Abstract
Background
Biomarkers have revolutionized the management of colorectal cancer (CRC), facilitating early detection, prevention, personalized treatment, and minimal residual disease (MRD) monitoring. This review explores current CRC screening strategies and emerging biomarker applications.
Main body.
We summarize the landscape of non-invasive CRC screening and MRD detection strategies, discuss the limitations of the current approaches, and highlight the promising potential of novel biomarker solutions. The fecal immunochemical test remained the cornerstone of CRC screening, but its sensitivity has been improved by assays that combined its performance with other stool analytes. However, their sensitivity for advanced adenomas and the patient compliance both remain suboptimal. Blood-based tests promise to increase compliance but require further refinement to compete with stool-based biomarker tests. The ideal scenario involves leveraging blood tests to increase screening participation, and simultaneously promote stool- and endoscopy-based screening among those who are compliant.
Once solely reliant on upfront surgery followed by stage and pathology-driven adjuvant chemotherapy, the treatment of stage II and III colon cancer has undergone a revolutionary transformation with the advent of MRD testing after surgery. A decade ago, the concept of using a post-surgical test instead of stage and pathology to determine the need for adjuvant chemotherapy was disruptive. Today, a blood test may be more informative of the need for chemotherapy than the stage at diagnosis.
Conclusion
Biomarker research is not just improving, but bringing a transformative change to CRC clinical management. Early detection is not just getting better, but improving thanks to a multi-modality approach, and personalized treatment plans are not just becoming a reality, but a promising future with MRD testing.
Keywords: Minimal residual disease, Screening, Colorectal adenomas, CtDNA, CfDNA, Liquid biopsy, Fecal immunochemical test, Colonoscopy, Molecular residual disease, Advanced adenoma (10/10)
Introduction
Though formally introduced in the 1950s, the term 'biomarker' embodies a concept deeply rooted in the history of medical research – objective indicators of physiological states observed externally. Over the years, biomarker studies have permeated various domains of healthcare, offering valuable diagnostic, prognostic, and predictive insights. Among these applications, colorectal cancer (CRC) stands as an exemplar field for biomarker exploration, owing to clinical needs, historical precedence, and contemporary innovations to improve the detection of pre-malignant lesions called advanced adenomas (AA). The role of biomarkers in the evolution of CRC research is a testament to their significance and impact in the field.
CRC holds a unique position as the only cancer routinely screened for in the general population. Unlike breast, cervical, and prostate cancer screening, which are targeted towards either men or women, or lung cancer screening, offered primarily to those with a significant smoking history, CRC screening is population wide. Furthermore, CRC is not only screenable but also preventable. While other cancers primarily rely on early detection to improve outcomes, CRC screening offers the unique ability to prevent disease altogether. This happens because CRC develops from well-defined precancerous lesions, the advanced adenomas (AAs). By detecting (and removing) the AAs, screening can interrupt the progression to invasive cancer. Therefore, unlike other cancers, which primarily rely on early detection to improve outcomes, CRC screening demonstrated the potential for both early-stage cancer detection and disease prevention. This seminal achievement of the twentieth century marked a paradigm shift, demonstrating the feasibility and efficacy of widespread screening based on the detection of a biomarker – fecal hemoglobin in this case. The affordability and accessibility of such tests further solidified the pivotal role of biomarker research in shaping clinical practice and improving public health.
The applications of biomarker research extend beyond screening. A recent breakthrough is the development of liquid biopsies to guide treatment decisions and post-surgical management of CRC. Liquid biopsies offer a significant advantage compared to traditional methods by assessing the molecular presence of tumors by measuring the circulating tumor DNA (ctDNA) in blood. This approach holds promise for improving CRC management in two key ways. Firstly, liquid biopsies may identify tumor recurrence before they become large enough to be visualized through standard imaging techniques. Secondly, ctDNA-based liquid biopsies are rapidly entering clinical practice as tools to detect molecular residual disease (MRD) after surgery. The ability to detect MRD is ushering in a new era of personalized CRC management, where treatment decisions may no longer hinge on cancer stage (the cornerstone of current strategies), but rather the presence of cancer-derived biomarkers in blood.
The uses of liquid biopsies, whether for AA screening or for MRD detection, present a common challenge – the need to capture subtle biological signals indicating the presence or progression of disease. This review provides a concise summary of the current landscape in CRC biomarker research. This review aims to elucidate the ongoing quest for innovative biomarker solutions in CRC management through a systematic evaluation of non-invasive CRC prevention and MRD detection strategies.
