Summary
Background
Insomnia is the most common sleep disorder in patients with epithelial ovarian cancer (EOC). We investigated the causal association between genetically predicted insomnia and EOC risk and survival through a two-sample Mendelian randomization (MR) study.
Methods
Insomnia was proxied using genetic variants identified in a genome-wide association study (GWAS) meta-analysis of UK Biobank and 23andMe. Using genetic associations with EOC risk and overall survival from the Ovarian Cancer Association Consortium (OCAC) GWAS in 66,450 women (over 11,000 cases with clinical follow-up), we performed Iterative Mendelian Randomization and Pleiotropy (IMRP) analysis followed by a set of sensitivity analyses. Genetic associations with survival and response to treatment in ovarian cancer study of The Cancer Genome Atlas (TCGA) were estimated controlling for chemotherapy and clinical factors.
Findings
Insomnia was associated with higher risk of endometrioid EOC (OR = 1.60, 95% CI 1.05–2.45) and lower risk of high-grade serous EOC (HGSOC) and clear cell EOC (OR = 0.79 and 0.48, 95% CI 0.63–1.00 and 0.27–0.86, respectively). In survival analysis, insomnia was associated with shorter survival of invasive EOC (OR = 1.45, 95% CI 1.13–1.87) and HGSOC (OR = 1.4, 95% CI 1.04–1.89), which was attenuated after adjustment for body mass index and reproductive age. Insomnia was associated with reduced survival in TCGA HGSOC cases who received standard chemotherapy (OR = 2.48, 95% CI 1.13–5.42), but was attenuated after adjustment for clinical factors.
Interpretation
This study supports the impact of insomnia on EOC risk and survival, suggesting treatments targeting insomnia could be pivotal for prevention and improving patient survival.
Funding
National Institutes of Health, National Cancer Institute. Full funding details are provided in acknowledgments.
Keywords: Mendelian randomization, Epithelial ovarian cancer, Insomnia, The ovarian cancer association consortium, Genome-wide association study, Patient survival
Research in context.
Evidence before this study
Sleep has been recognized as the third pillar of health alongside diet and exercise. Prior research has shed light on the potential adverse effects of sleep disorders on individuals both healthy and with cancer. Among these disorders, insomnia has been particularly spotlighted as a potential risk factor for various cancers, such as breast and ovarian cancers. Additionally, there is increasing evidence indicating that sleep disorders adversely affect cancer survival. However, the nature of these associations remains unclear, primarily due to the inherent limitations of observational studies.
Added value of this study
This study investigated the relationship between insomnia and ovarian cancer risk and survival through a Mendelian randomization (MR) approach by leveraging genome-wide association study information from the Ovarian Cancer Association Consortium (OCAC). The ovarian cancer study of The Cancer Genome Atlas (TCGA) was also used to confirm the association between insomnia and clinical outcomes. Ultimately, this study aims to provide a more thorough understanding of the potential causal relationship between insomnia and both the susceptibility and outcome for patients diagnosed with ovarian cancer.
Implications of all the available evidence
We observed that insomnia was linked with an increased risk of one histotype of ovarian cancer (endometrioid EOC) but seemed to decrease the risk for other histotypes (high-grade serous EOC (HGSOC) and clear cell EOC). Furthermore, for those with HGSOC, insomnia appeared to be associated with poorer patient survival. Altogether, our analysis provides compelling and independent evidence suggesting that sleep disorders could hold significant implications for both the prevention and management of ovarian cancer.
Introduction
Epithelial ovarian cancer (EOC) is a leading cause of cancer death in women throughout the world with nearly 13,000 deaths estimated to occur during 2022 in the United States alone.1 With a lack of clear symptoms at onset and a high recurrence rate, the early detection and prognosis of ovarian cancer are major clinical challenges. Identifying modifiable risk and prognostic factors is greatly needed. In recent years, sleep disorders have received much attention as risk factors for multiple cancers, including breast and ovarian.2, 3, 4, 5 Growing evidence suggests sleep disorders and disruption can also influence survival of ovarian6 and breast cancer.7 However, the nature of these associations is unclear.
Sleep and the circadian system are vitally linked to hormone production, metabolism, and immune regulatory mechanisms that play a role in the development and progression of cancer.8 Therefore, sleep has been considered as the third pillar of health along with nutrition and exercise. Notably, sleep disturbances occur at higher rates among patients with cancer than the general population, with insomnia (defined as difficulty in falling or maintaining asleep) ranking as the most prevalent sleep disorder in the oncological population, affecting up to 60% of those with EOC.9 In observational studies, insomnia and circadian disruption (usually assessed by night shift work) have been associated with an increased risk of invasive EOC and an even higher increased risk of early-stage ovarian cancer.5,10 However, it is important to acknowledge that such evidence is inconsistent across studies.11, 12, 13, 14 The interpretation and validation of these associations are limited given that sleep disorders correlate with a wide spectrum of environmental, psychiatric, and reproductive factors associated with EOC.15 Furthermore, sleep disorders tend to be more prevalent among patients receiving treatment such as chemotherapy and surgery,9 which may lead to spurious associations when evaluating survival outcomes. Understanding whether associations between insomnia and ovarian cancer risk and/or prognostic factors are potentially causal in nature could have significant implications for strategies to improve detection and survival.
