Abstract
Background
Despite advances in chronic myeloid leukemia (CML) genetics, the role of nitric oxide (NO) and hydrogen sulfide (H2S) gene mutations and their relationship to apoptotic genes is unclear. Therefore, this study investigated NO- and H2S-producing genes' mutations and their interactions with apoptotic genes using Sanger sequencing and next-generation sequencing (NGS).
Methodology
A complete blood count (CBC) was carried out to measure the total number of white blood cells, while IL-6 levels were assessed in both control and CML patients using an ELISA technique. Sanger sequencing was used to analyze mutations in the CTH and NOS3 genes, whereas NGS was applied to examine mutations on all chromosomes.
Results
White blood cell (WBC) and granulocyte counts were significantly higher in CML patients compared to controls (p<0.0001), and monocyte counts were similarly higher (p<0.05). Interleukin-6 (IL-6) levels were significantly elevated in CML patients than controls (p<0.0001), indicating a possible link to CML etiology or progression. Multiple mutations have been identified in both genes, notably in CTH exon 12 and the NOS3 genes VNTR, T786C, and G894T. This study also measured IL-6 concentrations using IL-6 assays, identifying its potential as a CML prognostic diagnostic. WBC counts, granulocyte counts, and mid-range absolute counts, or MID counts, were significantly higher in CML patients than in normal control individuals. NGS identified 1643 somatic and sex chromosomal abnormalities and 439 actively expressed genes in CML patients. The findings imply a genomic landscape beyond the BCR-ABL1 mutation in CML development compared to other databases.
Conclusion
In conclusion, this study advances the understanding of the genetic characteristics of CML by identifying mutations in the NO- and H2S-producing genes and their complex connections with genes involved in apoptosis. The comprehensive genetic profile obtained by Sanger sequencing and NGS provides possibilities for identifying novel targets for therapy and personalized treatments for CML, therefore contributing to developments in hematological diseases.
Keywords: next-generation sequencing (ngs), sanger sequencing, next generation sequencing (ngs), chronic myeloid leukaemia, nos3 gene, cth gene
Introduction
Chronic myeloid leukemia (CML), also known as chronic myelogenous leukemia, is a myeloproliferative neoplasm that involves uncontrolled myeloid cell growth [1]. CML differs from other myeloproliferative neoplasms because of the BCR-ABL1 fusion gene and Philadelphia chromosome (Ph) caused by t(9;22)(q34.1;q11.2) [2-4]. Proven (1805) genes from every leukemia subtype have been used to develop the database of leukemia gene literature, or dbLGL [5].
Hydrogen sulfide (H2S) is synthesized internally inside mammalian tissues by the enzymatic actions of cystathionine-β-synthase (CBS), cystathionine γ-lyase, and 3-mercaptopyruvate sulfurtransferase, which is located in the mitochondria. The mechanism regulates the vascular diameter, and protects the endothelium against oxidative stress, ischemia, reperfusion damage, and chronic inflammation, by activating potassium (K+) channels in vascular smooth muscle cells [6,7]. In mammals, nitric oxide synthase (NOS) in tissues generates NO. Neuronal, inducible, and endothelial NOS enzymes convert L-arginine to NO. Endothelial and neuronal NOS (eNOS and nNOS) are constitutive and calcium-dependent isoforms with small NO production. Conversely, the calcium-independent inducible NO synthase (iNOS) may be continually activated. The NO response depends on NO concentrations; lower NO concentrations stimulate cellular growth and suppress apoptosis, whereas greater NO concentrations stop the cell cycle and induce apoptosis [7-12].
Despite developments in CML genetics, little is known about NO and H2S gene mutations and their connections with apoptotic genes. To understand CML genetics, NO and H2S gene mutations and their interactions with apoptotic genes must be studied. This study will explain CML's molecular processes and the complex connection between these mutations and apoptotic genes. This study used Sanger sequencing and next-generation sequencing (NGS) to create a detailed genetic profile of CML, which could lead to novel therapeutic targets and personalized therapies for this hematological disorder.
Materials and methods
Sample collection
Blood samples were collected from 40 CML patients and 40 healthy individuals and placed into ethylene diamine tetraacetic acid (EDTA) tubes. Each tube received 3 ml of sodium citrate for haematology tests, and 3 ml was placed in gel tubes to induce coagulation and collect blood for interleukin (IL) measurement.
Complete blood count
The blood sample was analysed using a Coulter counter (Medonic M16M and M16 models; CLIAwaived Inc., CA, USA) to determine the total white blood cell (WBC), lymphocyte, and monocyte counts.
DNA extraction and quantification
The extraction of genomic DNA from blood samples collected from persons diagnosed with CML was performed using the genomic blood DNA isolation kit (Hibrigen, Turkey) according to the manufacturer's instructions, with some modifications. In summary, blood samples were obtained and promptly handled within a specified time period to avoid DNA deterioration. After extracting the DNA, we assessed both the amount and the quality of the isolated genomic DNA. The DNA concentration was measured using a nanodrop spectrophotometer at a wavelength of 260 nm. In addition, the quality of the extracted DNA was assessed by determining the A260/A280 ratio. A ratio between 1.8 and 2.0 indicates that the DNA is free of contaminants and lacks any protein or other impurities. Only DNA samples with A260/A280 ratios within the acceptable range were selected for downstream applications, ensuring high-quality genomic DNA for further molecular analysis.
Determination of genotype
Three genetic variants within the NOS3 gene and one variant of the CTH gene were studied. Individual amplification of DNA for each variant was performed using polymerase chain reaction (PCR), followed by gel electrophoresis and sequencing analysis. DNA sequencing plays a vital role in understanding genetic diversity and uncovering potential health and disease susceptibility implications. The PCR product underwent sequencing, particularly Sanger sequencing. Initially, the sample sequence was processed at the Kahramanmaraş Sütçü Imam University, ÜSKIM Laboratory, following purification and amplification with specific primers for both directions. Subsequently, a sequencing library was created using the Applied Biosystems ABI 3100 AVANT DNA Sequencer (Thermo Fisher Scientific Inc., Waltham, MA) to enable thorough sequencing analysis. The resulting extension file (AB1) was then scrutinised using Mutation Surveyor software, version 5.2.0 (SoftGenetics, State College, PA) to detect any mutations or variations in the target sequence.
NGS has transformed genomics by granting scientists unparalleled access to extensive genetic information. An essential stage in this procedure involves preparing the sequencing library, which entails converting the desired DNA into a suitable format for the sequencing platform. For this reason, we transferred the DNA samples to the Istanbul Laboratory, in Istanbul, Turkey. After checking the quality and purity of the DNA samples through nanodrop analysis, we proceeded to the next library preparation step. The library preparation process typically commences with fragmentation of the target DNA, followed by adapter ligation and PCR amplification. During the library preparation and sequencing process, numerous sequence artefacts negatively affect raw data quality for downstream analyses. Therefore, quality control and preprocessing of the raw data are crucial steps to ensure the accuracy and reliability of the sequencing results. Various tactics, such as paired-end and mate-pair sequencing, can be applied, which help the assembly of short sequences into contigs and scaffolds. After preparing the library through the standard protocols, we conducted the sequencing step using the DNBSEQ-G400 flexible genome sequencer (MGI Tech Co., Ltd, Thailand), created based on a new flow cell system that could flexibly assist a range of various sequencing modes. The raw data was analyzed using the SAMtools software (Sanger Institute, Cambridgeshire, UK), and then compared with external databases (such as gnomAD, COSMIC, and cBioPortal) to annotate and visualize the results. The subsequent data analysis involved several steps: quality control, read mapping, variant calling, and annotation.
IL-6 measurement
IL-6 levels in the study samples were measured with a particular kit (catalogue no. DE4640; Demeditec Diagnostics, Kiel, Germany). The concentration was calculated using the Stat Fax ELISA reader (Awareness Technology, Inc., Palm City, FL), and statistical analysis was performed using GraphPad Prism, version 10 (GraphPad Software, Inc./Dotmatics, Boston, MA) after establishing a standard curve using MyAssays software (MyAssays Ltd., Brighton, UK). All measurements were taken in triplicate following the manufacturer's recommendations to ensure accuracy. Furthermore, thorough quality control methods were implemented throughout the experimentation phase to validate the results acquired.
Statistical analysis
Comparisons between patients with CML and healthy individuals were performed using an unpaired t-test, and values were presented as means±SEMs. The graphics, computations, and statistical analyses were generated using GraphPad Prism, version 10. A p-value of <0.05 was considered statistically significant.
Ethical considerations
Ethical considerations concerning the collection of human blood samples for research purposes were addressed in accordance with the Declaration of Helsinki. The study was approved by the Human Ethics Research Committee of the College of Science, Salahaddin University-Erbil (under reference number 4/5/439). All patients provided an informed consent to allow their blood samples to be examined.
Results
Complete blood count
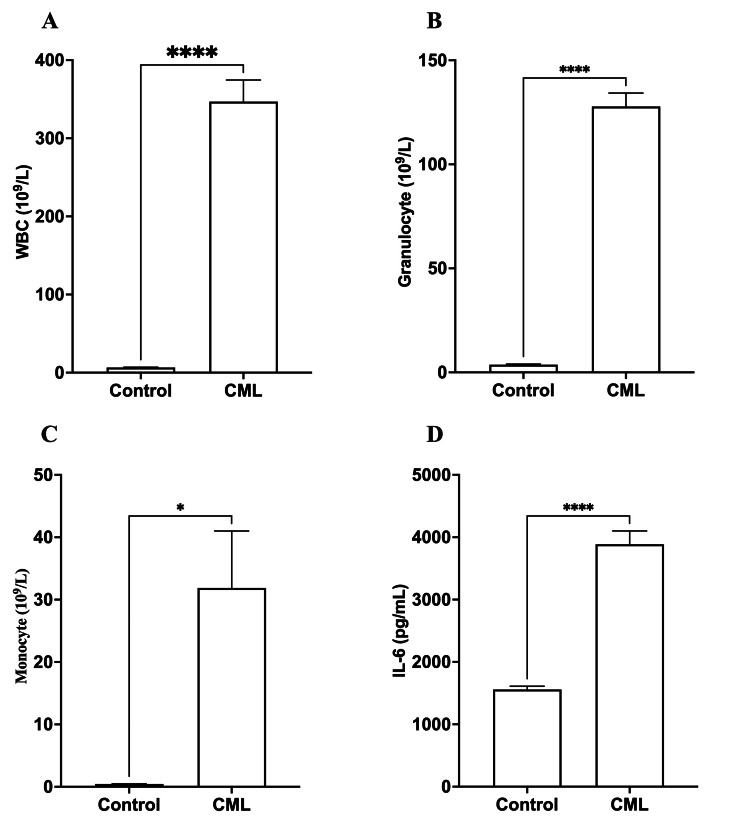
The results indicated significant differences between the control group and CML patients in all examined parameters, including WBC count, granulocyte count, monocyte count, and IL-6 levels. Patients with CML showed significantly higher (p<0.0001) WBC and granulocyte counts than the control group. In addition, monocyte counts were considerably greater (p<0.05) in individuals with CML. Still, the difference was not as noticeable as in WBC and granulocyte counts, as shown in Table 1 and Figures 1A-1C.
Table 1. A comparison of hematological parameters and IL-6 levels between controls and chronic myeloid leukemia (CML) patients.
