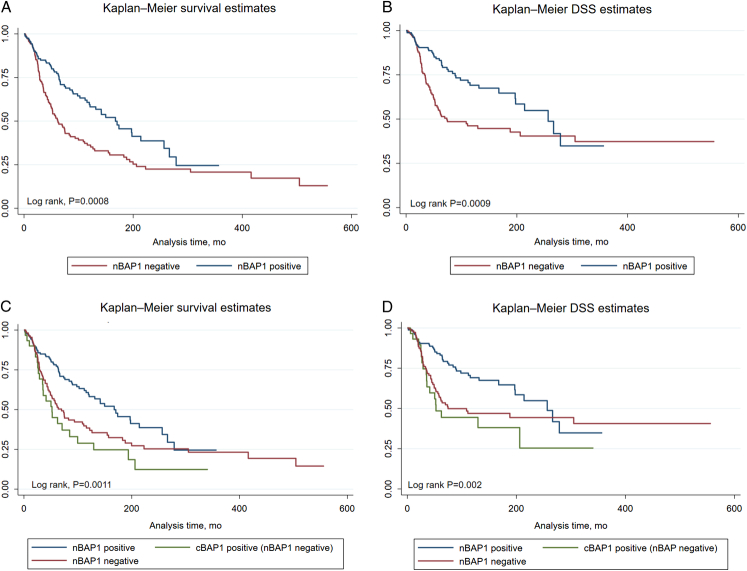
The prognostic role of nBAP1 expression by IHC was investigated. Kaplan-Meier OS and DSS curves of nBAP1-positive (n=164) with nBAP1-negative expression (n=144) highlight that nBAP1 negative is significantly associated with poor OS and DSS (Log rank, P=0.0008 and 0.0009) (Figs. 1A, B). Further detailed analysis shows the poorest OS and DSS in patients with cBAP1 positive (nBAP1 negative) and nBAP1 negative compared with nBAP1 positive (Log rank, P=0.0011 and 0.002) (Figs. 1C, D). There was no significant difference in OS between cBAP1 positive (nBAP1 negative) and nBAP1 negative (Log rank, P=0.237).
FIGURE 1.
Kaplan-Meier OS and DSS curves stratified by (A and B) nBAP1-positive and nBAP1-negative expression and (C and D) nBAP1-positive, nBAP1-negative, and cBAP1-positive (nBAP1 negative) expression by IHC. Log-rank tests were used to compare OS and DSS across groups. DSS indicates disease-specific survival; OS, overall survival.
Prognostic Role of nBAP1 Expression in UM
We stratified patients based on their Chr 3 disomy or monosomy status and assessed the impact of nBAP1 expression on OS and DSS. nBAP1 expression does not significantly impact the OS of patients with Chr3 monosomy (Log rank P=0.060); however, the median OS is 174 months versus 49 months for nBAP1 positive versus nBAP1 negative, indicating a better OS for patients expressing nBAP1 with M3 (Fig. 2A). The same trend is observed for DSS (Log rank, P=0.03) (Fig. 2B). Patients with D3 and nBAP1 negative had significantly poorer OS compared with those with nBAP1 positive, demonstrating the impact of nBAP1 expression in D3 patients (Log rank, P=0.033) (Fig. 2C); however, D3 status had no impact on DSS in patients with nBAP1-positive or nBAP1-negative expression (Log rank, P=0.194) (Fig. 2D).
FIGURE 2.
Impact of nBAP1 expression based on chromosome 3 status. Kaplan-Meier curves in patients stratified based on chromosome 3 monosomy (A) OS (B) DSS, and Kaplan-Meier curves in patients stratified based on chromosomal 3 disomy (C) OS and (D) DSS and nBAP1 expression. DSS indicates disease-specific survival; OS, overall survival.
Univariate analysis was performed to identify significant clinical and histopathological factors associated with OS and DSS. In this analysis, spindle cell type, AJCC pT1, and TCGA Class A were positive indicators for improved OS and DSS, whereas cell type (mixed and epithelioid), AJCC pT3 and pT4, TCGA class D, metastasis, Chr8qG, Chr3 monosomy, nBAP1 negative, and age>63 years were significantly associated with poorer OS and DSS (Figs. 3A, B; Supplementary Table 1, Supplemental Digital Content 2, http://links.lww.com/PAS/B740). Multivariate stepwise regression analysis revealed that age above 63 years (hazard ratio [HR] 1.74; 95% CI [1.05-2.86]; P=0.029), nBAP1 negative (HR 2.04; 95% CI [1.24-3.33]; P=0.004), and metastasis (HR 8.51; 95% CI [4.71-15.41]; P<0.001) were independent variables significantly associated with OS (Supplementary Table 1, Supplemental Digital Content 2, http://links.lww.com/PAS/B740; and Supplementary Table 2, Supplemental Digital Content 3, http://links.lww.com/PAS/B741).