In this study, we apply two-sample Mendelian randomization (MR) to investigate the potential causal relationship between insomnia and ovarian cancer risk and survival. MR is becoming an established method for estimating the causal effect of a modifiable exposure on the etiology of a disease or a clinical outcome, by using only summary statistics from large GWAS studies. The approach uses genetic markers as instrumental variables (IVs) to randomly assign lifetime risk of an exposure, such as insomnia, to create pseudo exposure. MR has increasingly been applied in cancer populations and epidemiological studies as it does not require longitudinal collection of exposure and outcome from the same study. The approach is also less prone to residual confounding, reverse causation and various biases.16 Thus far, MR studies have demonstrated that insomnia may be causally associated with an increased risk of type 2 diabetes, higher body mass index (BMI), and coronary heart disease.15 Here, we perform MR based on summary statistics derived from the largest GWAS of insomnia and the Ovarian Cancer Association Consortium (OCAC), which comprises data from over 66,000 women. Given the fact that EOC is a highly heterogeneous disease, our analysis is also stratified based on histological subtypes or histotypes. Data from the ovarian cancer study of the Cancer Genome Atlas (TCGA) was also analyzed to further evaluate the potential impact of insomnia on patient survival. Sensitivity analyses and multivariable MR analyses are also considered in our study to ensure the validity and reliability of our findings.
To the best of our knowledge, no existing epidemiological study has systematically investigated the potential causal relationship between insomnia and both the risk and survival outcomes of ovarian cancer. Leveraging the most comprehensive GWAS datasets available in this domain, our study seeks to offer a cutting-edge perspective on the implications of sleep disorders for patients with cancer, specifically in the context of ovarian cancer prevention and therapeutic approaches.
Methods
Ethics
This work was approved by the Institutional Review Board of Brigham and Women's Hospital and complies with all relevant ethical regulations.
Overview
We leveraged existing genotyping studies to perform two-sample Mendelian randomization analyses with summary level data. In discovery analysis, we used summary statistics from two large-scale genome-wide association studies (GWASs) for EOC and insomnia. In validation analysis, we further obtained individual level genotype and clinical data from an independent clinical cohort and performed GWAS for overall survival and therapy response outcomes. Then, the association summary statistics were used for validation MR. The overall study design is shown in Fig. 1.
Fig. 1.
Analysis flowchart for investigating potential causal relations between insomnia and ovarian cancer (EOC) outcomes.
Ovarian cancer risk and survival outcomes
For discovery analysis, genome-wide summary statics of EOC outcomes were obtained from previously published genome-wide association analysis (GWAS) in 66,450 women of European ancestry that was carried out by the Ovarian Cancer Association Consortium (OCAC).17 Analyses included 25,509 EOC cases and 40,941 controls with risk associations estimated for all invasive EOC and by histotype including 13,037 high grade serous (HGSOC), 1012 low-grade or borderline serous (LGSOC), 2810 endometroid, 1149 mucinous (invasive or borderline), and 1366 clear cell carcinomas defined by medical records. In brief, GWAS were performed for each genotyping project separately, adjusting for study and genotype principal components, and then pooled using a fixed effect meta-analysis using custom written software as previously described [20]. GWAS of overall survival was conducted in a subset of 11,311 EOC cases with clinical follow-up, regardless of chemotherapy and surgical intervention, and adjusted for EOC histotype or limited to HGSOC cases only as previously described.18
Insomnia exposure and instrumental variables
Genetic instruments of insomnia were selected based on the most recent and largest GWAS estimated in individuals of European ancestry from the UK Biobank (UKB) and 23andMe. In the study, insomnia was assessed using single or multiple questions and dichotomized as described before (total 593,724 cases and 1,771,286 controls).19 GWAS were performed in each study adjusting for age, sex, and genotype principal components, and then meta-analyzed using METAL software. 554 independent SNPs that were genome-wide significantly associated with insomnia were included as IVs (p < 5e-8; Supplementary Table S1). Because no significant sex heterogeneity was reported in UKB,15 we used summary statistics of combined males and females to increase statistical power and instrument variable (IV) strength. To reduce type 1 error, we performed linkage disequilibrium (LD) based clumping (r2 < 0.01) to retain independent IVs.
Genetic risk score analysis
We evaluated the combined (additive) association of the 554 insomnia SNPs with EOC outcomes using weighted genetic risk score (GRS) that was derived from summary level GWAS data with the grs.summary function in R package gtx (http://cran.nexr.com/web/packages/gtx/gtx.pdf). For each EOC outcome, the effect of GRS score was estimated as the combined effect of individual risk allele count weighted by their estimated effects in the ovarian GWAS.
Two-sample MR
As both insomnia and EOC are heterogenous traits involving different mechanisms, we applied Iterative Mendelian Randomization and Pleiotropy (IMRP) analysis as the primary method to remove pleiotropic outliers.20 Briefly, in each iterative step, IMRP estimates the biologically independent, often called horizontal, pleiotropic effect for each IV using an estimated causal association β between the exposure (insomnia) and outcome (EOC), and updates β excluding significant pleiotropic variants until no additional pleiotropic variants are left. This approach is computationally effective and does not need to simulate global β distribution.21 EOC risk associations were estimated from summary statistics for overall EOC risk and five histologic subtypes. EOC survival associations were estimated for three survival analyses (overall unadjusted, overall adjusted for histology, and high-grade serous only survival). For survival GWAS, we investigated the extent of index event bias which can occur when restricting to a case-only population and a confounder induces a false association with disease progression that is due to the association with disease incidence.22 To detect the presence of index event bias, we selected 34 known EOC susceptibility SNPs to serve as negative controls. All known risk loci were not significant (p < 0.05) in the survival GWAS, indicating index event bias was likely not significant (Supplemental Table S2). Associations between insomnia and EOC outcomes were considered significant using a Bonferroni corrected p-value threshold that accounted for all nine EOC outcomes tested (P < 0.005). Significant associations were followed up for sensitivity and multivariate MR analyses.