Patients with CML had significantly greater (p<0.001) total WBC and granulocyte counts. Patients with CML had a significantly higher monocyte count (p<0.05). In CML patients, IL-6 levels were significantly higher (p<0.001).
| Parameters | Control | CML | p-value |
| WBC (109/L) | 6.56±0.38 | 346.9±27.66 | 0.0001 |
| Granulocyte (109/L) | 3.68±0.288 | 127.8±6.448 | 0.0001 |
| Monocyte (109/L) | 0.455±0.035 | 31.87±9.141 | 0.05 |
| IL-6 (pg/mL) | 1558±53.5 | 3888±212.8 | 0.0001 |
Figure 1. A comparison of hematological parameters and IL-6 levels between controls and chronic myeloid leukemia (CML) patients.
Patients with CML had significantly greater (p<0.001) total WBC (A) and granulocyte counts (B). (C) Patients with CML had a significantly higher monocyte count (p<0.05). (D) In CML patients, IL-6 levels were significantly higher (p<0.001).
*p<0.05; ****p<0.0001 vs. healthy individuals
IL-6 concentration
Patients with CML had markedly increased (p<0.0001) levels of IL-6 compared to control individuals, indicating that IL-6 may have a role in the onset or progression of CML, as shown in Table 1 and Figure 1D.
Sanger sequencing
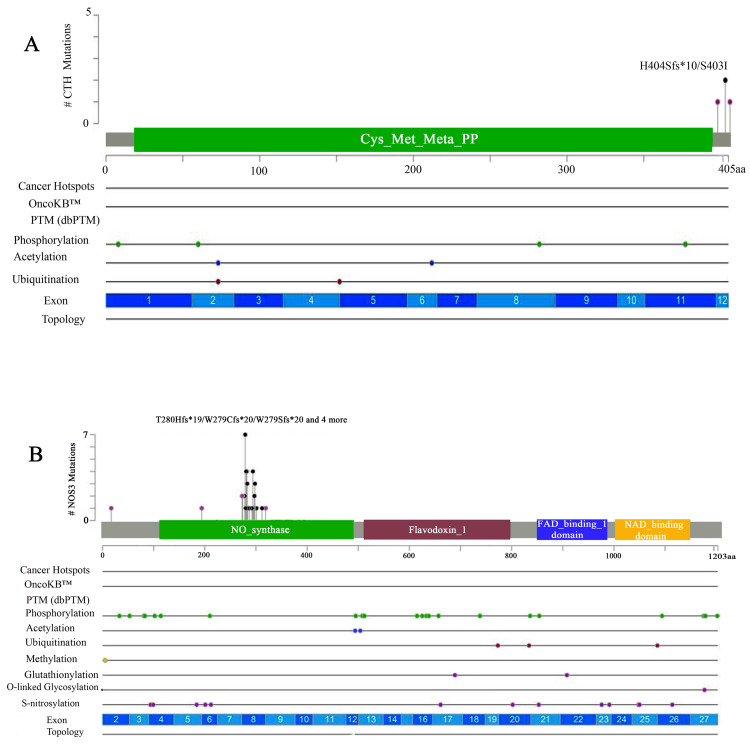
CML mutations were found in 40 NOS3 and CTH gene-sequenced CML patients compared to external databases (gnomAD, COSMIC, and cBioPortal). CTH determined exon 12 missense, substitution, inversion, and duplication mutations (Figure 2a). All missense genes (1:70904800) replicated in multiple patients, and heterozygous mutations (28400G>GT) led to amino acid changes (serine>isoleucine) (dbSNP:1021737), and all mutations were at the end of the cys_met_meta_pp domain, as shown in Figure 3A and Appendix A.
Figure 2. Sanger sequence analysis through the cBioPortal database.
(A) A lollipop mutational map showing the CTH gene mutation. (B) A lollipop mutational map showing the NOS3 gene (VNTR, T786C, and G894T).
PTM, post-translational modification
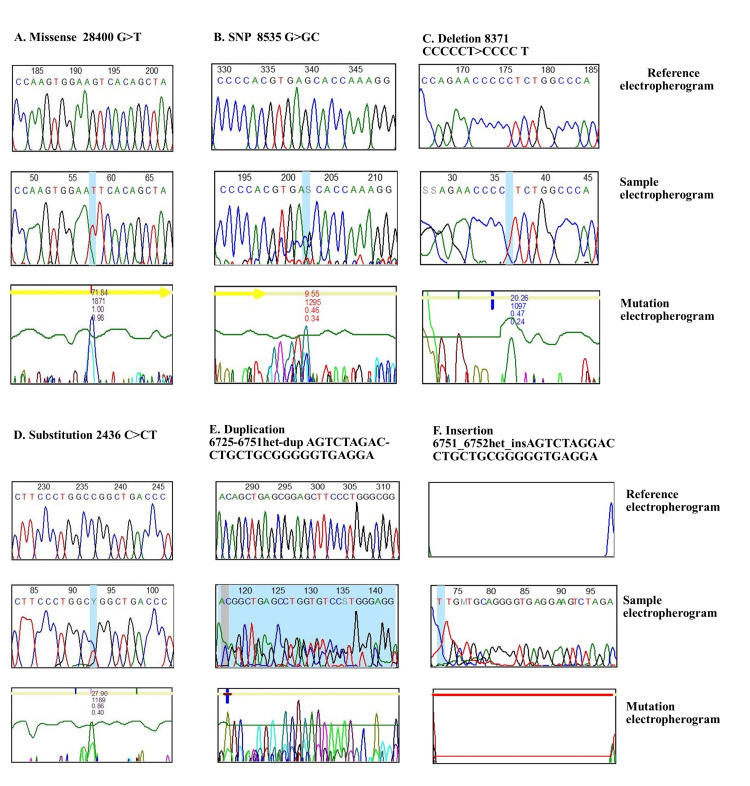
Figure 3. Electropherograms showing the mutational sample with reference.
(A) A Sanger sequence chromatogram for the CTH gene showing the missense mutation (dbSNP:1021737), and amino acid change (serine>isoleucine) in position (28400G>GT); (B) a Sanger sequence chromatogram for the NOS3 gene showing the mutation in the splice region on T786C that changes the nucleotide (8535G>GC); (C) a Sanger sequence chromatogram for the NOS3 gene showing the mutation in the splice region on T786C that changes the nucleotide (8371 C>T); (D) a Sanger sequence chromatogram for the NOS3 gene showing the substitution mutation (dbSNP:2070744) that changes the nucleotide (2436C>T) located on G894T; (E) a Sanger sequence chromatogram for the NOS3 gene showing the duplication mutation 6725_6751het_dupAGTCTAGACCTGCTGCG GGGGTGAGGA) located in VNTR; (F) a Sanger sequence chromatogram for the NOS3 gene showing the duplication mutation 6751_6752het_INSAGTCTAGGACCTGCTGCGGGGGTGAGGA) located in VNTR.
Additionally, the NOS3 gene, which was sequenced using three primers (VNTR 4a/b, T786C, and G894T), found numerous mutations in different locations on the gene (Figure 2b) when compared to external databases (gnomAD, COSMIC, and cBioPortal). The T786C and G894T mutations were located in the NOS3 gene domain, whereas the VNTR change occurred in intron 3 of all T786C patients. These mutations included missense, substitution, synonymous, splice region, and intron mutations. In addition, the (dbSNP:1799983) variant present in many samples had a missense mutation that changed the nucleotides (8468T>TG) on the position of (7:150696111), which replicated in many samples. The other three mutations in the splice region were (7:150696187, 7:150696176, and 7:150696178) and the variants (8533, 8535G>GC, and 8544G>GA) (Figure 3b; Appendix B). However, the G894T primers were sequenced, and different types of variations were estimated, including modifications to nucleotides that mutated sequences, and substitution mutations in all patients. The 21 (dbSNP:2070744) was found through the nucleotide variants (2436C>T) on (7:150690079) (Figure 3d; Appendix C). The VNTR modification was also on NOS3, and all variations that altered nucleotides included mutation types such as substitution, duplication, and insertion. The variant (dbSNP:3918168) is produced by a nucleotide change (6714G>GA) at location (7:150694357). This variant also resulted in duplication and insertion mutations, including (6725_6751het_dupAGTCTAGACCTGCTGCG GGGGTGAGGA) at locations (7:150694368_7:150694394). The VNTR also had an insertion mutation due to a changed nucleotide (6751_6752) (Figure 3e; Appendix D).
Next-generation sequencing
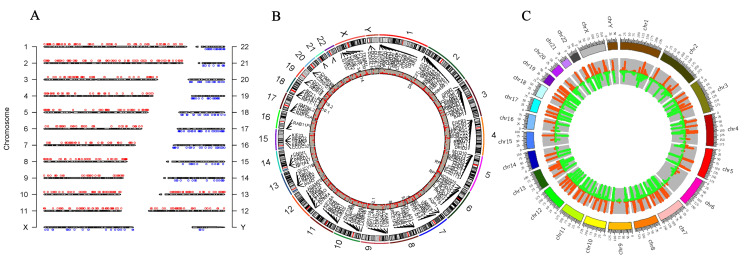
Next-generation whole-genome sequencing identified 1643 somatic and sex chromosomal abnormalities and 439 gene expressions in CML patients. The results were cross-referenced to the gnomAD, COSMIC, and cBioPortal databases. Patients with CML expressed 439 genes. Figure 4A shows how all chromosomes contribute to CML. Specifically, the X chromosome carries 96 of the 106 sex differences. Ninety-four intron alterations occur during gene expression, including upregulation and downregulation. Figure 4C shows the genes CXorf36, ASB11, ZRSR2, and TENM1. The remaining two mutations (out of 96) are unidentified. There are 10 chromosomal Y variants in four genes' intronic regions. Furthermore, chromosome 1 has 98 mutations. There are 69 mutations in 29 expressed genes, and 29 remain unidentified. Among the 69 mutations, CHD1L's frameshift-deletion mutation and PIK3CD's splice region variation stand out. Finally, 67 of the 69 variations are introns. Furthermore, chromosome 2 had 163 alterations, with 95 in 44 expressed genes. The non-transcription region (AC012363.8) had 14 mutations at the same location as MTND4P26, whereas EMILIN1 gained a missense mutation. There were also mutations in the GCA gene's intron and 3' untranslated region (3'UTR). There were 53 more unidentified mutations, including 15 in non-coding areas (Table 2).
Table 2. Variation distribution on genes and chromosomes.
| Chromosome no. | Mutation no. | Gene no. |
| X | 96 | 12 |
| Y | 10 | 4 |
| 1 | 98 | 30 |
| 2 | 158 | 44 |
| 3 | 121 | 28 |
| 4 | 81 | 14 |
| 5 | 87 | 27 |
| 6 | 43 | 14 |
| 7 | 141 | 25 |
| 8 | 101 | 17 |
| 9 | 68 | 17 |
| 10 | 72 | 27 |
| 11 | 83 | 30 |
| 12 | 91 | 23 |
| 13 | 41 | 13 |
| 14 | 25 | 13 |
| 15 | 36 | 8 |
| 16 | 35 | 15 |
| 17 | 68 | 25 |
| 18 | 44 | 12 |
| 19 | 53 | 19 |
| 20 | 29 | 6 |
| 21 | 23 | 8 |
| 22 | 44 | 11 |
Figure 4. Next-generation sequencing (NGS) analysed through the SRplot database.
