ABSTRACT
Neuroectoderm-derived tumors characteristically express gangliosides such as GD3 and GD2. Many studies have reported that gangliosides GD3/GD2 enhance malignant phenotypes of cancers. Recently, we reported that human gliomas expressing GD3/GD2 exhibited enhanced malignant phenotypes. Here, we investigated the function of GD3/GD2 in glioma cells and GD3/GD2-expressing glioma-derived exosomes. As reported previously, transfectant cells of human glioma U251 MG expressing GD3/GD2 showed enhanced cancer phenotypes compared with GD3/GD2-negative controls. When GD3/GD2-negative cells were treated with exosomes secreted from GD3/GD2-positive cells, clearly increased malignant properties were observed. Furthermore, increased phosphorylation of signaling molecules was detected after 5–15 min of exosome treatment, ie, higher tyrosine phosphorylation of platelet-derived growth factor receptor, focal adhesion kinase, and paxillin was found in treated cells than in controls. Phosphorylation of extracellular signal-regulated kinase-1/2 was also enhanced. Consequently, it is suggested that exosomes secreted from GD3/GD2-positive gliomas play important roles in enhancement of the malignant properties of glioma cells, leading to total aggravation of heterogenous cancer tissues, and also in the regulation of tumor microenvironments.
Key Words: glioma, exosome, ganglioside, malignancy
INTRODUCTION
Gliomas, a type of brain tumor, generally originate from glial cells or their precursor cells.1 Gliomas mostly exhibit a highly invasive activity, and expand outward by infiltrating normal brain tissues.2 Gangliosides, sialic acid-containing glycosphingolipids are expressed dominantly in nervous tissues. Complex gangliosides are generally expressed during the later stages of brain formation, while simple gangliosides are major structures at the early stage of embryogenesis.3 Some gangliosides like Neu5Acα2,8Neu5Acα2,3Galβ1,4Glc-ceramide (GD3) and GalNAcβ1,4(Neu5Acα2,8Neu5Acα2,3)Galβ1,4Glc-ceramide (GD2) have been reported to be tumor-associated antigens in neuro-ectoderm-derived cancers,4,5 such as melanomas6 and neuroblastomas.7 Other human cancers like small cell lung cancer,8,9 breast cancer,10 and osteosarcoma11 also express GD3 and/or GD2. There are reports of ganglioside expression on gliomas,12,13 but the roles of gangliosides in gliomas are not well-known. Previously, we reported that human gliomas express GD3/GD2, and they are involved in enhanced malignant properties.14 We also analyzed ganglioside expression in murine gliomas using the replication-competent avian leukemia virus splice acceptor (RCAS)/GFAP-transgenic (Gtv-a) system, in which expression of GD3/GD2 was shown in induced gliomas.15 However, details of their roles and action mechanisms have not been sufficiently investigated.
Regarding extracellular vesicles (EVs), exosomes are released by various cells including cancer cells.16 These exosomes contain many constituents of cells, ie, DNA, RNA, microRNA,17 lipids, and cytosolic and cell-surface proteins,16 and alter the biological nature of recipient cells.18,19 Exosomes are considered to be involved in intercellular communication.20 There have been interesting studies on exosomes released from cancers, ie, on regulation of the cancer microenvironment,21,22 and on cancer metastasis.23,24
Gliomas are resistant to the current therapy and difficult to control due to their invasive nature, leading to a very poor prognosis.25,26 Therefore, it is very important to clarify the roles of GD3/GD2-expressing glioma-derived exosomes in regulation of malignant properties of gliomas and surrounding normal cells to develop novel strategies for glioma treatment. Findings observed in this study would greatly contribute in the novel therapeutic approaches targeting GD3/GD2-expressing gliomas.
MATERIALS AND METHODS
Cell lines and cell culture
The cell lines used in this study were GT16, GT18, GT29, CV2, CV3, and CV5.14 GD3/GD2-positive cell lines (GT16, GT18, and GT29) were generated by transfecting the human glioma cell line, U-251MG (from JCRB Cell Bank, Osaka, Japan) with GD3 synthase (ID: 6489) cDNA and neo-resistant gene. The control cell lines (CV2, CV3, and CV5) were also generated from the U-251MG cell line by transfecting the neo-resistant gene only. The cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 7.5% fetal calf serum (FCS) and G418 (400 μg/mL) at 37 °C in a humidified atmosphere containing 5% CO2.
Antibodies and reagents
Anti-GD3 monoclonal antibody (mAb), R2427 was provided by L.J. Old at the Memorial Sloan Kettering Cancer Center (New York). Anti-GD2 mAb, 220–51, was generated in our laboratory.27 The other antibodies and reagents used in this research were obtained from various commercial sources as follows: fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (H+L), Cat. No. 55514, from Cappel (Durham, NC, USA), and horseradish peroxidase (HRP)-conjugated anti-mouse IgG (7076S) and HRP-conjugated anti-rabbit IgG (7074S) from Cell Signaling Technology (Danvers, MA, USA). Mouse anti-phosphotyrosine antibody, PY20 (sc-508) and mouse Tsg101 (C-2) (sc-7964) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-CD9 (014-27763) and anti-CD63 mAbs (012-27063) were from Fujifilm Wako (Osaka, Japan). Anti-CD81 mAb (66866-1-Ig), and anti-Alix antibody (12422-1-AP) were from Protein Tech (San Diego, California, USA). Tim4-beads (297-79701), PS Capture Exosome Flow Cytometry Kit (CC050), and 2-amino-2 hydroxymethyl-1,3-propanediol (201-06273) were obtained from Fujifilm Wako (Osaka, Japan). The ImmunoStar LD (290–69,904) detection kit, G418 (076-05962) were also from Wako. Matrigel (354234) was from BD Bioscience (San Jose, CA, USA), and collagen-1 (CC050) and Tween20 (P2287) were from Sigma-Aldrich. Protease Inhibitor Mixture (539131) was from Calbio-chem (San Diego, CA, USA), and cell lysis buffer (9803S) was from Cell Signaling.