Sensitivity analyses
There are three essential assumptions in MR: 1) the association between IVs and the exposure are sufficiently strong; 2) there is no direct association between IVs and the outcome except through the effects on exposure (i.e., no horizontal pleiotropy that occurs when a variant influences the two traits through independent physiological mechanisms); 3) the IVs are independent to confounders for the associations between the exposure and outcome.23 Sensitivity analyses were conducted to evaluate the robustness of our primary results to the assumptions of MR methods and the IVs selected. We performed multiple MR testing methods that each has its unique benefit under the different MR assumptions. These included the inverse variance weighted (IVW), MR-Egger, weighted median,24, 25, 26, 27 MR-RAP, and MR-Mix tests using the TwoSampleMR R package28 (https://mrcieu.github.io/TwoSampleMR/). IVW is the mostly commonly used MR approach, which may exhibit weak instrument bias. Weighted median test has better power when <50% estimation comes from invalid IVs. MR-Egger regression detects bias from horizontal pleiotropy effects (between the IV and outcome) based on deviation of the regression intercept from the origin (p < 0.05) and provides estimates of the causal association using the slope coefficient.27 MR-RAPS (Robust Adjusted Profile Score) is robust to many weak instruments and systematic and idiosyncratic pleiotropy.29 MR-Mix is robust to invalid IVs due to correlated pleiotropy, which occurs when a proportion of IVs have correlated effects on the exposure and outcome.30 We also performed the MR Steiger directionality test to remove IVs acting through potential reverse causality from EOC to insomnia, which were indicated if the variance explained in the EOC outcome was larger than the variance explained in the insomnia exposure by the IV.28 A consistent direction of MR estimates using these different methods supports the reliability of the causal estimate.
Additional sensitivity analyses were performed to test the validity of IV used for MR analysis. We evaluated IV strength (association between IV and exposure) using the mean Cragg–Donald F statistics after excluding pleiotropic outliers, which were calculated as: , where R2 is the variance of exposure explained by IVs, n is the sample size of exposure, and k is the number of IVs. A mean F > 10 indicates sufficient IV strength for MR analysis. We also evaluated the heterogeneity of IV effects across EOC outcomes using Cochran's Q (for IVW) and Rücker's Q’ (for MR-Egger) statistics23,31 to assess the robustness of IV association. In addition, we performed a leave-one-out analysis to test if the MR-estimated association was driven by an individual SNP. Finally, we performed a secondary IVW analysis using a separate IVs set that consisted of SNPs that were genome-wide significant in the female-specific insomnia GWAS (Supplementary Table S1).
Multivariate MR
To investigate whether the significant effects of insomnia genetic variants on EOC outcomes were confounded or mediated by other risk factors including sleep duration, chronotype (an indicator of circadian diurnal preference), BMI, and reproductive age, we performed multivariate MR analysis including genetic effects of each risk factor as covariates.32 Genetic effects were extracted from published GWASs of European ancestry samples, including self-reported sleep duration in the UK Biobank (N = 446,118)33; self-reported chronotype in the UK Biobank (N = 697,828),33 BMI in the GIANT consortium studies (N = 681,275)34; age at menarche, and age at menopause estimated in the ReproGen consortium studies (N = 182,416 and 69,360, respectively)35,36; smoking, depression and sex hormone-binding globulin in women in the UK Biobank provided by IEU OpenGWAS (N > 460,000).37
Independent validation analysis
To validate the association between insomnia and EOC survival, we further performed two-sample MR using publicly available individual-level of genotype and clinical data for HGSOC patients from TCGA in two steps. We first performed GWAS in 323 patients with genotype data that received chemotherapy, had surgical debulking, and were followed for response the chemotherapy and overall survival. Cox proportional hazard modeling was used for overall survival analyses. Logistic regression was used to estimate the association with response to treatment (RTT; complete vs. incomplete). All models used additive genetic models and adjusted for the first two principal components of European ancestry. All adjusted models included age, stage (early/advanced) and residual disease (optimal, <10 mm of disease vs. suboptimal, >10 mm of disease) as covariates. Fully adjusted overall survival models additionally included RTT as a covariate. We then perform two-sample MR between insomnia and TCGA EOC outcomes using the same instruments in discovery analysis.
Literature-mined entity-based analysis connecting sleep disorders and ovarian cancer
In order to further explore the plausible broader biological mechanisms linking sleep disorders to ovarian cancer, we mined the literature using EpiGraphDB, a biomedical knowledge graph database. This platform is designed to facilitate data mining of epidemiology relationships and literary-minded estimates for various health conditions, as well as molecular and lifestyle traits. Specifically, we queried EpiGraphDB to extract the semantic triple associated with the GWAS IDs “ukb-d-SLEEP” (sleep disorder) and “ieu-a-1120” (ovarian cancer).
Role of funders
The funders did not play a role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Results
A total of 554 independent SNPs significantly associated with insomnia (P < 5e-8) in the meta-analysis GWAS of the UK Biobank and 23andMe were selected as IVs (Supplementary Table S1). The weighted GRS of IVs constructed using summary level data were significantly associated with reduced risk of clear cell EOC (beta per SD change in GRS = −0.670, SE = 0.290, p = 0.021), increased OR of overall EOC survival time without and with adjusting for histology (beta SD change in GRS = 0.301 and 0.269, SE = 0.126 and 0.126, p = 0.017 and 0.033, respectively), and reduced survival time of HGSOC (OR per GRS sd = 1.35, 95% CI 1.01–1.80) (Supplementary Table S3).