(A) The chromosome distribution map illustrates the highest concentration of chromosomes; (B) a two-dimensional Circos plot displaying four columns, with the first indicating chromosomes, the second showing starting coordinates, the third indicating end coordinates, and the fourth representing fold change reflecting gene upregulation and downregulation during cancer progression and GC content variability; (C) an RCircos diagram (version 1.2.2, an R package for Circos 2D track plots) depicting gene names to showcase expressed genes and copy number variation, and includes information on chromosome location in the first three columns, along with gene name locations, while log2fc is displayed in another column.
Furthermore, chromosome 3 revealed 121 mutations in 28 expressed genes. These 34 mutations remain unidentified, while the other two were synonymous and located on the NPRL2 gene's PLCL2 non-coding transcript exon. The FLNB and TBCCD1 genes had two identical missense mutations. The remaining alterations were intron-based. Another 14 genes with 81 variations were found on chromosome 4. There were approximately 32 unidentified mutations, totaling 49. Two synonymous mutations and one missense FGFR3 mutation were identified. The remaining alterations affected genes located in introns. Of the 87 mutations on chromosome 5, 40 were unidentified mutations. Figure 4b shows 47 of the 87 variations on 27 expressed genes. This variant had a 3'UTR mutation and a CDH9 gene missense mutation. The RP11-232L2.1 gene had exons that did not code for proteins. PPP2R2B's 5' untranslated region (5'UTR) and GM2A's frameshift mutation were also identified; intronic mutations occurred. There were 14 active genes on chromosome 6. These genes contained 43 variations, including 24 unidentified mutations. The remaining 19 mutations were distributed among 14 genes, with six occurring in similar numbers on RP11-288G3.4 and HLA-V's non-coding transcript exons. Mutations to the HLA-DOA splice region and the TULP4 3'UTR were also identified. The remaining genes were introns.
Chromosome 7 had a total of 141 different variations. Out of 141 occurrences, 79 were characterized by unidentified mutations, while 62 were associated with 25 specific genes. This investigation identified three mutations in the 3'UTR of the AQP1 gene. We also detected two MUC12 and SMO missense mutations, two STRIP2 3'UTR mutations, and a CEP41 mutation. All the remaining ones were introns.
Furthermore, chromosome 8 contained a total of 101 genetic variations; 71 variations were not known and 30 variants had an impact on 17 genes being expressed. Both genes included equal quantities of non-coding transcript exons SMARCE1P4 and RP11-468O2.1. The 5'UTR of the CTSB gene had one mutation, whereas the other mutations were located in the introns. Of the 68 variants found on chromosome 9, 17 were linked to actively expressed genes. There were at least 24 variants that contained mutations whose identity was not known, whereas 44 variations had mutations that had been identified. The 44 variations consisted of two missense mutations in the KANK1 gene, three mutations in the 3'UTR of the CDKN2A gene, a mutation in a non-coding transcript exon of the CCL27 gene, a missense and synonymous mutation in the SURF6 gene, and three mutations in the 3'UTR of the MED22 gene. The most severe mutations were found in introns. The dataset contained a total of 72 mutations located on chromosome 10. There were 22 variations with unidentified alterations, whereas 50 variants were associated with 27 genes. Three mutations were detected in the 3'UTR of the VPS26A gene, whereas the other modifications were inside introns. Out of 83 variations, 26 were linked to mutations on chromosome 11 that were now unidentified. Of the 57 modifications, 30 were associated with expressed genes in intronic regions. However, there were three missense mutations in the PIDD1 gene and three missense and synonymous mutations in the MUC6 gene. Chromosome 12 contained a total of 91 genetic variants. Approximately 36 changes were associated with unidentified mutations, while the remaining mutations were associated with 24 functional genes. The KANSL2 gene harbored two synonymous alterations in its 3'UTR. ITGA7 and NEMP1 had missense mutations, whereas HOXC-AS3 had a mutation in a non-coding transcript exon.
Additionally, chromosome 13 had 41 variants. Twenty variants were related to unexplained mutations, whereas 13 to intron mutations in four expressed genes: TPTE2, PAN3, RXFP2, and LMO7. Chromosome 14 had 25 variants, of which two were unknown mutations. Except for synonymous NEK9 mutations, the remaining 23 variants corresponded to 13 intron-expressed genes. Chromosome 15 had 36 variants, four unexplained mutations, and nine related to expressed genes. All of these changes were intronic except for an MTHFS gene missense mutation. Unknown mutations accounted for 16 of the 35 variants on chromosome 16. The other 19 variants affected nine intron-region genes, including JPT2, TRAF7, and PLCG2.
Chromosome 17 had 68 variations. Among these variations, 13 were linked to unknown mutations, while 55 were related to 25 expressed genes. The ITGAE gene had a frameshift mutation, and the SMTNL2 and CHRNE 3'UTRs were also mutated. ULK2 had a synonymous mutation, whereas WIPF2 had both insertion and synonymous mutations. Intronic areas were mutated again. Chromosome 18 had 44 variations. The introns of 12 expressed genes (AKAIN1, CDH2, and CDH7) exhibited 33 variants. The remaining variations were unknown. Also, 26 of the 53 chromosome 19 mutations contained unknown mutations. An additional 27 variants were related to 19 expressed genes. The WDR18 gene had a synonymous mutation, but DOT1L and TDRD12 had two missense variations. Intron variation continued. Of the 29 chromosome 20 variants, 18 were associated with unknown mutations. This includes 11 changes to the intron regions of six expressed genes. The MIR646HG gene contained just one non-coding transcript exon mutation. Chromosome 21 had 23 variants, two related to unexplained mutations. Except for two C2CD2 and PDXK 3'UTR mutations, the other 21 variations were found in eight expressed genes. All of these variants were found in introns. Chromosome 22 had 44 mutations, 13 of which were unknown. Another 31 variations were linked to 11 BCR-expressed genes and a mutation in FOXRED2's intronic 3'UTR, as shown in Appendix E.
Discussion
Elevated WBC counts are commonly observed in individuals diagnosed with CML [13]. The cost-effective and direct approach for detecting CML involves utilizing differential analysis and CBC techniques [14]; the parameter of CBC generally changes during cancer incidence [15] and also after chemotherapy administration [16]. During the occurrence of cancer, an increase in the total WBC count is observed. It is possible that, following treatment, the WBC counts subsequently decrease. Due to this rationale, the WBC count obtained via the CBC test has emerged as a biomarker for the detection of leukemia. This study observed a high total WBC count, granulocyte count, and MID count.
IL-6 has been postulated as a potential prognostic marker for CML [17]. As a result, IL-6 levels may rise significantly throughout CML, exceeding the baseline rate. The acquired findings were statistically significant, demonstrating an increase in IL-6 levels with the onset of cancer.
The nucleotide sequences of the NOS3 and CTH genes were determined using Sanger sequencing. In CML patient samples, different changes were found in the CTH gene. These changes were all found outside the cys-met-meta-pp domain on exon 12. However, to our knowledge, no previous study has found a relationship between the CTH gene and CML, and this is the first study to show an extensive number of mutations in the CTH gene [18].
Furthermore, the NOS3 gene exhibited distinct mutations in the VNTR, T786C, and G894T genes in colorectal cancer [19]. Notably, all these variants were found within the NOS3 gene, except for specific variants in the VNTR. Together with the tyrosine kinase activator and BCR-ABL1 genes [20], these results show that the NOS3 gene is expressed in people with leukemia.
Many genetic disorders and syndromes have been identified in recent decades using NGS technologies. The utilization of NGS is rapidly becoming standardized as a diagnostic tool and for molecular patient monitoring, enabling the evaluation of treatment effectiveness [21]. The present study's findings indicate that 1643 variations were seen across the 22 chromosomes, including the XY chromosome. Furthermore, gene expression analysis revealed that 439 genes were actively expressed. Additionally, two genes were sequenced using the Sanger method, while one gene was identified using the ELISA technique. Nevertheless, the findings indicate that, apart from BCR-ABL1, several genes are linked to CML development.
A few study limitations may impact the ability to adapt to and understand the results. The study's sample size may not represent the CML population, limiting its external validity. The study's approach relies primarily on observational and genetic analysis, which may introduce biases or confounding factors that are not adequately controlled and addressed.
Conclusions
The study thoroughly investigated the genetic landscape of CML, revealing insights into the delicate interaction between NO, H2S, gene mutations, and apoptotic genes. The NOS3 and CTH gene mutations were identified using Sanger sequencing and NGS, indicating novel interactions with CML pathogenesis. The study found previously unknown mutations in the CTH gene and expanded the understanding of its role in CML. Additionally, various mutations in the NOS3 gene, such as the VNTR, T786C, and G894T variations, revealed CML's complex genetic landscape. The NGS study found 1643 somatic and sex chromosomal abnormalities and 439 actively expressed genes, revealing CML's genomic complexity beyond the well-known BCR-ABL1 mutation. These findings highlight the potential of NGS as a diagnostic and prognostic tool, providing insights into personalized treatment approaches for CML that extend beyond BCR-ABL1 targeting strategies.
Acknowledgments
We would like to thank the Nanakaly Hospital for Hematology & Oncology and the KMCA (Kurdistan Medical Control Agency) Directorate, Iraq, for providing the samples and machinery. We also thank Govand Qader and Twana Alkasalias for designing the primers and helping us in analyzing the sequencing results.
Appendices
Appendix A
Table 3. CTH gene variation with amino acid change in chronic myeloid leukemia (CML).