Isolation of exosomes from culture supernatants
Confluent cells, 70–80% in 15 cm dishes were washed three times with phosphate-buffered saline (PBS) and cultured with DMEM containing 1% ITS premix (Corning ITS premix universal culture supplement, Thermofisher Scientific, MA, USA) for 48 h. Culture supernatants were transferred in 50 mL falcon tubes and centrifuged at 4 °C and 500 g for 10 min. After centrifuging again at 4 °C and 20,000×g for 20 min, the supernatants were filtered using 0.22 μm Sartolab filters (RF-150) (Zartorius, Helsinki, Finland) and transferred into Beckman polypropylene ultracentrifuge tubes (Beckman Coulter, Brea, CA, USA). Then, the samples were centrifuged at 175,000 g and 4 °C for 84 min using Beckman SW32Ti rotor (Kent MI, USA). After discarding the supernatants and vortexing, sediments were suspended in cold PBS, and rotated again. After removal of the supernatants, the samples were vortexed, and 200 μL of cold PBS was added. The samples were then used immediately or stored at –80 °C. An exosome sample in PBS was used for measuring the protein amount using Pierce BCA Protein Assay Kit (23225, Thermofisher Scientific). To characterize the preparations, we performed immunoblotting (IB) of exosome markers, tetraspanins (CD9, CD63, CD81), Alix, and Tsg101.
Flow cytometry
Expression levels of the gangliosides GD3 and GD2 on the cell surface were analyzed by the Accuri C6 flow cytometer (BD Biosciences, USA), as previously described.28,29 Briefly, following the trypsinization of cells, the cells (5×105) were incubated with primary antibodies in PBS for 60 min on ice. After washing, cells were stained with secondary antibodies, FITC-conjugated goat anti-mouse IgG (H + L), Cappel, in PBS for 45 min on ice. Then, the relative expression levels were analyzed by a flow cytometer. Control samples were prepared using non-relevant mAbs with the same subclasses. The CFlow plus program was used for the data analysis. In the case of exosomes, the expression levels of tetraspanins, GD3, and GD2 on exosomes were analyzed by Tim4 beads. Exosomes were mixed with PS-capture Tim4 beads for 1 h at room temperature (RT) with light vortexing every 10 min. Exosome-bound Tim4 beads were washed twice using a magnetic stand (290-35591, FUJIFILM Wako), and primary antibodies were added and incubated for 1 h on ice. After washing, the beads were stained with secondary antibodies as described above, and exosomes bound to Tim4 beads were applied for flow cytometry as explained above.
MTT assay
Cells (3×103) were seeded in each well of 96-well plates with 100 μL of DMEM supplemented with FCS concentration of 4.0 and 7.5%. To examine the effect of exosomes on cell proliferation, exosomes were added just after the addition of cells.30 MTT solution (5 mg/mL 1×PBS, 20 μL) was added on days 0, 1, 2, 4, and 5, and incubated for 6 h in a 5% CO2 incubator at 37 °C. After 6 h, 150 μL of acidic 2-propanol (0.4% HCl and 0.1% NP-40 containing 2-propanol) was added to stop the reaction in each well, and it was vigorously pipetted to destroy cells. About 100 μL of cell lysates were transferred into a new 96-well plate and absorbance was measured at 595–620 nm by an automatic microplate reader (Thermofisher Scientific, Type: 357, Shanghai, China).
Invasion assay
Cell invasion activity was analyzed with the Boyden-chamber method, as described previously.31 Cell culture inserts (Transparent PET membrane, 24-well format, 8.0-μm pore size, Life Sciences, Durham, NC, USA) were used. For polymerization, Matrigel (a solubilized basement membrane preparation extracted from EHS mouse sarcoma, BD Bioscience, 20 μL) in cold PBS (200 μg/mL) was applied to the upper chamber of the cell culture inserts and incubated for 2 h at RT in a current clean bench. After removing PBS, the upper chamber was filled with 200 μL of serum-free DMEM and incubated for 1 h, and the lower chamber was filled with DMEM containing 7.5% FCS. GD3/GD2(+) and (–) cells (3×104) in 200 μL of serum-free DMEM were added in the upper chamber after removal of serum-free medium. To analyze the effect of exosomes on the invasion activity of cells, exosomes were added after the addition of cells and incubated for 24 h at 37 °C in a humidified atmosphere containing 5% CO2.32 After incubation, the numbers of cells that had migrated to the reverse side of the chamber were counted under a microscope (IX73P1F, Olympus, Tokyo, Japan) after being fixed and stained with Giemsa (Wako).
Migration assay
Two-dimensional migration activity of cells was measured with a wound healing scratch motility assay as described previously.33 The cells (3×105) were seeded in 6-cm dishes with 7.5% FCS-containing DMEM. After 70–80% confluence, cells were scratched with pipette tips (Pipette Tips RC UNV 250 μL 1000A/1). To observe the effect of exosomes on the migration activity of cells, 250 μL exosomes (4 μg/dish) were added after cell scratching. Individual cell migration/motility was observed under a microscope (IX73P1F, Olympus), and pictures of the scratched regions were taken at different time-points: 0, 4, 8, 12, 18, and 24 h. Then the wound-healing activities were measured and expressed as % to the initial wound lengths.
Cell adhesion assay
The adhesion activity of cells was measured using the real-time cell electronic sensing system (Wako), as described previously.34 An E-plate/microplate with electronic sensor was used to detect the cell adhesion activity. At the bottom of the microplates (E-Plate, 16X; ACEA Biosciences Inc., San Diego, CA, USA), microelectronic cell sensor arrays are integrated. The sensor provides information on the increased electrical resistance (cell index), indicating the increase in cell adhesion. E-plates were coated with collagen-1 [CL-1 (5 μg/mL in PBS, 100 μL/well)] at RT for 1 h, and blocked by 1% bovine serum albumin/7.5% FCS in DMEM (100 μL/well) at RT for 1 h. After blocking the micro-wells and removal of bovine serum albumin solution, cells (1×104) were seeded in each well containing the culture medium. To examine the effect of exosomes on cell adhesion activity, exosomes were added at time 0. The changes in cell adhesion were monitored continuously, and are expressed as the cell index (CI).
Preparation of cell and exosome lysates
The method of cell lysate preparation was described previously.31,35 Cultured cells on 6-cm dishes were washed three times with PBS, and then cell lysates were prepared by scraping the cells following the addition of lysis buffer (20 mM Tris-HCl, 1 mM EGTA, 1 mM Na2EDTA, 150 mM NaCl, 1% Triton X-100, 1 mM β-glycero-phosphate, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, and 1 μg/mL leupeptin; Cell Signaling), supplemented with 1 mM PMSF and Protease Inhibitor Mixture (Calbiochem). The collected lysates were centrifuged at 3000 rpm (Kubota 3740, Tokyo, Japan) for 5 min at 4 °C to remove insoluble cell debris. After repeated centrifugation, the supernatants were used to measure the protein concentration using the DC protein assay kit (Bio-Rad).