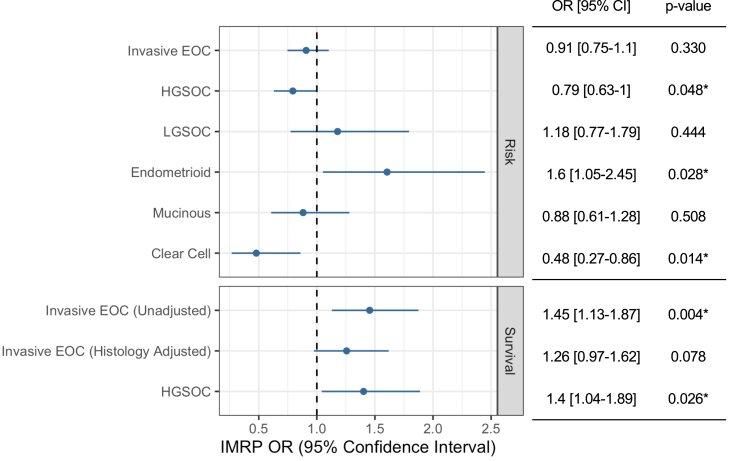
We excluded horizontal pleiotropic outliers for each EOC outcome detected by IMRP20 leaving at least 459 IVs for MR estimation for each EOC outcome (Supplementary Table S4). The primary results of two-sample MR analysis using summary level data between insomnia exposure and EOC outcomes are shown in Fig. 2. For risk in all invasive EOC, we did not observe an association with insomnia (OR = 0.91, 95% CI: 0.75–1.10). However, in histotype-specific analyses, we identified putative causal associations between insomnia with a reduced risk of HGSOC (OR = 0.79, 95% CI: 0.63–1.00) and clear cell EOC (OR = 0.48 95% CI: 0.27–0.86), and an increased risk of endometroid EOC (OR = 1.60, 95% CI 1.05–2.45). However, none of these associations were significant after accounting for multiple comparisons (p < 0.005).
Fig. 2.
Iterative Mendelian Randomization and Pleiotropy (IMRP) associations between insomnia and EOC outcomes.
Among EOC cases with clinical follow-up, regardless of treatment, insomnia was associated with decreased overall survival time (mortality OR = 1.45, 95% CI 1.13–1.87), which was significant after multiple testing correction (p < 0.005). This association was somewhat attenuated after adjustment for histology (OR = 1.26, 95% CI 0.97–1.62). Survival analysis limited to HGSOC cases showed a similar association between insomnia and HGSOC survival (OR = 1.40, 95% CI 1.04–1.89).
We performed a set of sensitivity analyses for the significant MR association between insomnia and overall EOC survival (IMRP p < 0.005). The IV-insomnia association mean F statistic was 49.8 (>10) showing sufficient strength of the instrument for MR analysis. The exposure variance explained by IVs as 1.0%. Consistent directions of MR associations were observed for the six EOC outcomes using alternative MR approaches (including IVW, MR-Egger, weighted median, MR-RAPs, and MR-CUE), suggesting reliable causal effects (Table 1). We did not observe significant evidence for the presence of horizontal pleiotropy, as indicated by the MR-Egger intercept test (p > 0.05), or remaining heterogeneity effects (IVW Cochran's Q and MR-Egger Rücker's Q′ p-values>0.05). Leave-one-out IVW analysis showed consistent association patterns (Supplementary Fig. S1). However, the MR Steiger test indicates potential causality in the reverse direction (p_Steiger = 2.03 x 10−19).
Table 1.
Sensitivity MR analyses for significant associations between insomnia and EOC overall survival.
| Method | #SNPs | IV strength | MR association |
Heterogeneity |
|||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | Q | P-value | |||
| IVW | 461 | Mean F = 49.2 | 1.42 | 1.1–1.85 | 0.007 | 354.9 | 0.999911 |
| MR Egger Intercept | 461 | 1.00 | 0.99–1 | 0.488 | |||
| MR Egger | 461 | Isq = 0.98 | 2.00 | 0.74–5.43 | 0.173 | 354.5 | 0.999905 |
| Weighted median | 461 | 1.35 | 0.93–1.94 | 0.112 | |||
| MR RAPS | 461 | 1.40 | 1.07–1.84 | 0.016 | |||
| MRMix | 461 | 1.16 | 0.95–1.42 | 0.144 | |||
| MR Steiger | 461 | 1.27 | 0.89–1.82 | 0.192 | |||
| IVW using female IVs | 238 | 1.92 | 1.1–3.32 | 0.021 | |||
IVW: inverse variance weighted; SE, standard error; OR, odds ratio; Q, Cochran's Q-value.
We next performed multivariable MR analysis32 to investigate whether significant univariate MR associations could be explained by other potential confounders. For EOC risk associations, estimates were largely unchanged (same direction of effect and p < 0.10) after adjusting for genetically predicted sleep duration, chronotype, BMI, smoking, depression, age at menopause and age at menarche (Table 2). For survival outcomes, the association between insomnia and poorer overall survival was largely unchanged. However, the MR association with HGSOC survival was not significant after adjustment for BMI (OR = 1.23, 95% CI 0.91–1.66), age at menopause (OR = 1.24, 95% CI: 0.92–1.67), and age at menarche (OR = 1.23, 95% CI: 0.91–1.67).