| Gene | Chromosome position | Mutation | Mutation genotype | Heterozygous/homozygous | Variants | Variant percentage | Amino acid change | External database |
| CTH | ||||||||
| 1:70904757 | Substitution | T>TA | Heterozygous | 28357T>TA | 5.0% | None | Not Found | |
| 1:70904800 | Missense | G>GT | Heterozygous | 28400G>GT | 35.0% | Serine>Isoleucine | dbSNP:1021737 | |
| 1:70904800 | Missense | G>GT | Heterozygous | 28400G>GT | 35.0% | Serine>Isoleucine | dbSNP:102173 | |
| 1:70905047 | Inversion | G>GA | Heterozygous | 28647G>GA | 5.3% | None | Not Found | |
| 1:70904800 | Missense | G>GT | Heterozygous | 28400G>GT | 35.0% | Serine>Isoleucine | dbSNP:1021737 | |
| 1:70904800 | Missense | G>GT | Heterozygous | 28400G>GT | 35.0% | Serine>Isoleucine | dbSNP:1021737 | |
| 1:70905019 | Duplication | A | Heterozygous | 28619het_dupA | 5.0% | None | Not Found | |
| 1:70904810 | Substitution | A>AG | Heterozygous | 28410A>AG | 5.0% | Glycin>Glycin | Not Found | |
| 1:70904811 | Substitution | T>TC | Heterozygous | 28411T>TC | 5.3% | None | Not Found | |
| 1:70905018 | Substitution | T>TA | Heterozygous | 28618T>TA | 5.0% | None | Not Found | |
| 1:70904770 | Substitution | C>CT | Heterozygous | 28370C>CT | 5.3% | None | Not Found | |
| 1:70904772 | Substitution | C>CG | Heterozygous | 28372C>CG | 5.0% | None | Not Found | |
| 1:70904800 | Missense | G>GT | Heterozygous | 28400G>GT | 35.0% | Serine>Isoleucine | dbSNP:1021737 | |
| 1:70904758 | Substitution | A>AT | Heterozygous | 28358A>AT | 4.8% | None | Not Found | |
| 1:70904800 | Missense | G>T | Homozygous | 28400G>T | 35.0% | Serine>Isoleucine | dbSNP:1021737 | |
| 1:70905045 | Inversion | T>TA | Heterozygous | 28645T>TA | 5.0% | None | Not Found | |
| 1:70905047 | Inversion | G>GA | Heterozygous | 28647G>GA | 5.3% | None | Not Found | |
| 1:70904810 | Substitution | G>GC | Heterozygous | 28410G>GC | 4.3% | Valine>Tyrosine | Not Found | |
| 1:70904800 | Missense | G>GT | Heterozygous | 28400G>GT | 35.0% | Serine>Isoleucine | Not Found | |
| 1:70905047 | Inversion | G>GA | Heterozygous | 28647G>GA | 5.3% | None | Not Found | |
| 1:70904758 | Substitution | A>AT | Heterozygous | 28358A>AT | 4.8% | None | Not Found | |
| 1:70905046 | Inversion | T>TA | Heterozygous | 28646T>TA | 5.0% | None | Not Found | |
| 1:70905047 | Inversion | G>GA | Heterozygous | 28647G>GA | 5.3% | None | Not Found | |
| 1:70904810 | Substitution | G>GC | Heterozygous | 28410G>GC | 4.3% | Valine>Tyrosine | Not Found | |
| 1:70904770 | Substitution | C>CT | Heterozygous | 28370C>CT | 5.3% | None | Not Found | |
| 1:70905019 | Duplication | A | Heterozygous | 28619het_dupA | 5.0% | None | Not Found | |
| 1:70905018 | Substitution | T>TA | Heterozygous | 28618T>TA | 5.0% | None | Not Found | |
| 1:70905046 | Inversion | T>TA | Heterozygous | 28646T>TA | 5.0% | None | Not Found | |
| 1:70904811 | Substitution | T>TC | Heterozygous | 28411T>TC | 5.3% | None | Not Found | |
| 1:70904758 | Substitution | A>AT | Heterozygous | 28358A>AT | 4.8% | None | Not Found | |
| 1:70905018 | Substitution | T>TA | Heterozygous | 28618T>TA | 5.0% | None | Not Found | |
| 1:70905045 | Inversion | T>TA | Heterozygous | 28645T>TA | 5.0% | None | Not Found | |
| 1:70904811 | Substitution | T>TC | Heterozygous | 28411T>TC | 5.3% | None | Not Found | |
| 1:70905047 | Inversion | G>GA | Heterozygous | 28647G>GA | 5.3% | None | Not Found | |
| 1:70904810 | Substitution | G>GC | Heterozygous | 28410G>GC | 4.3% | Valine>Tyrosine | Not Found | |
| 1:70904772 | Substitution | C>CG | Heterozygous | 28372C>CG | 5.0% | None | Not Found | |
| 1:70904811 | Substitution | T>TC | Heterozygous | 28411T>TC | 5.3% | None | Not Found | |
| 1:70905018 | Substitution | T>TA | Heterozygous | 28618T>TA | 5.0% | None | Not Found | |
| 1:70905045 | Inversion | T>TA | Heterozygous | 28645T>TA | 5.0% | None | Not Found | |
| 1:70904772 | Substitution | C>CG | Heterozygous | 28372C>CG | 5.0% | None | Not Found |
Appendix B
Table 4. eNOS (786) variation with different types of mutations and amino acid change in chronic myeloid leukemia (CML).
| Gene | Chromosome position | Mutation | Mutation genotype | Heterozygous/homozygous | Variants | Variant percentage | Amino acid change | External database |
| eNOS (786) | ||||||||
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696052 | No mutation | T>TG | Heterozygous | 8409T>TG | 8.7% | Trp>Gly | Not found | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696187 | Splice region | G>GA | Heterozygous | 8544G>GA | 4.3% | None | Not Found | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696038 | Substitution | G>GC | Heterozygous | 8395G>GC | 4.3% | Cys>Ser | Not Found | |
| 7:150696039 | Substitution | C>A | Homozygous | 8396C>A | 8.7% | Cys>Ser | Not Found | |
| 7:150696052 | Substitution | T>G | Homozygous | 8409T>G | 4.3% | Trp>Gly | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696100 | Substitution | G>GC | Heterozygous | 8457G>GC | 13.0% | Ala>Pro | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696038 | Substitution | G>GC | Heterozygous | 8395G>GC | 4.3% | Cys>Ser | Not Found | |
| 7:150696039 | Synonymous | C>A | Homozygous | 8396C>A | 8.7% | Cys>Ser | Not Found | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696014 | Intron/deletion | C>T | Homozygous | 8371C>T | 25.0% | None | Not found | |
| 7:150696098 | Missense | A>AG | Heterozygous | 8455A>AG | 39.1% | Gln>Arg | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Glu | dbSNP:1799983 | |
| 7:150696054 | Substitution | G>GC | Heterozygous | 8411G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696055 | Missense | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696098 | Missense | A>AC | Heterozygous | 8455A>AC | 4.3% | Gln>Pro | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696061 | Substitution | G>GA | Heterozygous | 8418G>GA | 4.3% | Gly>Arg | Not Found | |
| 7:150696058 | Substitution | C>CT | Heterozygous | 8415C>CT | 8.7% | Pro>Leu | Not Found | |
| 7:150696059 | Substitution | C>CT | Heterozygous | 8416C>CT | 8.7% | Pro>Leu | Not Found | |
| 7:150696055 | Substitution | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696098 | Missense | A>AC | Heterozygous | 8455A>AC | 17.4% | Gln>Pro | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Trp>Cys | dbSNP:1799983 | |
| 7:150696054 | Substitution | G>GC | Heterozygous | 8411G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696055 | Substitution | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696061 | Substitution | G>GA | Heterozygous | 8418G>GA | 4.3% | Gly>Arg | Not Found | |
| 7:150696098 | Missense | A>AC | Heterozygous | 8455A>AC | 17.4% | Gln>Pro | Not Found | |
| 7:150696099 | Substitution | G>GC | Heterozygous | 8456G>GC | 4.3% | Gln>His | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696052 | Substitution | T>TC | Heterozygous | 8409T>TC | 4.3% | Trp>Arg | Not Found | |
| 7:150696053 | Substitution | G>GC | Heterozygous | 8410G>GC | 4.3% | Trp>Ser | Not Found | |
| 7:150696055 | Missense | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696058 | Substitution | C>CT | Heterozygous | 8415C>CT | 8.7% | Pro>Ser | Not Found | |
| 7:150696060 | Substitution | A>AT | Heterozygous | 8417A>AT | 4.3% | Pro>Pro | Not Found | |
| 7:150696061 | Substitution | G>GA | Heterozygous | 8418G>GA | 4.3% | Gly>Arg | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696022 | Substitution | C>CA | Heterozygous | 8379C>CA | 4.3% | None | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696030 | Substitution | A>AT | Heterozygous | 8387A>AT | 4.8% | None | Not Found | |
| 7:150696032 | Substitution | A>AT | Heterozygous | 8389A>AT | 4.8% | None | Not Found | |
| 7:150696053 | Substitution | G>GC | Heterozygous | 8410G>GC | 4.3% | Trp>Ser | Not Found | |
| 7:150696055 | Missense | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696060 | Missense | A>AT | Heterozygous | 8417A>AT | 4.3% | Pro>Pro | Not Found | |
| 7:150696061 | Missense | G>GA | Heterozygous | 8418G>GA | 4.3% | Gly>Arg | Not Found | |
| 7:150696064 | Missense | A>AG | Heterozygous | 8421A>AG | 4.5% | Asn>Gly | Not Found | |
| 7:150696065 | Missense | A>AG | Heterozygous | 8422A>AG | 4.2% | Asn>Ser | Not Found | |
| 7:150696093 | Substitution | G>GC | Heterozygous | 8450G>GC | 4.3% | Leu>Leu | Not Found | |
| 7:150696103 | Missense | C>T | Homozygous | 8460C>T | 25.0% | Pro>Ser | Not Found | |
| 7:150696107 | Missense | A>AG | Heterozygous | 8464A>AG | 4.5% | Asp>Gly | Not Found | |
| 7:150696108 | Substitution | T>TG | Heterozygous | 8465T>TG | 91.3% | Asp>Glu | Not Found | |
| 7:150696110 | Missense | A>AC | Heterozygous | 8467A>AC | 4.3% | Asp>Ala | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Ala | dbSNP:1799983 | |
| 7:150696118 | Substitution | G>GT | Heterozygous | 8475G>GT | 4.3% | Glu>Glu | Not Found | |
| 7:150696152 | Substitution | A>AC | Heterozygous | 8509A>AC | 4.5% | Glu>Ala | Not Found | |
| 7:150696054 | Substitution | G>GC | Heterozygous | 8411G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696055 | Missense | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696098 | Missense | A>AC | Heterozygous | 8455A>AC | 17.4% | Gln>Pro | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696014 | Intron/deletion | C>T | Homozygous | 8371C>T | 25.0% | None | Not Found | |
| 7:150696054 | Substitution | G>GC | Heterozygous | 8411G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696055 | Missense | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696058 | Substitution | C>CT | Heterozygous | 8415C>CT | 8.7% | Pro>Ser | Not Found | |
| 7:150696059 | Substitution | C>CT | Heterozygous | 8416C>CT | 8.7% | Pro>Leu | Not Found | |
| 7:150696098 | Missense | A>AC | Heterozygous | 8455A>AC | 17.4% | Gln>Pro | Not Found | |
| 7:150696099 | Substitution | G>GC | Heterozygous | 8456G>GC | 4.3% | Gln>Pro | Not Found | |
| 7:150696111 | Missense | T>G | Homozygous | 8468T>G | 8.7% | Trp>Gly | dbSNP:1799983 | |
| 7:150696049 | Synonymous | G>GC | Heterozygous | 8406G>GC | 4.3% | Gly>Arg | Not Found | |
| 7:150696050 | Substitution | G>GC | Heterozygous | 8407G>GC | 4.3% | Gly>Ala | Not Found | |
| 7:150696052 | Substitution | T>C | Homozygous | 8409T>C | 4.3% | Trp>Arg | Not Found | |
| 7:150696058 | Substitution | C>CT | Heterozygous | 8415C>CT | 8.7% | Pro>Ser | Not Found | |
| 7:150696059 | Substitution | C>T | Homozygous | 8416C>T | 25.0% | Pro>Leu | Not Found | |
| 7:150696063 | Substitution | A>AG | Heterozygous | 8420A>AG | 4.5% | Gly>Gly | Not Found | |