In the case of exosomes, the isolated exosomes in PBS were used to prepare exosome lysates. Lysis buffer (2x) was added to lyse exosomes at 1:1 ratio on ice for 5 min, and well mixed. The protein concentration of the lysates was measured as described above.
IB
After preparing the lysates, the proteins were separated by SDS–PAGE as described previously.31,35 A sample buffer consisting of 125 mM Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, 0.1% bromophenol blue, 4% 2-mercaptoethanol was mixed with lysates at a 1:1 ratio, and boiled for 5 min at 95 °C before protein separation. The separated proteins in gels were transferred onto an Immobilon-P membrane (EMD Millipore, Burlington, MA, USA), and blots were blocked for 1 h/overnight with 5% skim milk in PBS with 0.05% Tween-20 (PBST). The reaction with the primary antibody was performed for 1 h at RT/overnight at 4 °C. After washing with PBST, the reaction with the secondary antibody was performed for 1 h at RT. After washing, bands of proteins were visualized using ImmunoStar LD detection kits (Wako), and band images were analyzed with Amersham Imager (Model: 680 software version 2.0, GE Healthcare, Uppsala, Sweden). Anti-β-actin antibodies were used to ensure equal amount of protein loading.
Statistical analyses
Statistical analyses were done as previously described.30 Data are presented as the mean ± SD. The data were analyzed by an unpaired Student’s two-tailed t-test, and two-way ANOVA with the Tukey post-hoc test to compare mean values. These results were indicated in each figure legend. P-values of <0.05 were considered significant. Significance was analyzed using R software (version 3.6.3).
RESULTS
Expression of gangliosides on cell surfaces and EVs
GD3/GD2(+) cell lines (GT16, GT18, and GT29) were established by transfecting a human glioma cell line, U-251MG (GD3/GD2 non-expressing), with GD3 synthase cDNA (Fig. 1A), and GD3/GD2(–) cell lines (CV2, CV3, and CV5) were vector controls.14 The surface expression of GD3/GD2 in each group was shown in Fig. 1B.
Fig. 1.

Biosynthesis pathway of gangliosides and expression levels of GD3 and GD2 on GD3 synthase cDNA transfected cells and on control cells
Fig. 1A: The ganglioside synthetic pathway and cDNAs of synthetic enzymes used to establish transfectant cells.
Fig. 1B: Cell surface expression of GD3/GD2 on the established GD3/GD2(+) and GD3/GD2(–) clones as analyzed by flow cytometry.
The expression of tetraspanins (CD9, CD63, and CD81) was also compared between GT16 and cell lines, showing no clear differences in flow cytometry (Fig. 2A). EVs derived from GT16 and CV2 cells were analyzed with Tim4 beads flow cytometry to investigate expression levels of gangliosides and tetraspanins. GT16 cell-derived EVs expressed GD3 and GD2, but CV2 cell-derived EVs did not. Similar expression of CD81 and CD63 was detected on both types of EVs, while that of CD9 was much higher on EVs from GT16 cells (Fig. 2B).
Fig. 2.

Expression of GD3, GD2 and tetraspanins on transfectant cells and EVs
Fig. 2A: Expression of tetraspanins on GD3/GD2+ GT16 cells and on GD3/GD2– CV2 cells as analyzed by flow cytometry. Anti-CD81, anti-CD63, and anti-CD9 mAbs were used as primary antibodies, and FITC-labeled secondary antibody was employed.
Fig. 2B: Expression of tetraspanins and gangliosides on exosomes was analyzed using Tim4 beads flow cytometry as described in 2A. Detection of exosome markers in transfected cells and EVs isolated from them.
Fig. 2C: IB of cell lysates (CV2 and GT16) and exosome lysates (derived from CV2 cells and GT16 cells) was performed using specific primary antibodies. IB was performed using antibodies reactive with EV markers, eg, CD9, CD81, Alix, and Tsg101.
Fig. 2D: The results of IB of cell lysates and EVs lysates with anti-CD63 mAb. Representative results from repeated experiments (at least 3 times) are presented.
EVs: extracellular vesicles
IB: Immunoblotting
exo: exosome
Expression of EV marker proteins in cell lines and EVs
IB of lysates from cells and EVs derived from GT16 and CV2 cells was performed using mAbs for EV markers. Similar levels of Alix and Tsg101 were detected in the lysates from GT16 and CV2 cells, while the GT16 cell lysate contained slightly higher levels of CD9 and CD81 than CV2 cell lysates (Fig. 2C). Higher expression of CD63 was observed in the CV2 cell lysate (Fig. 2D). Lysates from GT16-derived EVs showed higher levels of CD9 than those from CV2 cells (Fig. 2C), while CD63, CD81, Alix and Tsg101 showed almost equivalent bands in two kind EVs (Fig. 2C, D).
Effects of GD3/GD2 and EVs on cell phenotypes
In order to analyze the effects of EVs derived from GD3/GD2(+) and GD3/GD2(–) cells on the phenotypes of GD3/GD2(–) and (+) glioma cells, the proliferation, invasion, migration, and adhesion activities of cells were investigated.
Proliferation rate
To investigate the proliferation rate of GD3/GD2-expressing and non-expressing cell groups, the MTT assay was carried out in the presence or absence of EVs. In 7.5% FCS-containing DMEM, almost equivalent cell growth of all cell lines was found (Fig. 3A) as reported.14 However, a higher growth rate of GT16 cells than CV2 cells was found in 4% FCS-containing DMEM (Fig. 3A). Under this condition, CV2 cells showed significantly increased cell growth after treatment with GT16 cell-derived EVs in a dose-dependent manner (Fig. 3B). On the other hand, when EVs from CV2 cells were added to the culture medium of GT16 cells, the growth rate was reduced (Fig. 3B).
Fig. 3.