Table 2.
Multivariate MR associations between insomnia and EOC overall survival adjusting for potential confounders.
| Covariate | OR | 95% CI | P-value |
|---|---|---|---|
| Sleep duration | 1.42 | 1.12–1.82 | 0.004 |
| Chronotype | 1.44 | 1.13–1.83 | 0.003 |
| BMI | 1.39 | 1.06–1.83 | 0.018 |
| Current Smoking | 1.45 | 1.14–1.84 | 0.002 |
| Alcohol intake frequency | 1.45 | 1.14–1.84 | 0.002 |
| Depression | 1.45 | 1.14–1.84 | 0.003 |
| Age at menopause (in women) | 1.44 | 1.1–1.89 | 0.008 |
| Age at menarche (in women) | 1.43 | 1.09–1.88 | 0.010 |
| Sex hormone binding globulin (in women) | 1.44 | 1.14–1.82 | 0.002 |
To explore the association between insomnia and clinical outcomes further, we performed validation MR analysis for overall survival and response to therapy in 323 patients with HGSOC that received standard chemotherapy following surgical debulking. Clinical characteristics are provided in Supplemental Table S5. For both overall survival and response to therapy outcomes, we conducted GWAS analysis and then performed two-sample MR using the summary statistics. Insomnia was associated with worse survival of HGSOC (mortality OR = 2.48, 95% CI 1.13–5.42) which was still suggestive of association after adjustment for age of diagnosis, stage, and residual disease (OR = 2.09, 95% CI 0.94–4.62). When including adjustment for response to therapy, the estimated association between insomnia and survival was comparable (OR = 1.96, 95% CI 0.85–4.52). Insomnia was not associated with treatment response (OR = 0.87, 95% CI 0.22–3.43), although the confidence intervals were wide (Table 3).
Table 3.
Associations between insomnia and TCGA EOC outcomes using Inverse Variance Weighted Mendelian Randomization.
| Outcome | Modela | SNPs | OR | 95% CI | P-value |
|---|---|---|---|---|---|
| Survival | Unadjusted | 449 | 2.48 | 1.13–5.42 | 0.023 |
| Survival | Adjusted | 449 | 2.09 | 0.94–4.62 | 0.068 |
| Survival | Full | 449 | 1.96 | 0.85–4.52 | 0.114 |
| Treatment Response | Unadjusted | 449 | 0.90 | 0.25–3.27 | 0.875 |
| Treatment Response | Adjusted | 449 | 0.87 | 0.22–3.43 | 0.844 |
Models were unadjusted (no covariates), adjusted (included age, stage, and residual disease covariates), and for survival model we also ran a full model (included adjusted covariates plus a covariate for treatment response).
The bar plot in the bottom panel of Fig. 3 displays the frequency counts of overlapping terms identified through the entity-based literature-mined connecting sleep disorders and ovarian cancer, focusing on those with frequencies of three or more. Each of these entities, encompassing proteins, genes, hormones, or chemicals, represent a key point in a potential mechanism connecting the two traits. The entities with high frequency counts (with large numbers of supporting literatures) included melatonin, leptin, AKT1, and Proto-Oncogene Proteins c-akt. Melatonin is a well-known hormone related to sleep disorders and has important roles in cellular signaling and metabolic regulation. Leptin, a hormone primarily associated with energy balance and hunger regulation, has been implicated in cancer biology due to its roles in promoting cell proliferation and inhibiting apoptosis. Akt signalling pathway, a critical regulator of cell survival and growth, often becomes dysregulated in cancer, leading to enhanced tumour development and resistance to cell death. Therefore, the activation of leptin and Akt signalling pathways may suggest a potential synergistic effect on enhancing ovarian cancer cell survival and proliferation. Notably, there are multiple entities involved in cancer immune mechanisms, including TNF, IL-10, and G-Protein-Coupled Receptors (GPCRs), as well as in cancer proliferation, such as VEGFA, Prolactin, PPAR gamma, and Mitogen-Activated Protein Kinases (MAPKs). The network plot illustrated in Fig. 3 elucidates a primary pathway linking sleep disorders to ovarian cancer through leptin, with the involvement of other intermediate biological terms, suggesting a potential mechanism of interest for future studies.
Fig. 3.
Plausible biological mechanisms linking sleep disorders to ovarian cancer.
Discussion
In this study, we performed mendelian randomization to understand the potential for causal associations between insomnia and ovarian cancer risk and survival. We leveraged the largest genetic epidemiology studies for sleep disorders and EOC to investigate their relationship using a two-sample design that reduces sample selection bias. Our approach with IMRP reduced the impact of biologic (horizontal) pleiotropy on our association estimates and increased statistical power. Through a suite of MR diagnostic and sensitivity tests we found our findings to be relatively robust, and we further explored potential confounding effects. Finally, we leveraged the ovarian cancer study of TCGA to validate our survival analysis and explore more deeply potential clinical confounders of prognosis. Our results reveal significant associations between the genetic predisposition to insomnia and EOC survival that appear to be largely influenced by BMI and partially by clinical characteristics.