FIGURE 3.
Forest plot of the univariate Cox regression analysis for overall survival (A) and disease-specific survival (B) in all patients.
DISCUSSION
Current prognostic methods for identifying the 50% of UM patients who will metastasize depend on histologic parameters and abnormalities of chromosome 3 and 8, the latter either alone or in combination with TCGA genetic classification. However, these require cytogenetics and molecular analyses. In this study, we aim to define a prognostic role for nBAP1 protein expression assessment via rapid IHC in a cohort of 308 patients with extended follow-up data.
BAP1 is a chromatin-associated protein located in the nucleus of the cell when assessed by IHC.23 Mutations in the nuclear localization signal or the ubiquitin carboxyl-terminal hydrolase domain of the gene will impair its ability to localize to the nucleus and inactivate BAP1’s tumor suppressor functional ability.24,25 Negative nBAP1 immunohistochemical staining has been reported to be a reliable surrogate for BAP1 gene mutations.4 High-risk UM phenotypes are generally linked to mutated BAP1 or loss of BAP1 expression in the nucleus.18 Harbour et al15 previously described the central role of both germline and somatic BAP1 inactivating gene mutations linked to the development of metastasis in UM patients. BAP1 mutations have been reported in ~30% to 40% of UM patients and in over 80% of metastatic UM patients.14,15,26 Loss of nBAP1 protein expression by IHC has been shown to correlate with BAP1 mutation in the prediction of metastatic risk27,28; however, nongenetic mechanisms are associated with nBAP1 loss in a small subset of patients.29
We observed that nBAP1 protein expression was lost in 47% of all cases, with M3 observed in 68% and a 51% incidence of metastasis among these patients. Loss of nBAP1 was significantly associated with poorer OS and DSS. Although the Kaplan-Meier plots clearly dichotomize nBAP1 negative with poorer OS and DSS, the graphs converge after extended follow-up time of 25 years (300 months). This shows the possible role of other factors, such as the SF3B1 and EIF1AX genes, which are associated with the development of late metastases in patients with D3 status.30
In our study, we observed a total of 17% (53/308) of patients with AJCC pT1, of which 17% (9/53) developed metastasis, with 55% (4/9) of these nBAP1 negative. This is in line with other studies such as See et al31 but higher than observed by Mazloumi et al12 where they found that only 3.2% of T1 patients developed metastasis; however, our study has considerably longer follow-up with a median follow-up for pT1 of 119.7 versus 44.9 months.
Interestingly, we observed some cases with positive cytoplasmic BAP1 (cBAP1) with loss of nBAP1 expression as in other studies.29 Although our numbers were small 9.7% (30/308), these patients had worse OS and DSS with a median DSS of 52.5 versus 74.6 months for nBAP1 negative; however, this was not significant. See et al31 found that cBAP1 expression was nonsignificantly higher in gene expression profiling (GEP) class 2 but had no impact on the prediction of metastasis.
In our study, we found that nBAP1 loss was associated with M3 in our cohort. In patients with M3, nBAP1 protein expression status did not significantly affect OS, but nBAP1 loss was significantly associated with poorer DSS, a similar finding to Farquhar et al.29 Importantly, D3 tumors resulted in significantly worse OS but not DSS in patients with nBAP1 negative compared with nBAP1 positive. Our findings agree with previously published studies highlighting BAP1 protein expression as an efficient method of determining survival disadvantage.19,27–29
Many recent publications have shown that combining information from AJCC and chromosomal status provides greater accuracy than either system independently.13,32–34 Mazloumi et al12 compared the TCGA molecular classification and AJCC tumor staging system in 642 patients with choroidal and ciliary UM enrolled in their center over a 10-year period. Both systems correlated well, with more advanced tumors having a worse chromosomal profile. However, the TCGA classification was found to be superior to AJCC in predicting 5-year risk of distant metastases. In our retrospective study with long-term follow-up, we found that AJCC TNM staging, Chr 3 and Chr 8 status separately and combined as TCGA subgroups, nBAP1 protein expression, older age (>63 years), presence of metastasis, and cell type were all significantly associated with OS in univariate analysis. Of these, only nBAP1-negative protein expression, older age, and presence of metastasis remained significant in multivariate analysis.