Cell growth analysis of GD3/GD2(+) and (–) cells and effects of exosomes on their growth
Fig. 3A: The cell proliferation rate was compared between GD3/GD2(+) cells and GD3/GD2– cells in medium containing 4.0 and 7.5% FCS by the MTT assay. The MTT assay was performed by seeding cells in 96-well plates, and by measuring absorbance at 595–620 nm. Relative absorbance was plotted. Three each of sample group cells were examined by two-way ANOVA. GD3/GD2(+) cells exhibited a higher cell growth when cultured under an FCS concentration of 0–4%, but not in 7.5%. *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. 3B: Effects of EVs on growth of GT16 and CV2 cells were analyzed in 4% FCS-DMEM in the presence or absence of EVs. CV2 cells were treated with GT16 cell-derived EVs (0.2 and 1.0 μg), and showed an increased proliferation rate. In contrast, GT16 cells were treated with CV2 cell-derived EVs (0.2 and 1.0 μg), resulting in the suppression of cell growth. Each analysis was performed in triplicate, and the mean ± SD is presented. The data on days 0, 1, 2, 4, 5 and 6 were analyzed by two-way ANOVA with a Tukey post hoc test. *P < 0.05, **P < 0.01, and ***P < 0.001.
exo: exosome
FCS: fetal calf serum
DMEM: Dulbecco’s modified Eagle medium
Invasion activity
The cell invasion activity examined by the Boyden-chamber assay revealed that GD3/GD2(+) cells showed significantly higher invasion activity than GD3/GD2(–) cells (Fig. 4A, B) as previously reported.14 Then, invasion activity of GD3/GD2(–) cells was analyzed after the addition of EVs released from GD3/GD2(+) cells, showing that the invasion activity increased with the addition of EVs (Fig. 4C, D). On the other hand, GD3/GD2– cell-derived EVs suppressed the invasion activity of GT16 cells (Fig. 4E, F).
Fig. 4.
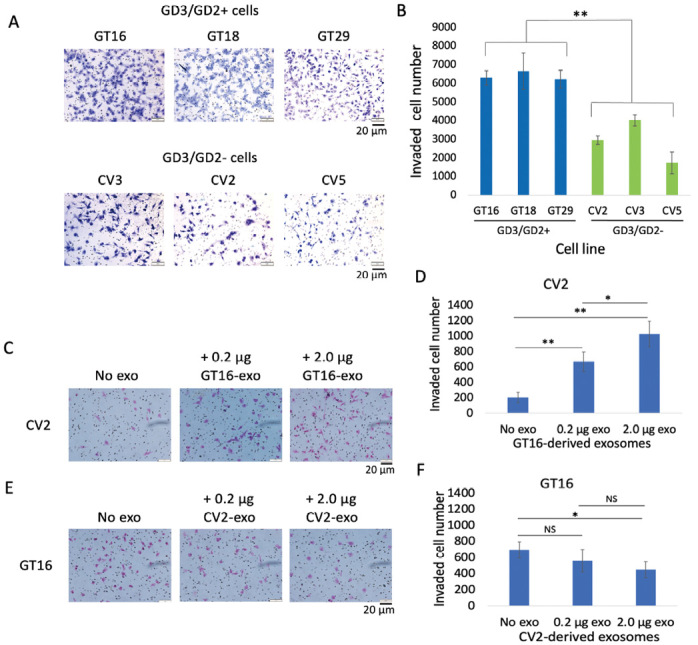
Invasion activity of GD3/GD2(+) and (–) cells, and effects of EVs
Fig. 4A: Microscopic images of invaded cells of both groups (GD3/GD2+ and –). Invasion activity was examined by the Boyden-chamber assay. The upper chamber was coated with matrigel. After 24 hrs of incubation, invaded cells on the reverse side of the membranes were counted under microscope following Giemsa staining.
Fig. 4B: The counted cell numbers (4A) were plotted where GD3/GD2(+) cells showed significantly higher invasion activity (**P < 0.01) than GD3/GD2(–) cells. The results are presented as means ± SD, and the data were analyzed with an unpaired Student’s two-tailed t test.
Fig. 4C: Invasion activity of CV2 cells was analyzed after treatment with GT16 cell-derived EVs (0.2 and 2.0 μg). EVs were added after seeding the cells.
Fig. 4D: The invaded cell numbers (4C) were plotted, showing that GT16 cell-derived EVs enhanced the invasion of CV2 in a dose dependent manner. The unpaired Student’s two-tailed t test was performed for evaluation of the results 4C. *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. 4E: The invasion activity of GD3/GD2+ GT16 cells were analyzed after treatment with CV2 cell-derived EVs (0.2 and 2.0 μg), and counted under microscope.
Fig. 4F: The counted cell numbers (4E) were plotted and showing a suppressive effect.
The analysis was performed in triplicate, and the mean ± SD is presented. The data were analyzed with an unpaired Student’s two-tailed t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
EVs: extracellular vesicles
exo: exosome
Migration activity
Effects of GD3/GD2 and EVs on 2-D migration of cells were investigated with the wound healing scratch assay. The GD3/GD2(+) cells showed higher migration activity than the controls (Fig. 5A, B). When CV2 cells were cultured with EVs from GT16 cells, they showed greater tendency to heal the wound than the non-EV control (Fig. 5C, D). However, no effect of CV2 cell-derived EVs on the motility of GT16 cells was observed (Fig. 5C, E).
Fig. 5.

GD3/GD2(+) cells exhibited higher migration activity, and exosomes derived from them enhanced the migration activity of GD3/GD2(–) cells
Fig. 5A: Migration activities of cells were analyzed by the wound healing scratch assay. CV5 and GT18 are representative cell lines from GD3/GD2(–) and GD3/GD2(+) cell groups, respectively. Cells were cultured in 7.5% FCS-containing medium and scratched at around 70–80% confluency. Individual cell migration was observed under microscope and pictures were taken of the scratched regions at the time points indicated.
Fig. 5B: Graphical presentation of the migration assay of cells. Wound areas are presented as a percentage of the initial wound size (100%). The mean values ± SD (n=3) were plotted for each time point. GD3/GD2(+) cells showed significantly (*P < 0.05) higher motility than the controls.
Fig. 5C: Migration activities of CV2 cells and GT16 cells were examined after the addition of EVs (4 μg) derived from GT16 cells and CV2 cells, respectively. Effects of exosomes on wound healing were examined at different time points as indicated.
Fig. 5D: The spaces of wound healing were measured and plotted as described in 5C (left). GD3/GD2(+) cell-derived exosomes significantly (**P < 0.01) increased the migration activity of GD3/GD2(–) cells.