| 7:150696064 | Substitution | A>AG | Heterozygous | 8421A>AG | 4.5% | Asn>Ala | Not Found | |
| 7:150696065 | Substitution | A>AC | Heterozygous | 8422A>AC | 4.5% | Asn>Thr | Not Found | |
| 7:150696077 | Substitution | A>AC | Heterozygous | 8434A>AC | 4.3% | Asp>Ala | Not Found | |
| 7:150696052 | Substitution | T>TC | Heterozygous | 8409T>TC | 4.3% | Cys>Arg | Not Found | |
| 7:150696053 | Substitution | G>GC | Heterozygous | 8410G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696054 | Substitution | G>C | Heterozygous | 8411G>C | 8.7% | Trp>Cys | Not Found | |
| 7:150696059 | Substitution | C>T | Heterozygous | 8416C>T | 25.0% | Pro>Leu | Not Found | |
| 7:150696061 | Substitution | G>GA | Heterozygous | 8418G>GA | 4.3% | Gly>Arg | Not Found | |
| 7:150696069 | Substitution | T>TC | Heterozygous | 8426T>TC | 4.2% | Gly>Gly | Not Found | |
| 7:150696096 | Substitution | G>GC | Heterozygous | 8453G>GC | 4.3% | Leu>Leu | Not Found | |
| 7:150696099 | Substitution | G>C | Heterozygous | 8456G>C | 8.7% | Gln>His | Not Found | |
| 7:150696100 | Substitution | G>C | Heterozygous | 8457G>C | 8.7% | Ala>Pro | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Ala | dbSNP:1799983 | |
| 7:150696176 | Splice region | G>GC | Heterozygous | 8533G>GC | 10.0% | None | Not Found | |
| 7:150696177 | Substitution | A>AC | Heterozygous | 8534A>AC | 4.5% | None | Not Found | |
| 7:150696178 | Splice region | G>GC | Heterozygous | 8535G>GC | 10.0% | None | Not Found | |
| 7:150696052 | Substitution | T>TC | Heterozygous | 8409T>TC | 4.3% | Cys>Arg | Not Found | |
| 7:150696053 | Substitution | G>GC | Heterozygous | 8410G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696054 | Substitution | G>C | Heterozygous | 8411G>C | 8.7% | Trp>Cys | Not Found | |
| 7:150696061 | Substitution | G>GA | Heterozygous | 8418G>GA | 4.3% | Gly>Arg | Not Found | |
| 7:150696062 | Substitution | G>GA | Heterozygous | 8419G>GA | 4.3% | Gly>Lys | Not Found | |
| 7:150696096 | Substitution | G>GC | Heterozygous | 8453G>GC | 4.3% | Leu>Leu | Not Found | |
| 7:150696099 | Substitution | G>C | Heterozygous | 8456G>C | 8.7% | Gln>His | Not Found | |
| 7:150696100 | Substitution | G>C | Heterozygous | 8457G>C | 8.7% | Ala>Pro | Not Found | |
| 7:150696052 | Substitution | T>TC | Heterozygous | 8409T>TC | 4.3% | Trp>Arg | Not Found | |
| 7:150696053 | Substitution | G>GC | Heterozygous | 8410G>GC | 4.3% | Trp>Ser | Not Found | |
| 7:150696055 | Missense | A>AC | Heterozygous | 8412A>AC | 13.0% | Thr>Pro | Not Found | |
| 7:150696054 | Substitution | G>GC | Heterozygous | 8411G>GC | 13.0% | Trp>Cys | Not Found | |
| 7:150696059 | Substitution | C>T | Homozygous | 8416C>T | 25.0% | Pro>Leu | Not Found | |
| 7:150696061 | Substitution | G>GT | Heterozygous | 8418G>GT | 4.3% | Gly>Gly | Not Found | |
| 7:150696096 | Substitution | G>GC | Heterozygous | 8453G>GC | 4.3% | Leu>Leu | Not Found | |
| 7:150696098 | Missense | A>AC | Heterozygous | 8455A>AC | 17.4% | Gln>Pro | Not Found | |
| 7:150696099 | Substitution | G>C | Heterozygous | 8456G>C | 8.7% | Gln>His | Not Found | |
| 7:150696111 | Missense | T>TG | Heterozygous | 8468T>TG | 91.3% | Asp>Ala | dbSNP:1799983 |
Appendix C
Table 5. eNOS (894) variation with mutations in chronic myeloid leukemia (CML).
| Gene | Chromosome position | Mutation | Mutation genotype | Heterozygous/homozygous | Variants | Variant percentage | Amino acid change | External database |
| eNOS (894) | ||||||||
| 7:150690102 | Other | G>GT | Heterozygous | 2459G>GT | 4.0% | None | Not found | |
| 7:150690119 | Other | C>CG | Heterozygous | 2476C>CG | 3.8% | None | Not found | |
| 7:150690120 | Other | G>GT | Heterozygous | 2477G>GT | 4.0% | None | Not found | |
| 7:150690121 | Substitution | G>GT | Heterozygous | 2478G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 4.2% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>CT | Heterozygous | 2436C>CT | 4.0% | None | dbSNP:2070744 | |
| 7:150690118 | Substitution | G>GT | Heterozygous | 2475G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 4.2% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 4.2% | None | dbSNP:2070744 | |
| 7:150689998 | Substitution | G>GT | Heterozygous | 2355G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 4.2% | None | dbSNP:2070744 | |
| 7:150689996 | Substitution | C>CT | Heterozygous | 2353C>CT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150689998 | Substitution | G>GT | Heterozygous | 2355G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>CT | Homozygous | 2436C>CT | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>CT | Homozygous | 2436C>CT | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150690102 | Substitution | G>GT | Heterozygous | 2459G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150689998 | Substitution | G>GT | Heterozygous | 2355G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150690060 | Substitution | C>CA | Heterozygous | 2417C>CA | 4.3% | None | Not found | |
| 7:150690062 | Substitution | T>TA | Heterozygous | 2419T>TA | 7.0% | None | Not found | |
| 7:150690063 | Substitution | C>CA | Heterozygous | 2420C>CA | 4.3% | None | Not found | |
| 7:150690065 | Substitution | A>AG | Heterozygous | 2422A>AG | 6.2% | None | Not found | |
| 7:150690067 | Substitution | C>CG | Heterozygous | 2424C>CG | 100.0% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150689998 | Substitution | G>GT | Heterozygous | 2355G>GT | 4.0% | None | Not found | |
| 7:150689998 | Substitution | G>GT | Heterozygous | 2355G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>CT | Homozygous | 2436C>CT | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>CT | Homozygous | 2436C>CT | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150690047 | Substitution | G>GC | Heterozygous | 2404G>GC | 25.0% | None | Not found | |
| 7:150690048 | Substitution | C>T | Homozygous | 2405C>T | 4.0% | None | Not found | |
| 7:150690062 | Substitution | T>TG | Heterozygous | 2419T>TG | 4.2% | None | Not found | |
| 7:150690078 | Substitution | C>CA | Heterozygous | 2435C>CA | 4.3% | None | Not found | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150689998 | Substitution | G>GT | Heterozygous | 2355G>GT | 4.0% | None | Not found | |
| 7:150690079 | Substitution | C>CT | Heterozygous | 2436C>CT | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 | |
| 7:150690079 | Substitution | C>T | Homozygous | 2436C>T | 91.3% | None | dbSNP:2070744 |
Appendix D
Table 6. eNOS (VNTR) variation with different types of mutations in chronic myeloid leukemia (CML).
| Gene | Chromosome position | Mutation | Mutation genotype | Heterozygous/homozygous | Variants | Variant percentage | Amino acid change | External database |
| eNOS (VNTR) | ||||||||
| 7:150694357 | Substitution | G>GA | Heterozygous | 6714G>GA | 16.7% | None | dbSNP:3918168 | |
| 7:150694570 | Substitution | C>CA | Heterozygous | 6927C>CA | 4.3% | None | Not Found | |
| 7:150694571 | Substitution | C>CA | Heterozygous | 6928C>CA | 4.3% | None | Not Found | |
| 7:150694598 | Substitution | C>CG | Heterozygous | 6955C>CG | 7.1% | None | Not Found | |
| 7:150694617 | Substitution | C>CA | Heterozygous | 6974C>CA | 4.3% | None | Not Found | |
| 7:150694619 | Substitution | C>CA | Heterozygous | 6976C>CA | 4.3% | None | Not Found | |
| 7:150694620 | Substitution | C>CA | Heterozygous | 6977C>CA | 4.3% | None | Not Found | |
| 7:150694621 | Substitution | T>TA | Heterozygous | 6978T>TA | 4.5% | None | Not Found | |
| 7:150694622 | Substitution | G>GA | Heterozygous | 6979G>GA | 16.7% | None | Not Found | |
| 7:150694624 | Substitution | G>GA | Heterozygous | 6981G>GA | 16.7% | None | Not Found | |
| 7:150694357 | Substitution | G>GA | Heterozygous | 6714G>GA | 16.7% | None | dbSNP:3918168 | |
| 7:150694368_ 7:150694394 | Duplication | AGTCTA GACCTG CTGCGG GGGTGA GGA | Heterozygous | 6725_6751het_ dupAGTCTAG ACCTGCTGCG GGGGTGAGGA | 5.6% | None | Not Found | |
| 7:150694395 | Substitution | C>CA | Heterozygous | 6752C>CA | 4.3% | None | Not Found | |
| 7:150694606 | Substitution | A>G | Homozygous | 6963A>G | 7.1% | None | Not Found | |
| 7:150694619 | Substitution | C>CA | Heterozygous | 6976C>CA | 4.3% | None | Not Found | |
| 7:150694620 | Substitution | C>CA | Heterozygous | 6977C>CA | 4.3% | None | Not Found | |
| 7:150694357 | Substitution | G>GA | Heterozygous | 6714G>GA | 16.7% | None | dbSNP:3918168 | |
| 7:150694394_ 7:150694395 | Insertion | AGTCTA GGACCT GCTGCG GGGGTG AGGA | Heterozygous | 6751_6752het_ insAGTCTAGG ACCTGCTGCGG GGGTGAGGA | 5.6% | None | Not Found | |
| 7:150694619 | Substitution | C>CA | Heterozygous | 6976C>CA | 4.3% | None | Not Found | |
| 7:150694620 | Substitution | C>CA | Heterozygous | 6977C>CA | 4.3% | None | Not Found | |
| 7:150694621 | Substitution | C>CA | Heterozygous | 6978T>TA | 4.5% | None | Not Found | |
| 7:150694620 | Substitution | C>CA | Heterozygous | 6977C>CA | 4.3% | None | Not Found | |
| 7:150694622 | Substitution | G>GA | Heterozygous | 6979G>GA | 16.7% | None | Not Found | |
| 7:150694623 | Substitution | T>TA | Heterozygous | 6980T>TA | 4.5% | None | Not Found | |
| 7:150694620 | Substitution | C>CA | Heterozygous | 6977C>CA | 4.3% | None | Not Found | |
| 7:150694622 | Substitution | G>GA | Heterozygous | 6979G>GA | 16.7% | None | Not Found | |
| 7:150694346_ 7:150694347 | Insertion | SACCTG MTGCA GGGGT GAGGA GTCTA | Heterozygous | 6703_6704ins SACCTGMTG CAGGGGTGA GGAGTCTA | 10.5% | None | Not Found | |
| 7:150694337 | Substitution | A>AT | Heterozygous | 6694A>AT | 5.3% | None | Not Found | |
| 7:150694348 | Substitution | A>AT | Heterozygous | 6705A>AC | 7.1% | None | Not Found | |
| 7:150694348_ 7:150694349 | Insertion | TTGMTG CAGGGG TGAGGA AGTCTAG A | Heterozygous | 6705_6706ins TTGMTGCAG GGGTGAGGA AGTCTAGA | 10.5% | None | Not Found | |
| 7:150694325 | Substitution | G>GC | Heterozygous | 6682G>GC | 5.3% | None | Not Found | |
| 7:150694328 | Substitution | G>GT | Heterozygous | 6685G>GT | 7.1% | None | Not Found | |
| 7:150694385_ 7:150694386 | Insertion | TGGAGC CTGCCCA GTATAGA ACTGCTG CGG | Heterozygous | 6742_6743het_ insTGGAGCCT GCCCAGTATA GAACTGCTGC GG | 5.6% | None | Not Found | |
| 7:150694318 | Substitution | T>TC | Heterozygous | 6675T>TC | 5.4% | None | Not Found | |
| 7:150694321 | Substitution | A>C | Homozygous | 6678A>C | 7.1% | None | Not Found | |
| 7:150694328 | Substitution | G>GT | Heterozygous | 6685G>GT | 4.3% | None | Not Found |
Appendix E
Table 7. NGS with all variations and mutation types and expressed genes in chronic myeloid leukemia (CML).