Importance of advanced adenomas as endpoints for colorectal cancer screening programs
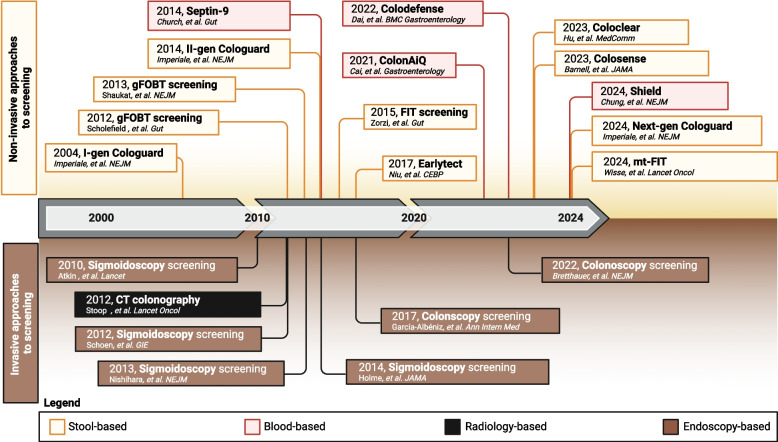
The identification and removal of AAs, the precursor lesions of CRC, is a key strategy for reducing both the mortality and the burden of this disease. Yet, CRC remains a significant public health threat. Despite being a potentially preventable malignancy, CRC remains the world's third most commonly diagnosed and second-leading cause of cancer-related deaths. Numerous approaches have been undertaken to reduce CRC mortality via screening, aiming to optimize patient compliance, boost screening participation, and ultimately reduce both CRC-specific mortality and overall incidence (Fig. 1).
Fig. 1.
A legacy of prevention
Performance of fecal immunochemical testing in detecting advanced adenomas
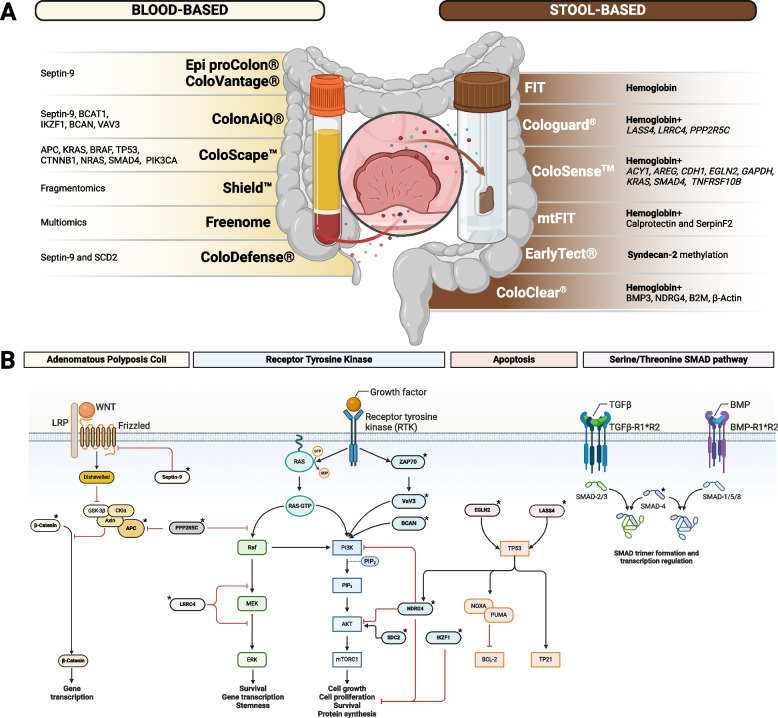
Fecal Immunochemical Testing (FIT) remains, to this day, one of the most efficient and cost-effective tools to combat CRC [1, 2]. This benefit is primarily attributed to a "stage shifting" effect, where FIT detects cancer at earlier, more treatable stages [3, 4]. However, because its sensitivity for AAs is low, FIT bears primarily on mortality and significantly less on cancer incidence [5–8]. Moreover, the fact that it requires the individuals to collect a sample of feces themselves can be a deterrent to compliance [9, 10]. Therefore, despite the unquestionable successes of FIT, studies have also explored whether an endoscopy-first approach may be superior. Endoscopic screening clearly maximizes the sensitivity for early-stage CRC and AA, thus lowering both mortality (by detecting CRC at an earlier stage) and incidence (by detecting and removing AAs before their malignant transformation) [11–26]. However, the contentious results of the NordICC trial have yet again highlighted the challenges of an endoscopy-first approach [27]. Concerns around invasiveness, higher costs, lower participation rates, and poorer patient compliance remain [27–36]. The low participation rate in colonoscopic screening (54% in the US population) underscores the critical need to promote adherence and access to screening [34]. In other words, screening programs are only as effective as they are popular among the population, and non-invasive tests promote participation among screening-reluctant individuals [37–41]. In recent years, significant advances have been made towards the development of a new generation of CRC screening tests that are more patient-friendly, simple, and cost-effective to address patient convenience and the physician’s needs while aligning with the goals of healthcare systems (Fig. 2A) [42–44]. Various biomarkers have been tested, and, interestingly, most of these analytes belong to or interact with one of five main pathways: the canonical Wnt/APC, the K-RAS, the mTOR, the TP53, and the SMAD pathways (Fig. 2B), which collectively regulate gene transcription, survival, stemness, proliferation, and apoptosis. Striving for a balance between sensitivity, specificity, and compliance, various screening approaches have reached acceptable sensitivity values for CRC [45]. However, persistent sensitivity issues remain, particularly concerning early-stage CRCs and AAs (Table 1) [40, 44, 46–65].
Fig. 2.