Fig. 5E: The wound healing spaces as described in 5C (right) were measured at mentioned time points and plotted. No effects of EVs from CV2 cells on invasion of GT16 cells were noted.
exo: exosome
EVs: extracellular vesicles
Adhesion activity
The adhesion of cells to collagen-1 was investigated using real-time cell electronic sensing system. The GD3/GD2(+) cells showed higher adhesion to collagen-1 than the GD3/GD2(–) cells (Fig. 6A). Then, the effect of EVs on cell adhesion was analyzed. When CV2 cells were plated with EVs from GT16 cells, cell adhesion was increased after 1 h depending on the amount of EVs (Fig. 6B). In contrast, CV2 cell-derived EVs suppressed the adhesion activity of GT16 cells (Fig. 6C) from an early time after addition of EVs.
Fig. 6.

Effects of GD3/GD2 and GD3/GD2(+) cell-derived exosomes on cell adhesion
Fig. 6A: Adhesion activity of GD3/GD2(+) and (–) cells to collagen-I examined by the real time cell sensing system. Cells (1×104) were seeded in collagen-1-precoated wells containing 100 μL of culture medium and incubated. The data at 17 h (A, upper) and 6 h (A, lower) of incubation were shown. GD3/GD2 enhanced the adhesion activity of cells.
Fig. 6B: Adhesion activity of CV2 cells was investigated after treatment with GT16 cell-derived exosomes. Exosomes (0.2 and 2.0 μg) were added at 0 h, and the changes in cell adhesion are expressed as the cell index. GT16 cell-derived exosomes increased the adhesion activity of CV2 cells. The data until 6 h of incubation (B, lower) were analyzed with an unpaired Student’s two-tailed t test. **P < 0.01, and ***P < 0.001.
Fig. 6C: GT16 cells were investigated after treatment with CV2 cell-derived exosomes (0.2 and 2.0 μg), resulting in the suppression of adhesion activity in a dose dependent manner. The data until 6 h of incubation (C lower) were also analyzed with an unpaired Student’s two-tailed t test. **P < 0.01, and ***P < 0.001.
exo: exosome
Effects of GD3/GD2(+) cell-derived EVs on the phosphorylation of signaling molecules of GD3/GD2(–) cells during their growth
After being cultured in the FCS-free medium, cells were cultured in 4% FCS-containing DMEM. Then, cell lysates were prepared along the time course indicated in Fig. 7A, and used for IB with anti-phospho-tyrosine mAb, PY20. Increased tyrosine-phosphorylated proteins were observed in GT16 cells (Fig. 7B), suggesting that they are focal adhesion kinase (FAK), paxillin, and platelet-derived growth factor receptor β. The intensity of the three major bands in Fig. 7B were measured and plotted in Fig. 7C. The identity of these tyrosine-phosphorylated bands (FAK, paxillin, extracellular signal-regulated kinase-1/2) was reported previously.14 To investigate the effects of EVs derived from GT16 cells on the growth signals in CV2 cells, IB was performed using CV2 cell lysates treated with serum-containing DMEM with or without GT16 cell-derived EVs (Fig. 7D). When cultured in 4% FCS-containing DMEM, a few weak bands were detected by PY20 at 5 min of stimulation (Fig. 7E Left). Strong bands were detected 15 min after the addition of GT16 cell-derived EVs (Fig. 7E Right). The intensity of the three major bands in Fig. 7E was measured and plotted in Fig. 7F.
Fig. 7.

Cell growth signals during cell growth
Tyrosine-phosphorylated protein levels in GD3/GD2(–) cells were increased after treatment with GD3/GD2(+) cell-derived exosomes.
Fig. 7A: A schema of lysate preparation during cell growth of GD3/GD2(–) cells and GD3/GD2(+) GT16 cells in 4% FCS-DMEM at a various time points of plating after 48 h starvation of serum.
Fig. 7B: The prepared lysates (7A) were used for immunoblotting (IB) with PY20. Higher tyrosine phosphorylation was observed in PDGFRβ, FAK, and paxillin of GT16 cells than those in CV2.
Fig. 7C: Band intensities at 188, 125, and 68-kDa in 7B were measured using Amersham Imager 680 software version 2.0, and plotted.
Fig. 7D: A schema to prepare lysates during CV2 cell growth in 4% FCS-DMEM in the presence or absence of exosomes derived from GT16 cells at different time points after plating. Cells were prepared as in 7A. Then cells were cultured in the presence or absence of exosomes (4 μg) derived from GT16 cells in 4% FCS-DMEM, and incubated as indicated. After incubation, the cells were lysed.
Fig. 7E: The prepared lysates described in 7D were subjected to IB with PY20.
Fig. 7F: Band intensities in 7E were measured and plotted in 7F. Higher tyrosine phosphorylation was observed after 15 min of EV treatment. Representative results from repeated experiments (at least 3 times) are presented.
FCS-DMEM: fetal calf serum-Dulbecco’s modified Eagle medium
EV: extracellular vesicle
PDGFRβ: platelet-derived growth factor receptor β
FAK: focal adhesion kinase
IB: immunoblotting
Erk-1: extracellular signal-regulated kinase-1
Erk-2: extracellular signal-regulated kinase-2
exo: exosome
Phosphorylation of signaling molecules during cell adhesion
To investigate the effects of GD3/GD2 on intracellular signaling during cell adhesion, we performed IB of CV2 and GT16 cell lysates using PY20 (Fig. 8A, B). After starvation and detaching, cells were placed in collagen-1-precoated dishes with plain DMEM, and cell lysates were prepared as shown in Fig. 8A, and subjected to IB. Phospho-tyrosine bands at 5 min were similar in both cell types in Fig. 8B, but after 5 min, bands were stronger in GT16 than CV2 cells. The two major bands in Fig. 8B were scanned and plotted in Fig. 8C. Erk1 and 2 also showed stronger phosphorylation in GT16. The effects of EVs from GT16 cells on intracellular signaling during adhesion of CV2 cells were examined by preparing CV2 cell lysates in the presence or absence of GT16 cell-derived EVs followed by IB using PY20 (Fig. 8D). Higher phosphorylation in FAK and paxillin was detected in CV2+ GT16-EVs samples after 15 min of treatment (Fig. 8E), and the measured band intensities were plotted in Fig. 8F.
Fig. 8.

Adhesion signals during cell adhesion to collagen-I
Tyrosine-phosphorylated protein levels in GD3/GD2(–) cells were increased after treatment with GD3/GD2(+) cell-derived exosomes.