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| X | 96 | 94 | 2 | CXorf36 | 7 | INTRON | 0 |
| GYG2 | 1 | INTRON | 0 | ||||
| ASB11 | 1 | INTRON | 0 | ||||
| CA5B | 2 | INTRON | 0 | ||||
| ZRSR2 | 3 | INTRON | 0 | ||||
| CASK | 1 | INTRON | 0 | ||||
| RP11-342D14. | 7 | INTRON | 0 | ||||
| CYSLTR1 | 9 | INTRON | 0 | ||||
| TENM1 | 23 | INTRON | 0 | ||||
| ENOX2 | 7 | INTRON | 0 | ||||
| LINC01201 | 1 | INTRON | 0 | ||||
| MTCP1 | 3 | INTRON | 0 | ||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| Y | 10 | 10 | DUX4L16 | 2 | INTRON | 0 | |
| DUX4L17 | 2 | INTRON | 0 | ||||
| DUX4L18 | 2 | INTRON | 0 | ||||
| MED14P1 | 4 | INTRON | 0 | ||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 1 | 98 | 69 | 29 | LINC01786 | 2 | INTRON | 0 |
| CAMTA1 | 2 | INTRON | 0 | ||||
| PIK3CD | 1 | INTRON/Splice region | 0 | ||||
| CASZ1 | 1 | INTRON | 0 | ||||
| C1orf127 | 1 | INTRON | 0 | ||||
| HMGCL | 1 | INTRON | 0 | ||||
| NCMAP | 1 | INTRON | 0 | ||||
| ZCCHC17 | 1 | INTRON | 0 | ||||
| AGO3 | 1 | INTRON | 0 | ||||
| PTPRF | 1 | INTRON | 0 | ||||
| PODN | 1 | INTRON | 0 | ||||
| DAB1 | 3 | INTRON | 0 | ||||
| AK5 | 1 | INTRON | 0 | ||||
| BCL10-AS1 | 1 | INTRON | 0 | ||||
| RP11-421L21.3 | 2 | INTRON | 0 | ||||
| RAP1A | 1 | INTRON | 0 | ||||
| MAGI3 | 2 | INTRON | 0 | ||||
| SPAG17 | 1 | INTRON | 0 | ||||
| CHD1L | 1 | Frameshift/deletion | |||||
| CHD1L | 1 | INTRON | 0 | ||||
| FCRL4 | 1 | INTRON | 0 | ||||
| RP11-550P17.5 | 1 | INTRON | 0 | ||||
| TBX19 | 1 | INTRON | 0 | ||||
| PAPPA2 | 1 | INTRON | 0 | ||||
| LHX4 | 4 | INTRON | 0 | ||||
| LHX4 | 2 | 3UTR | 0 | ||||
| ACBD6 | 9 | INTRON | 0 | ||||
| PLEKHA6 | 2 | INTRON | 0 | ||||
| LAMB3 | 2 | INTRON | 0 | ||||
| HHAT | 1 | INTRON | 0 | ||||
| HEATR1 | 1 | INTRON | 0 | ||||
| CHRM3 | 1 | INTRON | 0 | ||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 2 | 166 | 95 | 68 | DNAJC27-AS1 | 7 | INTRON | 0 |
| HAAO | 1 | INTRON | 0 | ||||
| SNTG2 | 1 | INTRON | 0 | ||||
| TSSC1 | 1 | INTRON | 0 | ||||
| RNF144A | 1 | INTRON | 0 | ||||
| FAM228A | 1 | INTRON | 0 | ||||
| EFR3B | 1 | INTRON | 0 | ||||
| EMILIN1 | 1 | 0 | Missense | ||||
| FAM179A | 2 | INTRON | 0 | ||||
| ALK | 1 | INTRON | 0 | ||||
| AC009499.1 | 2 | INTRON | 0 | ||||
| THADA | 1 | INTRON | 0 | ||||
| CAMKMT | 1 | INTRON | 0 | ||||
| PRKCE | 2 | INTRON | 0 | ||||
| AC007682.1 | 1 | INTRON | 0 | ||||
| ACYP2 | 1 | INTRON | 0 | ||||
| PNPT1 | 1 | INTRON | 0 | ||||
| ZNF638 | 4 | INTRON | 0 | ||||
| STARD7-AS1 | 1 | INTRON | 0 | ||||
| AC021188.4 | 1 | INTRON | 0 | ||||
| ANKRD36 | 1 | INTRON | 0 | ||||
| RANBP2 | 1 | INTRON | 0 | ||||
| MERTK | 1 | INTRON | 0 | ||||
| EPB41L5 | 2 | INTRON | 0 | ||||
| AC012363.4 | 13 | INTRON | 0 | ||||
| AC012363.8 | 1 | Non-coding transcription | 0 | ||||
| MTND4P26 | 14 | Non-coding transcription | 0 | ||||
| RALB | 2 | INTRON | 0 | ||||
| AC018866.1 | 1 | INTRON | 0 | ||||
| CNTNAP5 | 1 | INTRON | 0 | ||||
| FAM168B | 2 | INTRON | 0 | ||||
| ITGB6 | 1 | INTRON | 0 | ||||
| GCA | 1 | 3utr | 0 | ||||
| ABCB11 | 2 | INTRON | 0 | ||||
| MTX2 | 1 | INTRON | 0 | ||||
| LINC01473 | 2 | INTRON | 0 | ||||
| HECW2 | 1 | INTRON | 0 | ||||
| PLCL1 | 1 | INTRON | 0 | ||||
| LINC01877 | 2 | INTRON | 0 | ||||
| PTH2R | 1 | INTRON | 0 | ||||
| XRCC5 | 1 | INTRON | 0 | ||||
| RPL37A | 1 | INTRON | 0 | ||||
| UGT1A10 | 1 | INTRON | 0 | ||||
| UBE2F | 1 | INTRON | 0 | ||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 3 | 121 | 87 | 34 | PLCL2 | 2 | Synonymous | |
| GRM7 | 1 | INTRON | |||||
| SGO1-AS1 | 1 | INTRON | |||||
| RBMS3 | 1 | INTRON | |||||
| LINC00693 | 3 | INTRON | |||||
| KRBOX1 | 1 | INTRON | |||||
| CDCP1 | 1 | INTRON | |||||
| CCR5AS | 3 | INTRON | |||||
| NPRL2 | 1 | Non-coding transcript exon | |||||
| BAP1 | 1 | INTRON | |||||
| CACNA2D3 | 1 | INTRON | |||||
| IL17RD | 4 | INTRON | |||||
| FLNB | 54 | INTRON | |||||
| FLNB | 2 | Missense | |||||
| CFAP20DC | 1 | INTRON | |||||
| MAGI1 | 1 | INTRON | |||||
| SLC25A26 | 2 | INTRON | |||||
| FRMD4B | 21 | INTRON | |||||
| RAB7A | 6 | INTRON | |||||
| EPHB1 | 1 | INTRON | |||||
| CP | 1 | INTRON | |||||
| SSR3 | 1 | INTRON | |||||
| SI | 1 | INTRON | |||||
| PEX5L | 1 | INTRON | |||||
| LINC01206 | 1 | INTRON | |||||
| MAP3K13 | 1 | INTRON | |||||
| TBCCD1 | 1 | 0 | Missense | ||||
| TBCCD1 | 1 | 0 | Missense | ||||
| P3H2 | 1 | INTRON | |||||
| PAK2 | 1 | 3UTR | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 4 | 81 | 49 | 32 | FGFR3 | 2 | 0 | Synonymous |
| FGFR3 | 1 | 0 | Missense | ||||
| LRPAP1 | 1 | INTRON | |||||
| PPP2R2C | 1 | INTRON | |||||
| SORCS2 | 1 | INTRON | |||||
| SORCS2 | 1 | INTRON | |||||
| QDPR | 22 | INTRON | |||||
| RBM47 | 1 | INTRON | |||||
| FIP1L1 | 5 | INTRON | |||||
| DRID2 | 3 | INTRON | |||||
| RP11-729M20.1 | 7 | INTRON | |||||
| GSTCD | 2 | INTRON | |||||
| ZGRF1 | 1 | INTRON | |||||
| LINC01098 | 1 | INTRON | |||||
| TENM3 | 1 | INTRON | |||||
| SORBS2 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 5 | 87 | 47 | 40 | MYO10 | 3 | INTRON | |
| ADCY2 | 1 | INTRON | |||||
| SEMA5A | 2 | INTRON | |||||
| LINC02150 | 1 | INTRON | |||||
| CDH9 | 1 | 3'UTR | |||||
| CDH9 | 1 | 0 | Missense | ||||
| RP11-232L2.1 | 1 | 0 | Non-coding transcript exon | ||||
| LINC01340 | 2 | INTRON | |||||
| LINC02113 | 1 | INTRON | |||||
| FBXL17 | 1 | INTRON | |||||
| FER | 1 | INTRON | |||||
| NREP | 1 | INTRON | |||||
| YTHDC2 | 3 | INTRON | |||||
| KIF3A | 1 | INTRON | |||||
| PITX1-AS1 | 1 | INTRON | |||||
| ANKHD1 | 2 | INTRON | |||||
| PPP2R2B | 1 | 5' UTR | |||||
| GM2A | 1 | 0 | Frameshift | ||||
| GRIA1 | 1 | INTRON | |||||
| GEMIN5 | 2 | INTRON | |||||
| CLINT1 | 1 | INTRON | |||||
| CTC-535M15.2 | 1 | INTRON | |||||
| FBXW11 | 4 | INTRON | |||||
| CTB-32H22.1 | 1 | INTRON | |||||
| CPEB4 | 1 | INTRON | |||||
| NSG2 | 2 | INTRON | |||||
| COL23A1 | 8 | INTRON | |||||
| ZFP2 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 6 | 43 | 19 | 24 | LINC02525 | 1 | INTRON | |
| RP11-288G3.4 | 3 | 0 | Non-coding transcript exon | ||||
| CASC15 | 1 | INTRON | |||||
| ZKSCAN3 | 1 | INTRON | |||||
| HLA-V | 3 | 0 | Non-coding transcript exon | ||||
| HLA-DOA | 1 | 0 | Splice region | ||||
| PKHD1 | 1 | INTRON | |||||
| DST | 1 | INTRON | |||||
| EYS | 1 | INTRON | |||||
| KCNQ5 | 1 | INTRON | |||||
| PLAGL1 | 1 | INTRON | |||||
| TULP4 | 1 | 3'UTR | |||||
| FNDC1 | 1 | INTRON | |||||
| PDE10A | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 7 | 141 | 62 | 79 | AQP1 | 1 | INTRON | |
| 3 | 3'UTR | ||||||
| NME8 | 1 | INTRON | |||||
| VPS41 | 1 | INTRON | |||||
| DBNL | 1 | INTRON | |||||
| SEPT7P2 | 2 | INTRON | |||||
| AC004870.3 | 5 | INTRON | |||||
| SEC61G-DT | 1 | INTRON | |||||
| LANCL2 | 1 | INTRON | |||||
| CACNA2D1 | 1 | INTRON | |||||
| PPP1R9A | 1 | INTRON | |||||
| MUC12 | 1 | 0 | Missense | ||||
| CUX1 | 1 | INTRON | |||||
| PRKRIP1 | 10 | INTRON | |||||
| ALKBH4 | 4 | INTRON | |||||
| AC002463.3 | 1 | INTRON | |||||
| COMETT | 1 | INTRON | |||||
| CFTR | 1 | INTRON | |||||
| SMO | 3 | INTRON | |||||
| SMO | 1 | Missense | |||||
| STRIP2 | 2 | 3'UTR | |||||
| CEP41 | 1 | 3'UTR | |||||
| CALD1 | 2 | INTRON | |||||
| ZNF425 | 7 | INTRON | |||||
| ZNF398 | 5 | INTRON | |||||
| RP4-555L14.4 | 3 | INTRON | |||||
| PTPRN2 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 8 | 101 | 30 | 71 | ERICH1-AS1 | 1 | INTRON | |
| ARHGEF10 | 2 | INTRON | |||||
| FAM167A | 1 | INTRON | |||||
| CTSB | 1 | 5'UTR | |||||
| EPHX2 | 1 | INTRON | |||||
| DCTN6 | 2 | INTRON | |||||
| SMARCE1P4 | 1 | Non-coding transcript exon | |||||
| RP11-56A10.1 | 1 | INTRON | |||||
| CHD7 | 1 | INTRON | |||||
| NCOA2 | 10 | INTRON | |||||
| RP11-463D19.2 | 1 | INTRON | |||||