Harbour35 developed a 15 GEP, which was able to accurately distinguish between low metastatic risk (class 1 tumors) and high metastatic risk (class 2 tumors), providing a significant improvement in prognostic accuracy over Chr 3 status. They found that Chr 3 or BAP1 status alone did not provide prognostic information that was independent of GEP. Subsequently, additional studies have reported that nBAP1 protein expression correlates with GEP classification and is as accurate for predicting metastasis.18,31,36
Our findings indicate that BAP1 protein expression can be used to identify patients who are more likely to experience poorer OS or DSS, and may benefit from more intensive surveillance, possibly with cell free DNA (cfDNA) studies of BAP1 mutations.
CONCLUSIONS
The aim of this study was to determine how BAP1 protein expression impacts long-term survival outcomes for UM patients. The results of this study confirm the importance of BAP1 protein status in disease progression. Ascertaining a patient’s BAP1 status using IHC provides important prognostic information. Extended OS and DSS of UM patients are shown to be much more likely for a patient who has positive nBAP1 protein expression. We found that nBAP1 protein expression and routine histology are better predictors of patient outcome than molecular genetic testing using FISH assessment of Chr 3 and Chr 8 status. This has important implications for surveillance and treatment planning.
It is also very useful for the efficient clinical management of patients and pathology budget planning to have evidence that routine and rapid IHC provides better prognostic information than expensive and not readily available cytogenetics and molecular genetic analysis.
Supplementary Material
Footnotes
This work was supported by funding from the Cancer Clinical Research Trust (CCRT) and the Research Foundation of the Royal Victoria Eye and Ear Hospital, Dublin.
Conflicts of Interest and Source of Funding: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.ajsp.com.
Contributor Information
Susan Kennedy, Email: Susan.Kennedy@rveeh.ie.
Sally Owens, Email: sally.owens7@mail.dcu.ie.
Laura Ivers, Email: laura.ivers@dcu.ie.
Ciara Hegarty, Email: ciara.hegarty25@mail.dcu.ie.
Valerie O’Neill, Email: Valerie.ONeill@rveeh.ie.
Jose J. Berenguer-Pina, Email: jpina@svhg.ie.
Noel Horgan, Email: noelhorgan@gmail.com.
John Crown, Email: john.crown@ccrt.ie.
Naomi Walsh, Email: naomi.walsh@dcu.ie.
REFERENCES
- 1.Robertson AG, Shih J, Yau C, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahendraraj K, Lau CS, Lee I, et al. Trends in incidence, survival, and management of uveal melanoma: a population-based study of 7,516 patients from the Surveillance, Epidemiology, and End Results database (1973-2012). Clin Ophthalmol. 2016;10:2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rantala ES, Hernberg M, Kivelä TT. Overall survival after treatment for metastatic uveal melanoma: a systematic review and meta-analysis. Melanoma Res. 2019;29:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–1885. [DOI] [PubMed] [Google Scholar]
- 5.Nathan P, Hassel JC, Rutkowski P, et al. Overall survival benefit with tebentafusp in metastatic uveal melanoma. N Engl J Med. 2021;385:1196–1206. [DOI] [PubMed] [Google Scholar]
- 6.Rowcroft A, Loveday BPT, Thomson BNJ, et al. Systematic review of liver directed therapy for uveal melanoma hepatic metastases. HPB. 2020;22:497–505. [DOI] [PubMed] [Google Scholar]
- 7.Kujala E, Damato B, Coupland SE, et al. Staging of ciliary body and choroidal melanomas based on anatomic extent. J Clin Oncol. 2013;31:2825–2831. [DOI] [PubMed] [Google Scholar]
- 8.Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. [DOI] [PubMed] [Google Scholar]
- 9.Prescher G, Bornfeld N, Hirche H, et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. [DOI] [PubMed] [Google Scholar]
- 10.van Beek JGM, Koopmans AE, Vaarwater J, et al. The prognostic value of extraocular extension in relation to monosomy 3 and gain of chromosome 8q in uveal melanoma. Invest Ophthalmol Vis Sci. 2014;55:1284–1291. [DOI] [PubMed] [Google Scholar]
- 11.Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Genomic profile of 320 uveal melanoma cases: chromosome 8p-loss and metastatic outcome. Invest Ophthalmol Vis Sci. 2013;54:5721–5729. [DOI] [PubMed] [Google Scholar]