Fig. 8A: A schema for preparing cells to obtain lysates during cell adhesion at various time points. After culture of CV2 cells and GT16 cells in 6-cm dishes with regular medium, cells were starved in plain DMEM for 30 h. After cells were detached, the cell suspension was rotated at 37 °C for 1 h. Then, cells were placed in collagen-1-precoated plates in DMEM, and incubated for 0–60 min at 37 °C. After incubation, the cells were lysed.
Fig. 8B: The prepared lysates mentioned in 8A were used (4 μg/well) for IB using PY20, and images were taken.
Fig. 8C: Band intensities in 8B were measured and plotted. Bands of FAK and paxillin in GT16 cells were higher than CV2 cells after 5 min of incubation.
Fig. 8D: A schema of preparing CV2 cells to obtain lysates during their adhesion to collagen-I in the presence or absence of exosomes derived from GT16 cells at several time points. Cells were placed in a collagen-1-precoated plate (6-cm) in plain DMEM, and incubated for 0–60 min at 37 °C in the presence or absence of EVs (4 μg) derived from GT16 cells. After incubation, the cells were lysed.
Fig. 8E: The prepared lysates were subjected to SDS-PAGE (4 μg/well). Subsequently, IB was performed with PY20.
Fig. 8F: Band intensities in 8E were measured using Amersham Imager 680 software version 2.0, and plotted. Representative results from repeated experiments (at least 3 times) are presented.
IB: immunoblotting
DMEM: Dulbecco’s modified Eagle medium
FAK: focal adhesion kinase
Erk-1: extracellular signal-regulated kinase-1
Erk-2: extracellular signal-regulated kinase-2
exo: exosome
DISCUSSION
Functions of cancer-associated glycosphingolipids (GSLs) have been rigorously studied,4 especially after genetic engineering of carbohydrate expression in cultured cells and experimental animals became possible based on the cDNA cloning of glycosyltransferases.36 Although a number of cancer-associated GSLs have been studied, those of disialyl gangliosides such as GD3 and GD2 were major subjects in the recent cancer glycobiology research.37 We have reported changes in malignant phenotypes and intracellular signals in glyco-remodeled cancer cells, ie, malignant melanomas,31,34,38 lung cancers,39 osteo-sarcomas,11 and gliomas.14 GSLs exert their roles in the membrane microdomains, modulating the structures and functions of membrane molecules4 and modulating intracellular signaling, leading to more malignant phenotypes. Sometimes, enhanced EMT has been demonstrated.40,41
As for roles of cancer-associated GSLs in exosomes, there have been few reports to date. Recently, we reported functions of GD2 on exosomes in the aggravation of malignant melanomas.30 While there are many reports on exosomes from gliomas,42 no studies on glycolipids in exosomes in gliomas have been reported. In this study, we analyzed roles of GD3/GD2 in exosomes for the first time, showing their important roles in human gliomas that were reported to be effects of gangliosides on the cell surface.14 Consequently, exosomes released from GD3/GD2-expressing glioma cells enhanced cell growth, invasion, migration and cell invasion as shown in melanomas.31,34 Furthermore, exosomes derived from GD3/GD2-expressing gliomas induced increased phosphorylation levels of the platelet-derived growth factor receptor β, FAK, paxillin, and extracellular signal-regulated kinase-1/2, suggesting possible molecular basis for enhanced malignant phenotypes. The fact that only CD9 among tetraspanins showed higher levels on EVs from GT16 cells was very intriguing (Fig. 2B, C). But, their roles in exosome functions are not well understood, being expected further study.
Based on the detected signal activation profiles, it may be expected how exosomes target recipient cells and modulate their phenotypes. The facts that many signaling molecules underwent phosphorylation after 5–15 min of exosome treatment suggest that these effects are induced through some reactions on the target cell surface, but not by the expression of functional molecules introduced into the recipient cells.43 in the early phase. Cargo molecules included in the exosomes secreted from GD3/GD2-expressing gliomas should also play important roles at the later stages, leading to enhanced malignant phenotypes. The modes of action of exosomes as well as their fates in the cells remain to be investigated.
When compared with effects of exosomes derived from GD2-expressing melanoma cells on GD2-negative cells,30 enhancements of cell adhesion were less in glioma system. Furthermore, Tsg101, Flotillin-1 and tetraspanin levels were almost equivalent between GD3/GD2+ and GD3/GD2– glioma cells, while they increased in GD2+ melanoma-derived exosomes, suggesting that different mechanisms of exosomes are working among individual cancer types as reported in the function analysis of cancer-associated gangliosides.44
In this study, we analyzed the effects of exosomes derived from GD3/GD2-expressing gliomas on the nature of GD3/GD2-negative gliomas, showing clear functions in phenotypic changes and signal enhancement of target cells. However, cancer tissues generally comprise heterogenous cell populations, and exosomes may play roles in the aggravation of cancer cell features, leading to higher levels of malignant cell groups. Exosomes released from glioma cells may also play important roles in regulation of the tumor-microenvironment,45 and cancer metastasis as reported.46 So, roles of exosomes in regulation of the surrounding normal cell components in cancer tissues are equally important to understand their comprehensive significance and for the construction of novel anti-glioma strategies.
AUTHOR CONTRIBUTIONS
MAH, YK, RHB, KeF and KoF designed the study, and MAH, YukO, FY, MK, RHB, MM, NH, AT, YuhO, KK, and OT performed the study. YuhO, NH, and AT contributed new analytical tools, and YuhO, NH, YukO, KeF and KoF analyzed data. MAH and KoF wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
FUNDING
This study was supported by Grants-in-Aids from the Ministry of Education, Culture, Sports and Technology of Japan (MEXT; 19K22518, 19K07393, 21K06828, 21H02699, 21K15493) and by JST-CREST (Grant Number: JPMJCR17H2). This study was also supported by the Interdisciplinary Joint Research Program of the J-Glyconet cooperative network, which was accredited by the MEXT, Japan.