Although not significant after accounting for multiple comparisons, we observed heterogeneity in associations between insomnia and EOC risk based on histotype. Insomnia has been associated with higher risk of invasive serous and the characteristically more aggressive, high-grade type II ovarian cancers but not with noninvasive or less aggressive, low-grade type I ovarian cancers.5 Further, sleep problems are associated with increased incidence of several cancers but inversely associated with incidence and mortality of others.38 In the current study, we observed a suggestive association between insomnia and increased endometroid EOC risk but an inverse association of insomnia for HGSOC and clear cell risk (IMRP p < 0.05). EOC histotypes are known to have distinct pathogenesis and molecular features that are reflected by different associations with epidemiologic risk factors, response to standard chemotherapy, and prognoses which may be contributing to the differences seen in our study.17,39,40 Thus, the opposite associations with insomnia for endometrioid and clear cell histotypes could reflect their differing origin, clinical behaviour, and relationship with endometriosis.41 While the underlying biology relating insomnia to ovarian cancer risk is unclear, potential pathways have been proposed which largely center around disruption of immune function, induction of cancer-stimulatory cytokines, and proinflammatory responses that most likely effect ovarian cancer pathogenesis differentially by histotype.42
Persistent insomnia in EOC survivors has been associated with anxiety, depression, wellbeing and quality of life,9,43,44 and shorter survival.45,46 However, the causal link between insomnia and EOC survival has not been established. We analyzed EOC survival in two independent studies and identified causal associations between insomnia and all invasive EOC and HGSOC-specific survival (OR>1 and p < 0.05). In our discovery analysis, effects were largely unchanged after adjusting for sleep duration, chronotype, smoking, and depression but decreased after adjusting for BMI and reproductive age. Notably, the validation analysis in the ovarian cancer study of TCGA found insomnia was associated with an approximately two times higher rate of death for HGSOC that was relatively robust (OR∼2) after adjustment for clinical covariates and response to treatment, although not statistically significant. These results suggest that insomnia may impact EOC survival through pathways overlapped with obesity, possibly involving metabolism, energy balance and chronic inflammation. We cannot rule out the potential of reverse direction of effect, given the results of the MR Steiger test and that we could not test for reverse MR directly. However, our prior MR analyses have identified causal associations between insomnia with increased depressive symptoms and reduced subjective well-being, but not vice versa.15 Taken together with our current findings, insomnia may be an upstream risk factor for BMI and poor EOC survival. This is consistent with a recent observational study that found persistent insomnia in cancer survivors despite clinical and psychological improvements, especially in women.47 However, future mediation analyses are needed to validate this hypothesis.
Prevalent insomnia in patients with EOC may relate to pain, anxiety, hot flashes and night sweats, gastrointestinal, bladder, and breathing problems, and certain treatment such as chemotherapy and hormonal therapy. The importance of early insomnia management has been recognized in cancer survival. Cognitive-behavioural therapy for insomnia (CBT-i) has been considered first-line practice for insomnia in cancer survivors that contributes to improved physical and functional well-being and quality of life.44,48 In contrast, sleep medications seemingly have little effect on improving chronic insomnia or other adverse effects in patients with cancer.49,50 Future clinical trials are needed to understand the effects of insomnia treatments on EOC survival.
This study has several limitations. First, the insomnia GRS was constructed using significant variants associated with insomnia that do not fully capture the polygenic risk of insomnia. Polygenic risk score (PRS) using advance methods Polygenic Risk Score-Continuous Shrinkage51 with individual level data is warranted to understand the shared genetic etiology. Second, IVs were selected based on GWAS conducted in a population of both males and females to gain instrument strength and statistical power. Sensitivity analysis using IVs that were genome-wide significant in females suggested consistent associations but statistical significance was attenuated. In addition, the association statistics for several covariates (e.g., sleep duration, chronotype, BMI, smoking, and depression) were estimated in male and female populations. This may influence the validation of MVMR since heterogeneous effects may exist between male and female, particularly for BMI. Finally, the exposure classification and statistical power of this study was limited by the heterogeneity of the insomnia phenotype. Recent epidemiological studies using accelerometric data have identified subtypes of insomnia symptoms (e.g., associated with long vs short sleep duration, long-term vs short-term midawake or fragmented sleep, or due to irregular sleep and circadian misalignment) that are associated with distinct lifestyle and environmental risk factors, and cardiovascular and psychiatric outcomes.52 Future work of subtyping insomnia and leveraging objective sleep measures, such as actigraphy or polysomnography, are needed to better understand the relationship between insomnia and EOC to improve treatment. Additionally, to improve generalizability, further studies are needed in non-European populations, particularly African Americans and non-Hispanic populations which have a higher prevalence and severity of sleep disorders53 and poorer EOC survival.54
In summary, we performed the first MR study examining insomnia and EOC and found insomnia is associated with poorer survival after diagnosis. Future longitudinal analyses are needed to validate this finding and investigate the potential mediating effects of BMI. Elucidating the causal relationships of these modifiable behaviors could inform clinical prevention strategies and therapeutics for EOC.
Contributors
HW and XW conceptualized and supervise the study. HW and SR acquired the fundings. HW, BMR and XW curated data, designed the methodology and provided project administration. HW and BMR performed the formal statistical analysis. HW, BMR, RCR, JML, RS, BDG, BLF, SR, SST, and XW reviewed results and verified the conclusions. HW and XW provided overall supervision. HW, BMR, and XW wrote the original draft. HW, BMR, RCR, JML, RS, BDG, BLF, SR, SST and XW edited the original and revised manuscripts. All authors have read and approved the final version of the manuscript.