| OSGIN2 | 1 | INTRON | |||||
| CDH17 | 1 | INTRON | |||||
| RP11-468O2.1 | 1 | Non-coding transcript exon | |||||
| TRAPPC9 | 1 | INTRON | |||||
| DENND3 | 1 | INTRON | |||||
| LNCOC1 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 9 | 68 | 47 | 22 | KANK1 | 2 | Missense | |
| SLC24A1 | 1 | INTRON | |||||
| CDKN2A | 3 | 3'UTR | |||||
| CDKN2A | 3 | INTRON | |||||
| CCL27 | 1 | 0 | Non-coding transcript exon | ||||
| TRPM6 | 1 | INTRON | |||||
| VPS13A | 2 | INTRON | |||||
| PSAT1 | 1 | INTRON | |||||
| CENPP | 1 | INTRON | |||||
| GSN | 1 | INTRON | |||||
| NR6A1 | 2 | INTRON | |||||
| LMX1B | 1 | INTRON | |||||
| NIBAN2 | 2 | INTRON | |||||
| NIBAN2 | 1 | INTRON | |||||
| SLC25A25-AS1 | 1 | INTRON | |||||
| SURF6 | 13 | INTRON | |||||
| SURF6 | 1 | INTRON | |||||
| SURF6 | 1 | 0 | Missense | ||||
| SURF6 | 1 | 0 | Synonymous | ||||
| MED22 | 2 | 3'UTR | |||||
| MED22 | 1 | 3'UTR | |||||
| LL09NC01-254D11.1 | 1 | INTRON | |||||
| VAV2 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 10 | 72 | 50 | 22 | LARP48 | 2 | INTRON | |
| FRMD4A | 1 | INTRON | |||||
| ST8SIA6 | 2 | INTRON | |||||
| NEBL | 1 | INTRON | |||||
| ABL1 | 4 | INTRON | |||||
| C10orf68 | 2 | INTRON | |||||
| ZNF33B | 5 | INTRON | |||||
| AGAP9 | 1 | INTRON | |||||
| ASAH2 | 1 | INTRON | |||||
| ANK3 | 1 | INTRON | |||||
| JMJD1C | 1 | INTRON | |||||
| CTNNA3 | 4 | INTRON | |||||
| VPS26A | 3 | 3'UTR | |||||
| VPS26A | 2 | INTRON | |||||
| LINC02622 | 1 | INTRON | |||||
| NRG3 | 3 | INTRON | |||||
| CCSER2 | 2 | INTRON | |||||
| GRID1 | 3 | INTRON | |||||
| SNCG | 1 | INTRON | |||||
| FRA10AC1 | 1 | INTRON | |||||
| ENTPD1 | 2 | INTRON | |||||
| BTRC | 1 | INTRON | |||||
| SUFU | 2 | INTRON | |||||
| LINC02661 | 6 | INTRON | |||||
| VTI1A | 1 | INTRON | |||||
| ATRNL1 | 4 | INTRON | |||||
| ATE1 | 1 | INTRON | |||||
| TACC2 | 6 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 11 | 83 | 57 | 26 | HRAS | 1 | INTRON | |
| PIDD1 | 1 | INTRON | |||||
| PIDD1 | 3 | 0 | Missense | ||||
| MUC6 | 1 | 0 | Synonymous | ||||
| MUC6 | 1 | 0 | Missense | ||||
| CTSD | 3 | INTRON | |||||
| KIF18A | 2 | INTRON | |||||
| RCN1 | 7 | INTRON | |||||
| WT1 | 3 | INTRON | |||||
| ALX4 | 1 | INTRON | |||||
| OR9Q1 | 2 | INTRON | |||||
| AHNAK | 1 | INTRON | |||||
| SLC22A6 | 1 | INTRON | |||||
| EHBP1L1 | 4 | INTRON | |||||
| KDM2A | 2 | INTRON | |||||
| LINC02754 | 8 | INTRON | |||||
| CAPN5 | 2 | INTRON | |||||
| TENM4 | 1 | INTRON | |||||
| DLG2 | 3 | INTRON | |||||
| DISC1FP1 | 5 | INTRON | |||||
| HEPHL1 | 2 | INTRON | |||||
| DYNC2H1 | 1 | INTRON | |||||
| RP11-144G7.2 | 1 | INTRON | |||||
| C11orf65 | 1 | INTRON | |||||
| ALG6 | 1 | INTRON | |||||
| LINC02762 | 5 | INTRON | |||||
| PHLDB1 | 2 | INTRON | |||||
| CBL | 3 | INTRON | |||||
| USP2-AS1 | 6 | INTRON | |||||
| NECTIN1 | 1 | INTRON | |||||
| GRIK4 | 1 | INTRON | |||||
| RP11-744N12.3 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 12 | 91 | 55 | 36 | DYRK4 | 24 | INTRON | |
| ANO2 | 1 | INTRON | |||||
| ETV6 | 7 | INTRON | |||||
| PTPRO | 1 | INTRON | |||||
| IRAG2 | 1 | INTRON | |||||
| KIF21A | 1 | INTRON | |||||
| ADAMTS20 | 2 | INTRON | |||||
| OR8S1 | 3 | INTRON | |||||
| KANSL2 | 2 | 3'UTR | |||||
| KANSL2 | 6 | INTRON | |||||
| KANSL2 | 1 | Synonymous | |||||
| CCDC65 | 1 | INTRON | |||||
| CCDC65 | 1 | INTRON | |||||
| BIN2 | 1 | INTRON | |||||
| SLC4A8 | 1 | INTRON | |||||
| HOXC-AS3 | 1 | 0 | Non-coding transcript exon | ||||
| RP11-968A15.8 | 1 | INTRON | |||||
| ITGA7 | 1 | INTRON | |||||
| ITGA7 | 1 | 0 | Missense | ||||
| NEMP1 | 1 | 0 | Missense | ||||
| NEMP1 | 1 | INTRON | |||||
| TAFA2 | 1 | INTRON | |||||
| CAPS2-AS1 | 2 | INTRON | |||||
| E2F7 | 1 | INTRON | |||||
| ANKS1B | 1 | INTRON | |||||
| IGF1 | 14 | INTRON | |||||
| RNF10 | 1 | INTRON | |||||
| CABP1 | 1 | INTRON | |||||
| ADGRD1 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 13 | 41 | 20 | 21 | TPTE2 | 2 | INTRON | |
| PAN3 | 1 | INTRON | |||||
| RXFP2 | 1 | INTRON | |||||
| NBEA | 1 | INTRON | |||||
| ELF1 | 1 | INTRON | |||||
| GUCY1B2 | 1 | INTRON | |||||
| TPTE2P3 | 1 | INTRON | |||||
| LINC00458 | 1 | INTRON | |||||
| LMO7 | 1 | INTRON | |||||
| MYCBP2 | 5 | INTRON | |||||
| GPC6 | 1 | INTRON | |||||
| GPC6 | 1 | INTRON | |||||
| NALF1 | 2 | INTRON | |||||
| PCID2 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 14 | 25 | 20 | 5 | NF1P4 | 3 | INTRON | |
| SLC7A7 | 4 | INTRON | |||||
| PRKD1 | 1 | INTRON | |||||
| CDKL1 | 1 | INTRON | |||||
| CNIH1 | 2 | INTRON | |||||
| LINC02284 | 4 | INTRON | |||||
| NEK9 | 1 | Synonymous | |||||
| GALC | 1 | INTRON | |||||
| RIN3 | 2 | INTRON | |||||
| PAPOLA | 1 | INTRON | |||||
| MOK | 1 | INTRON | |||||
| RCOR1 | 1 | INTRON | |||||
| TDRD1 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 15 | 36 | 32 | 4 | CCDC32 | 1 | INTRON | |
| EBP42 | 1 | INTRON | |||||
| TRIM69 | 1 | INTRON | |||||
| WDR72 | 1 | INTRON | |||||
| KIF23 | 1 | INTRON | |||||
| MTHFS | 1 | INTRON | |||||
| MTHFS | 23 | INTRON | |||||
| MTHFS | 1 | Missense | |||||
| FANCI | 1 | INTRON | |||||
| CTD-2544M6.1 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 16 | 35 | 19 | 16 | RAB11FIP3 | 1 | INTRON | |
| 2 | |||||||
| JPT2 | 1 | INTRON | |||||
| NDUFB10 | 1 | INTRON | |||||
| TRAF7 | 1 | INTRON | |||||
| AJ003147.9 | 1 | INTRON | |||||
| LA16c-306E5.3 | 1 | INTRON | |||||
| SRL | 2 | INTRON | |||||
| 13 | |||||||
| 1 | |||||||
| VPS35 | 1 | INTRON | |||||
| ADCY7 | 1 | INTRON | |||||
| CMTM4 | 3 | INTRON | |||||
| RFWD3 | 1 | INTRON | |||||
| ADAMTS18 | 1 | INTRON | |||||
| WWOX | 1 | INTRON | |||||
| PLCG2 | 1 | INTRON | |||||
| SPG7 | 2 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 17 | 68 | 55 | 13 | RAP1GAP2 | 1 | INTRON | |
| ITGAE | 1 | Frameshift | |||||
| ITGAE | 1 | INTRON | |||||
| SMTNL2 | 1 | 3'UTR | |||||
| CHRNE | 1 | 3'UTR | |||||
| INCA1 | 1 | INTRON | |||||
| DNAH2 | 12 | INTRON | |||||
| DNAH2 | 2 | INTRON | |||||
| ULK2 | 1 | Synonymous | |||||
| LINC02002 | 1 | INTRON | |||||
| PIGS | 3 | INTRON | |||||
| MYO1D | 1 | INTRON | |||||
| ASIC2 | 1 | INTRON | |||||
| CDK12 | 1 | INTRON | |||||
| WIPF2 | 1 | Synonymous | |||||
| WIPF2 | 1 | Inframe insertion | |||||
| BRCA1 | 1 | INTRON | |||||
| RP11-1072C15.7 | 1 | INTRON | |||||
| MSI2 | 1 | INTRON | |||||
| TANC2 | 1 | INTRON | |||||
| CEP112 | 9 | INTRON | |||||
| ABCA9 | 1 | INTRON | |||||
| RAB37 | 2 | INTRON | |||||
| CYTH | 4 | INTRON | |||||
| CEP131 | 2 | INTRON | |||||
| SLC38A10 | 1 | INTRON | |||||
| SLC25A10 | 1 | INTRON | |||||
| AC139099.4 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 18 | 44 | 33 | 11 | RP11-172F10.1 | 1 | INTRON | |
| AKAIN1 | 1 | INTRON | |||||
| EBP41L3 | 1 | INTRON | |||||
| RP11-805F19.2 | 1 | INTRON | |||||
| CDH2 | 1 | INTRON | |||||
| KIAA1328 | 1 | INTRON | |||||
| MIR4527HG | 6 | INTRON | |||||
| MAPK4 | 1 | INTRON | |||||
| RP11-671P2.1 | 1 | INTRON | |||||
| RP11-795H16.3 | 1 | INTRON | |||||
| CDH7 | 1 | INTRON | |||||
| LINC00908 | 14 | INTRON | |||||
| LINC00908 | 1 | INTRON | |||||
| LINC00908 | 1 | INTRON | |||||
| LINC00908 | 1 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 19 | 53 | 41 | 12 | WDR18 | 1 | INTRON | |
| WDR18 | 1 | 0 | Synonymous | ||||
| DOT1L | 1 | 0 | Missense | ||||
| TLE2 | 1 | INTRON | |||||
| CACTIN | 1 | INTRON | |||||
| CHAF1A | 1 | INTRON | |||||
| LONP1 | 11 | INTRON | |||||
| NFIX | 2 | INTRON | |||||
| NFIX | 2 | INTRON | |||||
| NCAN | 1 | INTRON | |||||
| CTD-2043I16.1 | 2 | INTRON | |||||
| URI1 | 1 | INTRON | |||||
| TDRD12 | 2 | INTRON | |||||
| TDRD12 | 1 | 0 | Missense | ||||
| TDRD12 | 14 | 0 | |||||
| ZNF540 | 1 | INTRON | |||||
| PCAT19 | 1 | INTRON | |||||
| ZNF227 | 1 | INTRON | |||||