ACKNOWLEDGMENTS
We thank Ms Y. Imao, Y. Kitaura, H. Ohkuma, and T. Ito for technical assistance. We also thank M. Kojima for excellent secretarial assistance.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Abbreviations
- EVs
extracellular vesicles
- GD3
Neu5Acα2,8Neu5Acα2,3Galβ1,4Glc-ceramide
- GD2
GalNAcβ1,4(Neu5Acα2,8Neu5Acα2,3)Galβ1,4Glc-ceramide
- DMEM
Dulbecco’s modified Eagle medium
- FCS
fetal calf serum
- IB
immunoblotting
- PBS
phosphate-buffered saline
REFERENCES
- 1.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed]
- 2.Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16(12):1575–1584. doi: 10.1093/neuonc/nou147. [DOI] [PMC free article] [PubMed]
- 3.Schengrund CL. Gangliosides: Glycosphingolipids essential for normal neural development and function. Trends Biochem Sci. 2015;40(7):397–406. doi: 10.1016/j.tibs.2015.03.007. [DOI] [PubMed]
- 4.Regina Todeschini A, Hakomori SI. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim Biophys Acta. 2008;1780(3):421–433. doi: 10.1016/j.bbagen.2007.10.008. [DOI] [PMC free article] [PubMed]
- 5.Yeh SC, Wang PY, Lou YW, et al. Glycolipid GD3 and GD3 synthase are key drivers for glioblastoma stem cells and tumorigenicity. Proc Natl Acad Sci U S A. 2016;113(20):5592–5597. doi: 10.1073/pnas.1604721113. [DOI] [PMC free article] [PubMed]
- 6.Lloyd KO. Humoral immune responses to tumor-associated carbohydrate antigens. Semin Cancer Biol. 1991;2(6):421–431. [PubMed]
- 7.Schulz G, Cheresh DA, Varki NM, Yu A, Staffileno LK, Reisfeld RA. Detection of ganglioside GD2 in tumor tissues and sera of neuroblastoma patients. Cancer Res. 1984;44(12 Pt 1):5914–5920. [PubMed]
- 8.Yoshida S, Fukumoto S, Kawaguchi H, Sato S, Ueda R, Furukawa K. Ganglioside G(D2) in small cell lung cancer cell lines: enhancement of cell proliferation and mediation of apoptosis. Cancer Res. 2001;61(10):4244–4252. [PubMed]
- 9.Cheresh DA, Rosenberg J, Mujoo K, Hirschowitz L, Reisfeld RA. Biosynthesis and expression of the disialoganglioside GD2, a relevant target antigen on small cell lung carcinoma for monoclonal antibody-mediated cytolysis. Cancer Res. 1986;46(10):5112–5118. [PubMed]
- 10.Cazet A, Bobowski M, Rombouts Y, et al. The ganglioside G(D2) induces the constitutive activation of c-Met in MDA-MB-231 breast cancer cells expressing the G(D3) synthase. Glycobiology. 2012;22(6):806–816. doi: 10.1093/glycob/cws049. [DOI] [PubMed]
- 11.Shibuya H, Hamamura K, Hotta H, et al. Enhancement of malignant properties of human osteosarcoma cells with disialyl gangliosides GD2/GD3. Cancer Sci. 2012;103(9):1656–1664. doi: 10.1111/j.1349-7006.2012.02344.x. [DOI] [PMC free article] [PubMed]
- 12.Nakamura O, Iwamori M, Matsutani M, Takakura K. Ganglioside GD3 shedding by human gliomas. Acta Neurochir (Wien). 1991;109(1–2):34–36. doi: 10.1007/BF01405694. [DOI] [PubMed]
- 13.Wikstrand CJ, Fredman P, Svennerholm L, Bigner DD. Detection of glioma-associated gangliosides GM2, GD2, GD3, 3’-isoLM1 3’,6’-isoLD1 in central nervous system tumors in vitro and in vivo using epitope-defined monoclonal antibodies. Prog Brain Res. 1994;101:213–223. doi: 10.1016/s0079-6123(08)61951-2. [DOI] [PubMed]
- 14.Iwasawa T, Zhang P, Ohkawa Y, et al. Enhancement of malignant properties of human glioma cells by ganglioside GD3/GD2. Int J Oncol. 2018;52(4):1255–1266. doi: 10.3892/ijo.2018.4266. [DOI] [PubMed]
- 15.Ohkawa Y, Zhang P, Momota H, et al. Lack of GD3 synthase (St8sia1) attenuates malignant properties of gliomas in genetically engineered mouse model. Cancer Sci. 2021;112(9):3756–3768. doi: 10.1111/cas.15032. [DOI] [PMC free article] [PubMed]
- 16.Dai J, Su Y, Zhong S, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5(1):145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed]
- 17.Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed]
- 18.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed]
- 19.Spaull R, McPherson B, Gialeli A, et al. Exosomes populate the cerebrospinal fluid of preterm infants with post-haemorrhagic hydrocephalus. Int J Dev Neurosci. 2019;73:59–65. doi: 10.1016/j.ijdevneu.2019.01.004. [DOI] [PubMed]
- 20.Meldolesi J. Exosomes and Ectosomes in Intercellular Communication. Curr Biol. 2018;28(8):R435-R444. doi: 10.1016/j.cub.2018.01.059. [DOI] [PubMed]
- 21.Jin Y, Xing J, Xu K, Liu D, Zhuo Y. Exosomes in the tumor microenvironment: Promoting cancer progression. Front Immunol. 2022;13:1025218. doi: 10.3389/fimmu.2022.1025218. [DOI] [PMC free article] [PubMed]
- 22.da Costa VR, Araldi RP, Vigerelli H, et al. Exosomes in the Tumor Microenvironment: From Biology to Clinical Applications. Cells. 2021;10(10):2617. doi: 10.3390/cells10102617. [DOI] [PMC free article] [PubMed]
- 23.Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed]
- 24.Tian W, Liu S, Li B. Potential Role of Exosomes in Cancer Metastasis. Biomed Res Int. 2019;2019:4649705. doi: 10.1155/2019/4649705. [DOI] [PMC free article] [PubMed]
- 25.Seker-Polat F, Pinarbasi Degirmenci N, Solaroglu I, Bagci-Onder T. Tumor Cell Infiltration into the Brain in Glioblastoma: From Mechanisms to Clinical Perspectives. Cancers (Basel). 2022;14(2):443. doi: 10.3390/cancers14020443. [DOI] [PMC free article] [PubMed]