Data sharing statement
GWAS summary statistics of EOC are available from OCAC (http://ocac.ccge.medschl.cam.ac.uk/). GWAS summary statistics of insomnia are available on Sleep Disorders Knowledge Portal (https://sleep.hugeamp.org). All data collected and processed by TCGA is available at the Genomic Data Commons (GDC; https://portal.gdc.cancer.gov).
Declaration of interests
BDG reports fees from Sure Med Compliance and Elly Health. SR reports fees and support to attend an advisory meeting from Eli Lilly and unpaid leadership role as a board/group member at the National Sleep Foundation and Alliance of Sleep Apnea Partner. SST reports grants from the NIH, State of Florida, ACS, DOD and BMS, fees from Ponce Health Sciences University, Ovarian Cancer Research Alliance, UNC Lineberger Comprehensive Cancer Center and AACR, and leadership role as a board/group member at City of Hope, Alberta Cancer Center and Fred Hutchinson Cancer Center. All other authors declare no competing interests.
Acknowledgements
This work was supported in part by National Institutes of Health (R01HL153814 to H.W., R35HL135818 to S.R); and by the Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center and Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292). In addition, for the valuable comments and suggestions we thank Zoe Zhou and six anonymous reviewers.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105175.
Appendix A. Supplementary data
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Brzecka A., Sarul K., Dyła T., et al. The association of sleep disorders, obesity and sleep-related hypoxia with cancer. Curr Genom. 2020;21(6):444–453. doi: 10.2174/1389202921999200403151720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muzyka M., Tagliafico L., Serafini G., et al. Neuropsychiatric disorders and frailty in older adults over the spectrum of cancer: a narrative review. Cancers. 2022;14(1):258. doi: 10.3390/cancers14010258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richmond R.C., Anderson E.L., Dashti H.S., et al. Investigating causal relations between sleep traits and risk of breast cancer in women: mendelian randomisation study. BMJ. 2019;365 doi: 10.1136/bmj.l2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang X., Harris H.R., Hendryx M., et al. Sleep characteristics and risk of ovarian cancer among postmenopausal women. Cancer Prev Res. 2021;14(1):55–64. doi: 10.1158/1940-6207.CAPR-20-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter B.D., Diver W.R., Hildebrand J.S., Patel A.V., Gapstur S.M. Circadian disruption and fatal ovarian cancer. Am J Prev Med. 2014;46(3 Suppl 1):S34–S41. doi: 10.1016/j.amepre.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 7.Bach L., Kalder M., Kostev K. Depression and sleep disorders are associated with early mortality in women with breast cancer in the United Kingdom. J Psychiatr Res. 2021;143:481–484. doi: 10.1016/j.jpsychires.2020.11.036. [DOI] [PubMed] [Google Scholar]
- 8.Kelleher F.C., Rao A., Maguire A. Circadian molecular clocks and cancer. Cancer Lett. 2014;342(1):9–18. doi: 10.1016/j.canlet.2013.09.040. [DOI] [PubMed] [Google Scholar]
- 9.Palagini L., Miniati M., Massa L., et al. Insomnia and circadian sleep disorders in ovarian cancer: evaluation and management of underestimated modifiable factors potentially contributing to morbidity. J Sleep Res. 2021;31 doi: 10.1111/jsr.13510. [DOI] [PubMed] [Google Scholar]
- 10.Bhatti P., Cushing-Haugen K.L., Wicklund K.G., Doherty J.A., Rossing M.A. Nightshift work and risk of ovarian cancer. Occup Environ Med. 2013;70(4):231–237. doi: 10.1136/oemed-2012-101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dun A., Zhao X., Jin X., et al. Association between night-shift work and cancer risk: updated systematic review and meta-analysis. Front Oncol. 2020;10:1006. doi: 10.3389/fonc.2020.01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris M.A., MacLeod J., Kim J., et al. Use of a Canadian population-based surveillance cohort to test relationships between shift work and breast, ovarian, and prostate cancer. Ann Work Expo Health. 2020;64(4):387–401. doi: 10.1093/annweh/wxaa017. [DOI] [PubMed] [Google Scholar]
- 13.Leung L., Grundy A., Siemiatycki J., et al. Shift work patterns, chronotype, and epithelial ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2019;28(5):987–995. doi: 10.1158/1055-9965.EPI-18-1112. [DOI] [PubMed] [Google Scholar]
- 14.Poole E.M., Schernhammer E., Mills L., Hankinson S.E., Tworoger S.S. Urinary melatonin and risk of ovarian cancer. Cancer Causes Control. 2015;26(10):1501–1506. doi: 10.1007/s10552-015-0640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane J.M., Jones S.E., Dashti H.S., et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019;51(3):387–393. doi: 10.1038/s41588-019-0361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith G.D., Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33(1):30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 17.Phelan C.M., Kuchenbaecker K.B., Tyrer J.P., et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49(5):680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnatty S.E., Tyrer J.P., Kar S., et al. Genome-wide analysis identifies novel loci associated with ovarian cancer outcomes: findings from the ovarian cancer association consortium. Clin Cancer Res. 2015;21(23):5264–5276. doi: 10.1158/1078-0432.CCR-15-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe K., Jansen P.R., Savage J.E., et al. Genome-wide meta-analysis of insomnia prioritizes genes associated with metabolic and psychiatric pathways. Nat Genet. 2022;54(8):1125–1132. doi: 10.1038/s41588-022-01124-w. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X., Li X., Xu R., Wang T. An iterative approach to detect pleiotropy and perform Mendelian Randomization analysis using GWAS summary statistics. Bioinformatics. 2021;37(10):1390–1400. doi: 10.1093/bioinformatics/btaa985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell R.E., Hartley A.E., Walker V.M., et al. Strategies to investigate and mitigate collider bias in genetic and Mendelian randomisation studies of disease progression. PLoS Genet. 2023;19(2) doi: 10.1371/journal.pgen.1010596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362 doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartwig F.P., Davey Smith G., Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11) doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Q., Wang J., Hemani G., et al. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat. 2020;48(3):1742–1769. [Google Scholar]