| ZNF235 | 1 | INTRON | |||||
| ZNF285 | 1 | INTRON | |||||
| PVR | 1 | INTRON | |||||
| CBLC | 1 | INTRON | |||||
| MARK4 | 5 | INTRON | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 20 | 29 | 11 | 18 | C20orf27 | 1 | INTRON | |
| SLC23A2 | 1 | INTRON | |||||
| CHMP4B | 5 | INTRON | |||||
| DHX35 | 2 | INTRON | |||||
| SLC2A10 | 1 | INTRON | |||||
| MIR646HG | 1 | Non-coding transcript exon | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 21 | 23 | 21 | 2 | IFNGR2 | 8 | INTRON | |
| CHODL | 2 | INTRON | |||||
| IFNAR1 | 1 | INTRON | |||||
| TMEM50B | 3 | INTRON | |||||
| TMEM50B | 3 | INTRON | |||||
| GART | 1 | INTRON | |||||
| RUNX1 | 1 | INTRON | |||||
| C2CD2 | 1 | 3'UTR | |||||
| PDXK | 1 | 3'UTR | |||||
| Chr. | Chr. mutation | Mutation | Unknown mutation | Expressed gene | Mutation no. | Mutation position | Mutation type |
| 22 | 44 | 29 | 15 | TPTEP1 | 3 | INTRON | |
| SMPD4P1 | 2 | INTRON | |||||
| BCR | 1 | INTRON | |||||
| MYO188 | 1 | INTRON | |||||
| DEPDC5 | 1 | INTRON | |||||
| FOXRED2 | 1 | 3'UTR | |||||
| ELFN2 | 2 | INTRON | |||||
| XPNPEP3 | 1 | INTRON | |||||
| TCF20 | 15 | INTRON | |||||
| CYB5R3 | 1 | INTRON | |||||
| TAFA5 | 1 | INTRON |
Disclosures
Human subjects: Consent was obtained or waived by all participants in this study. Human Ethics Research Committee of the College of Science, Salahaddin University-Erbil issued approval 4/5/439.
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
Conflicts of interest: In compliance with the ICMJE uniform disclosure form, all authors declare the following:
Payment/services info: All authors have declared that no financial support was received from any organization for the submitted work.
Financial relationships: All authors have declared that they have no financial relationships at present or within the previous three years with any organizations that might have an interest in the submitted work.
Other relationships: All authors have declared that there are no other relationships or activities that could appear to have influenced the submitted work.
Author Contributions
Acquisition, analysis, or interpretation of data: Bahaaddin A. Saber
Drafting of the manuscript: Bahaaddin A. Saber, Abbas Salihi
Critical review of the manuscript for important intellectual content: Bahaaddin A. Saber, Abbas Salihi, Ashabil Aygan
Concept and design: Abbas Salihi, Ashabil Aygan
Supervision: Abbas Salihi, Ashabil Aygan
References
- 1.Sulfurtransferases and cystathionine beta-synthase expression in different human leukemia cell lines. Jurkowska H, Wróbel M, Jasek-Gajda E, Rydz L. Biomolecules. 2022;12:148. doi: 10.3390/biom12020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO 2016 definition of chronic myeloid leukemia and tyrosine kinase inhibitors. Haznedaroğlu İC, Kuzu I, İlhan O. Turk J Haematol. 2020;37:42–47. doi: 10.4274/tjh.galenos.2019.2019.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Measuring the symptom burden associated with the treatment of chronic myeloid leukemia. Williams LA, Garcia Gonzalez AG, Ault P, et al. Blood. 2013;122:641–647. doi: 10.1182/blood-2013-01-477687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.BCR-ABL1 tyrosine kinase complex signaling transduction: challenges to overcome resistance in chronic myeloid leukemia. Amarante-Mendes GP, Rana A, Datoguia TS, Hamerschlak N, Brumatti G. Pharmaceutics. 2022;14:215. doi: 10.3390/pharmaceutics14010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.dbLGL: an online leukemia gene and literature database for the retrospective comparison of adult and childhood leukemia genetics with literature evidence. Liu Y, Luo M, Jin Z, Zhao M, Qu H. Database (Oxford) 2018;2018:0. doi: 10.1093/database/bay062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cystathionine β-synthase in physiology and cancer. Zhu H, Blake S, Chan KT, Pearson RB, Kang J. Biomed Res Int. 2018;2018:3205125. doi: 10.1155/2018/3205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasotransmitters in the tumor microenvironment: impacts on cancer chemotherapy (review) Salihi A, Al-Naqshabandi MA, Khudhur ZO, et al. Mol Med Rep. 2022;26:233. doi: 10.3892/mmr.2022.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The yin and yang of nitric oxide in cancer progression. Burke AJ, Sullivan FJ, Giles FJ, Glynn SA. Carcinogenesis. 2013;34:503–512. doi: 10.1093/carcin/bgt034. [DOI] [PubMed] [Google Scholar]
- 9.Molecular mechanisms of nitric oxide in cancer progression, signal transduction, and metabolism. Somasundaram V, Basudhar D, Bharadwaj G, et al. Antioxid Redox Signal. 2019;30:1124–1143. doi: 10.1089/ars.2018.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitric oxide (NO) and NO synthases (NOS)-based targeted therapy for colon cancer. Wang H, Wang L, Xie Z, Zhou S, Li Y, Zhou Y, Sun M. Cancers (Basel) 2020;12:1881. doi: 10.3390/cancers12071881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.In vitro anticancer activity of hydrogen sulfide and nitric oxide alongside nickel nanoparticle and novel mutations in their genes in CRC patients. Housein Z, Kareem TS, Salihi A. Sci Rep. 2021;11:2536. doi: 10.1038/s41598-021-82244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan-cancer analysis of NOS3 identifies its expression and clinical relevance in gastric cancer. Zou D, Li Z, Lv F, et al. Front Oncol. 2021;11:592761. doi: 10.3389/fonc.2021.592761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preleukemic phase of chronic myelogenous leukemia: morphologic and immunohistochemical characterization of 7 cases. Aye le L, Loghavi S, Young KH, et al. Ann Diagn Pathol. 2016;21:53–58. doi: 10.1016/j.anndiagpath.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Acute myeloid leukemia in the real world: why population-based registries are needed. Juliusson G, Lazarevic V, Horstedt AS, et al. Blood. 2012;119:3890–3899. doi: 10.1182/blood-2011-12-379008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baseline blood count levels increase odds of cytopenia among CML patients in Kenya: a case control study. McLigeyo A, Rajab J, Oyiro P, et al. BMC Cancer. 2022;22:128. doi: 10.1186/s12885-021-09162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Interleukin 6 and disease transformation in chronic myeloid leukemia: a northeast Indian population study. Sharma K, Singh U, Rai M, Shukla J, Gupta V, Narayan G, Kumar S. J Cancer Res Ther. 2020;16:30–33. doi: 10.4103/jcrt.JCRT_137_17. [DOI] [PubMed] [Google Scholar]
- 17.Hydrogen sulfide, an endogenous stimulator of mitochondrial function in cancer cells. Szabo C. Cells. 2021;10:220. doi: 10.3390/cells10020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A relationship between endothelial nitric oxide synthetase gene variants and substance use disorder. Pehlivan S, Aydin PC, Nursal AF, Pehlivan M, Oyaci Y, Yazici AB. Endocr Metab Immune Disord Drug Targets. 2021;21:1679–1684. doi: 10.2174/1871530320666201013154917. [DOI] [PubMed] [Google Scholar]
- 19.Clinical interpretation of genomic variations. Sayitoğlu M. Turk J Haematol. 2016;33:172–179. doi: 10.4274/tjh.2016.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chronic myeloid leukemia, from pathophysiology to treatment-free remission: a narrative literature review. Rinaldi I, Winston K. J Blood Med. 2023;14:261–277. doi: 10.2147/JBM.S382090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Identification of prognostic and susceptibility markers in chronic myeloid leukemia using next generation sequencing. Shokeen Y, Sharma NR, Vats A, et al. Ethiop J Health Sci. 2018;28:135–146. doi: 10.4314/ejhs.v28i2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]