- 26.Verdugo E, Puerto I, Medina MÁ. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun (Lond). 2022;42(11):1083–1111. doi: 10.1002/cac2.12361. [DOI] [PMC free article] [PubMed]
- 27.Zhao J, Furukawa K, Fukumoto S, et al. Attenuation of interleukin 2 signal in the spleen cells of complex ganglioside-lacking mice. J Biol Chem. 1999;274(20):13744–13747. doi: 10.1074/jbc.274.20.13744. [DOI] [PubMed]
- 28.Bhuiyan RH, Kondo R, Yamaguchi T, et al. Expression analysis of 0-series gangliosides in human cancer cell lines with monoclonal antibodies generated using knockout mice of ganglioside synthase genes. Glycobiology. 2016;26(9):984–998. doi: 10.1093/glycob/cww049. [DOI] [PubMed]
- 29.Yesmin F, Bhuiyan RH, Ohmi Y, et al. Aminoglycosides are efficient reagents to induce readthrough of premature termination codon in mutant B4GALNT1 genes foumd in families of hereditary spastic paraplegia. J Biochem. 2020;168(2):103–112. doi: 10.1093/jb/mvaa041. [DOI] [PubMed]
- 30.Yesmin F, Furukawa K, Kambe M, et al. Extracellular vesicles released from ganglioside GD2-expressing melanoma cells enhance the malignant properties of GD2-negative melanomas. Sci Rep. 2023;13(1):4987. doi: 10.1038/s41598-023-31216-4. [DOI] [PMC free article] [PubMed]
- 31.Yesmin F, Bhuiyan RH, Ohmi Y, et al. Ganglioside GD2 Enhances the Malignant Phenotypes of Melanoma Cells by Cooperating with Integrins. Int J Mol Sci. 2021;23(1):423. doi: 10.3390/ijms23010423. [DOI] [PMC free article] [PubMed]
- 32.Ohkawa Y, Momota H, Kato A, et al. Ganglioside GD3 Enhances Invasiveness of Gliomas by Forming a Complex with Platelet-derived Growth Factor Receptor α and Yes Kinase. J Biol Chem. 2015;290(26):16043–16058. doi: 10.1074/jbc.M114.635755. [DOI] [PMC free article] [PubMed]
- 33.Dong Y, Ikeda K, Hamamura K, et al. GM1/GD1b/GA1 synthase expression results in the reduced cancer phenotypes with modulation of composition and raft-localization of gangliosides in a melanoma cell line. Cancer Sci. 2010;101(9):2039–2047. doi: 10.1111/j.1349-7006.2010.01613.x. [DOI] [PMC free article] [PubMed]
- 34.Ohmi Y, Kambe M, Ohkawa Y, et al. Differential roles of gangliosides in malignant properties of melanomas. PLoS One. 2018;13(11):e0206881. doi: 10.1371/journal.pone.0206881. [DOI] [PMC free article] [PubMed]
- 35.Bhuiyan RH, Ohmi Y, Ohkawa Y, et al. Loss of Enzyme Activity in Mutated B4GALNT1 Gene Products in Patients with Hereditary Spastic Paraplegia Results in Relatively Mild Neurological Disorders: Similarity with Phenotypes of B4galnt1 Knockout Mice. Neuroscience. 2019;397:94–106. doi: 10.1016/j.neuroscience.2018.11.034. [DOI] [PubMed]
- 36.Nagata Y, Yamashiro S, Yodoi J, Lloyd KO, Shiku H, Furukawa K. Expression cloning of beta 1,4 N-acetylgalactosaminyltransferase cDNAs that determine the expression of GM2 and GD2 gangliosides. J Biol Chem. 1992;267(17):12082–12089. [PubMed]
- 37.Furukawa K, Ohmi Y, Hamamura K, et al. Signaling domains of cancer-associated glycolipids. Glycoconj J. 2022;39(2):145–155. doi: 10.1007/s10719-022-10051-1. [DOI] [PubMed]
- 38.Hamamura K, Furukawa K, Hayashi T, et al. Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proc Natl Acad Sci U S A. 2005;102(31):11041–11046. doi: 10.1073/pnas.0503658102. [DOI] [PMC free article] [PubMed]
- 39.Aixinjueluo W, Furukawa K, Zhang Q, et al. Mechanisms for the apoptosis of small cell lung cancer cells induced by anti-GD2 monoclonal antibodies: roles of anoikis. J Biol Chem. 2005;280(33):29828–29836. doi: 10.1074/jbc.M414041200. [DOI] [PubMed]
- 40.Ohkawa Y, Miyazaki S, Hamamura K, et al. Ganglioside GD3 enhances adhesion signals and augments malignant properties of melanoma cells by recruiting integrins to glycolipid-enriched microdomains. J Biol Chem. 2010;285(35):27213–27223. doi: 10.1074/jbc.M109.087791. [DOI] [PMC free article] [PubMed]
- 41.Nazha B, Inal C, Owonikoko TK. Disialoganglioside GD2 Expression in Solid Tumors and Role as a Target for Cancer Therapy. Front Oncol. 2020;10:1000. doi: 10.3389/fonc.2020.01000. [DOI] [PMC free article] [PubMed]
- 42.Russo MN, Whaley LA, Norton ES, Zarco N, Guerrero-Cázares H. Extracellular vesicles in the glioblastoma microenvironment: A diagnostic and therapeutic perspective. Mol Aspects Med. 2023;91:101167. doi: 10.1016/j.mam.2022.101167. [DOI] [PMC free article] [PubMed]
- 43.Xu W, Yang Z, Lu N. From pathogenesis to clinical application: insights into exosomes as transfer vectors in cancer. J Exp Clin Cancer Res. 2016;35(1):156. doi: 10.1186/s13046-016-0429-5. [DOI] [PMC free article] [PubMed]
- 44.Furukawa K, Hamamura H, Ohkawa Y, Ohmi Y, Furukawa K. Disialyl gangliosides enhance tumor phenotypes with differential modalities. Glycoconj J. 2012;29(8–9):579–584. doi: 10.1007/s10719-012-9423-0. [DOI] [PubMed]
- 45.Godlewski J, Krichevsky AM, Johnson MD, Chiocca EA, Bronisz A. Belonging to a network--microRNAs, extracellular vesicles, and the glioblastoma microenvironment. Neuro Oncol. 2015;17(5):652–662. doi: 10.1093/neuonc/nou292. [DOI] [PMC free article] [PubMed]
- 46.Naghibi AF, Daneshdoust D, Taha SR, et al. Role of cancer stem cell-derived extracellular vesicles in cancer progression and metastasis. Pathol Res Pract. 2023;247:154558. doi: 10.1016/j.prp.2023.154558. [DOI] [PubMed]