- 12.Mazloumi M, Vichitvejpaisal P, Dalvin LA, et al. Accuracy of The Cancer Genome Atlas Classification vs American Joint Committee on Cancer Classification for prediction of metastasis in patients with uveal melanoma. JAMA Ophthalmol. 2020;138:260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelmi MC, Bas Z, Malkani K, et al. Adding the cancer genome atlas chromosome classes to American Joint Committee on cancer system offers more precise prognostication in uveal melanoma. Ophthalmology. 2022;129:431–437. [DOI] [PubMed] [Google Scholar]
- 14.Louie BH, Kurzrock R. BAP1: not just a BRCA1-associated protein. Cancer Treat Rev. 2020;90:102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbour JW, Onken MD, Roberson EDO, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilarski R, Carlo MI, Cebulla C, et al. BAP1 tumor predisposition syndrome In: Adam MP, Mirzaa GM, Pagon RA, et al., eds. GeneReviews. University of Washington; 2016. [PubMed] [Google Scholar]
- 17.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Essen TH, van Pelt SI, Versluis M, et al. Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol. 2014;98:1738–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koopmans AE, Verdijk RM, Brouwer RWW, et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol. 2014;27:1321–1330. [DOI] [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann Surg Oncol. 2010;17:1471–1474. [DOI] [PubMed] [Google Scholar]
- 21.McLean IW, Foster WD, Zimmerman LE, et al. Modifications of Callender’s classification of uveal melanoma at the Armed Forces Institute of Pathology. Am J Ophthalmol. 1983;96:502–509. [DOI] [PubMed] [Google Scholar]
- 22.Gleeson G, Larkin A, Horgan N, et al. Evaluation of chromogenic in situ hybridization for the determination of monosomy 3 in uveal melanoma. Arch Pathol Lab Med. 2014;138:664–670. [DOI] [PubMed] [Google Scholar]
- 23.Szalai E, Wells JR, Ward L, et al. Uveal melanoma nuclear BRCA1-associated protein-1 immunoreactivity is an indicator of metastasis. Ophthalmology. 2018;125:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas S, Pütter C, Weber S, et al. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study. Br J Cancer. 2012;106:1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbone M, Yang H, Pass HI, et al. BAP1 and cancer. Nat Rev Cancer. 2013;13:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013;45:116–126. [DOI] [PubMed] [Google Scholar]
- 27.Kalirai H, Dodson A, Faqir S, et al. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111:1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah AA, Bourne TD, Murali R. BAP1 protein loss by immunohistochemistry: a potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology. 2013;45:651–656. [DOI] [PubMed] [Google Scholar]
- 29.Farquhar N, Thornton S, Coupland SE, et al. Patterns of BAP1 protein expression provide insights into prognostic significance and the biology of uveal melanoma. J Pathol Clin Res. 2018;4:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M, Maßhöfer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.See TR, Stålhammar G, Phillips S, et al. BAP1 immunoreactivity correlates with gene expression class in uveal melanoma. Ocul Oncol Pathol. 2020;6:129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogrusöz M, Jager MJ. Genetic prognostication in uveal melanoma. Acta Ophthalmol. 2018;96:331–347. [DOI] [PubMed] [Google Scholar]
- 33.Bagger MM. Intraocular biopsy of uveal melanoma risk assessment and identification of genetic prognostic markers. Acta Ophthalmol. 2018;96(suppl A112):1–28. [DOI] [PubMed] [Google Scholar]
- 34.Negretti GS, Gurudas S, Gallo B, et al. Survival analysis following enucleation for uveal melanoma. Eye. 2022;36:1669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harbour JW. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile In: Thurin M, Marincola FM, eds. Molecular Diagnostics for Melanoma: Methods and Protocols. Methods in Molecular Biology. Humana Press; 2014:427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glasgow BJ, McCannel TA. Correlation of immunocytochemistry of BRCA1-associated protein 1 (BAP1) with other prognostic markers in uveal melanoma. Am J Ophthalmol. 2018;189:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]