- 30.Qi G., Chatterjee N. Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nat Commun. 2019;10(1):1941. doi: 10.1038/s41467-019-09432-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden J., Spiller W., Del Greco M.F., et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47(6):2100. doi: 10.1093/ije/dyy265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess S., Thompson S.G. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dashti H.S., Jones S.E., Wood A.R., et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10(1):1100. doi: 10.1038/s41467-019-08917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yengo L., Sidorenko J., Kemper K.E., et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27(20):3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry J.R., Day F., Elks C.E., et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514(7520):92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day F.R., Ruth K.S., Thompson D.J., et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294–1303. doi: 10.1038/ng.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elsworth B., Lyon M., Alexander T., et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv. 2020;2020.08.10 [Google Scholar]
- 38.Sillah A., Watson N.F., Peters U., et al. Sleep problems and risk of cancer incidence and mortality in an older cohort: the Cardiovascular Health Study (CHS) Cancer Epidemiol. 2022;76 doi: 10.1016/j.canep.2021.102057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lheureux S., Gourley C., Vergote I., Oza A.M. Epithelial ovarian cancer. Lancet. 2019;393(10177):1240–1253. doi: 10.1016/S0140-6736(18)32552-2. [DOI] [PubMed] [Google Scholar]
- 40.Lheureux S., Braunstein M., Oza A.M. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69(4):280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 41.Bergamini A., Mangili G., Ambrosi A., et al. Endometriosis-related ovarian cancers: evidence for a dichotomy in the histogenesis of the two associated histotypes. Diagnostics. 2023;13(8):1425. doi: 10.3390/diagnostics13081425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berisha A., Shutkind K., Borniger J.C. Sleep disruption and cancer: chicken or the egg? Front Neurosci. 2022;16 doi: 10.3389/fnins.2022.856235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beesley V.L., Webber K., Nagle C.M., et al. When will I feel normal again? Trajectories and predictors of persistent symptoms and poor wellbeing after primary chemotherapy for ovarian cancer. Gynecol Oncol. 2020;159(1):179–186. doi: 10.1016/j.ygyno.2020.07.029. [DOI] [PubMed] [Google Scholar]
- 44.Ross T.L., DeFazio A., Friedlander M., et al. Insomnia and its association with quality of life in women with ovarian cancer. Gynecol Oncol. 2020;158(3):760–768. doi: 10.1016/j.ygyno.2020.06.500. [DOI] [PubMed] [Google Scholar]
- 45.Gaitskell K., Hermon C., Barnes I., et al. Ovarian cancer survival by stage, histotype, and pre-diagnostic lifestyle factors, in the prospective UK Million Women Study. Cancer Epidemiol. 2022;76 doi: 10.1016/j.canep.2021.102074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagle C.M., Dixon S.C., Jensen A., et al. Obesity and survival among women with ovarian cancer: results from the Ovarian Cancer Association Consortium. Br J Cancer. 2015;113(5):817–826. doi: 10.1038/bjc.2015.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schieber K., Niecke A., Geiser F., et al. The course of cancer-related insomnia: don't expect it to disappear after cancer treatment. Sleep Med. 2019;58:107–113. doi: 10.1016/j.sleep.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Squires L.R., Rash J.A., Fawcett J., Garland S.N. Systematic review and meta-analysis of cognitive-behavioural therapy for insomnia on subjective and actigraphy-measured sleep and comorbid symptoms in cancer survivors. Sleep Med Rev. 2022;63 doi: 10.1016/j.smrv.2022.101615. [DOI] [PubMed] [Google Scholar]
- 49.Dirksen S.R., Epstein D.R. Efficacy of an insomnia intervention on fatigue, mood and quality of life in breast cancer survivors. J Adv Nurs. 2008;61(6):664–675. doi: 10.1111/j.1365-2648.2007.04560.x. [DOI] [PubMed] [Google Scholar]
- 50.Palesh O., Scheiber C., Kesler S., Mustian K., Koopman C., Schapira L. Management of side effects during and post-treatment in breast cancer survivors. Breast J. 2018;24(2):167–175. doi: 10.1111/tbj.12862. [DOI] [PubMed] [Google Scholar]
- 51.Ge T., Chen C.-Y., Ni Y., Feng Y.-C.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katori M., Shi S., Ode K.L., Tomita Y., Ueda H.R. The 103,200-arm acceleration dataset in the UK Biobank revealed a landscape of human sleep phenotypes. Proc Natl Acad Sci U S A. 2022;119(12) doi: 10.1073/pnas.2116729119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X., Wang R., Zee P., et al. Racial/ethnic differences in sleep disturbances: the multi-ethnic study of atherosclerosis (MESA) Sleep. 2015;38(6):877–888. doi: 10.5665/sleep.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bandera E.V., Lee V.S., Rodriguez-Rodriguez L., Powell C.B., Kushi L.H. Racial/ethnic disparities in ovarian cancer treatment and survival. Clin Cancer Res. 2016;22(23):5909–5914. doi: 10.1158/1078-0432.CCR-16-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.