Available screening tests (A) and their molecular basis (B). Stars (*) indicate molecules that are evaluated in one or more assays
Table 1.
Sensitivity for advanced adenomas reported in various assays of commercial and academic development
|
Test’s name [Ref] [Company name] |
Tested in | Target(s) tested | Population tested and study design |
Sensitivity for AA (95% Confidence intervals) |
|---|---|---|---|---|
| FIT [59] | Feces | Hemoglobin |
Meta analysis of FITs vs colonoscopy for average-risk persons (31 studies with 120,255 participants) |
40% (33%—47%)A |
| FIT [66] | Feces | Hemoglobin |
Meta analysis of screening studies for FIT’s sensitivity to adenomas (10 studies with 63,473 participants) |
27% (23% – 31%) |
|
Cologuard [67] [Exact Sciences] |
Feces |
Hemoglobin + 3 mutations of KRAS + 10 mutations of APC + 8 mutation of TP53 |
Cologuard (vs. Hemoccult II) in asymptomatic, average-risk adults aged 50 or older (N = 2,507) |
12.1% (12.0% – 19.0%) |
|
Cologuard [68] [Exact Sciences] |
Feces |
Hemoglobin + 7 mutations of KRAS + 2 gene methylation status (NDGR4 and BMP3) + β‐Actin |
Cologuard (vs. FIT) in asymptomatic, average-risk adults aged 50–84 years (N = 9,989) |
42.4% (38.9% – 46.0%) |
|
Cologuard [69] [Exact Sciences] |
Feces |
Hemoglobin + 4 gene methylation status (LASS4, LRRC4, PPP2R5C, and ZDHHC1) |
Cologuard (vs. FIT) in asymptomatic, average-risk adults aged 40 or older (N = 20,176) |
43.4% (41.3% – 45.6%) |
|
ColoSense [70] [Geneoscopy] |
Feces |
Hemoglobin + Smoking status + 8 mRNA targets (ACY1, AREG, CDH1, EGLN2, GAPDH, KRAS, SMAD4, and TNFRSF10B) |
ColoSense (vs. FIT) in asymptomatic, average-risk adults aged 45 or older; blinded, prospective, cross-sectional (N = 8,920) |
45.9% (42% – 50%) |
|
Epi proColon [Epigenomics] Colovantage |
Blood | Septin-9 methylation status | Prospective study of adults aged 50 or older (N = 7,941) | 11.2% |
|
[Genomic Tree] Colosafe [Creative Biosciences] |
Feces | Gene methylation status of SCD2 |
Meta-analysis of blood Septin-9 (25 studies) |
15% (11% – 19%) |
|
ColonAiQ [74] [Singlera/Clinomics] |
Blood | 5 gene methylation status (Septin-9, BCAT1, IKZF1, BCAN, VAV3) |
Retrospective case–control study (N = 348) |
42% (33.1% – 51.5%) |
|
Coloscape [Diacarta] [75] |
Blood |
61 mutations in 8 genes (APC, KRAS, BRAF, TP53, CTNNB1, NRAS, SMAD4, and PIK3CA) + 7 methylated markers |
Retrospective case–control study (N = NA) B |
60% (17%−93%) |
|
Shield [76] [Guardant Health] |
Blood | Fragementomic evaluation of cfDNA | Prospective trial of 7861 participants |
13.2% (11.3% – 15.3%) |
| ColoDefense2.0 [VersaBio Technologies] [77] | Blood | 2 gene metylation status (Septin-9 and SCD2) |
Retrospective case–control study (N = 529) |
55.0% (38.6% – 70.4%) |
|
Coloclear [Prenetics] [78] |
Stool |
Haemoglobin + KRAS mutations + 4 gene methylation status (BMP3 promoter, NDRG4 promoter, β‐Actin, and B2M) |
ColoClear (vs. FIT) in a prospective screening study of high-risk CRC individuals (N = 4,758) |
63.5% (58.6% – 68.3%) |
|
Freenome [79] [Freenome] |
Blood | Multiomics | 48,995 screen-eligible adults aged 45–85, using colonoscopy as reference method |
12.5% (11.3% – 13.8%) |
AAt 10 ug/g threshold
BFull study results not yet available (abstract only)
Improvements to FIT by incorporating additional analytes to stool hemoglobin testing
FIT solidified its position as the benchmark screening test after decades of extensive testing in large randomized clinical trials [59, 60, 65, 80]. However, it had two main limitations: patient reluctance due to stool sample collection, leading to suboptimal compliance and low sensitivity for early-stage CRC and AAs [50, 58–62, 65]. Many studies, therefore, reasoned that a pragmatic approach towards progress seems to lie in incremental enhancements to screening methodologies – making small and strategic improvements to FIT rather than completely novel breakthrough innovations. To enhance FIT's detection of early-stage CRC and particularly AAs, additional molecular targets have been incorporated alongside hemoglobin. These targets include DNA mutations, methylated genes, mRNAs, noncoding RNAs (e.g., miRNAs, lncRNAs, circRNAs), and other proteins. However, only a handful of these approaches have reached clinical trial testing and met the primary endpoints to be granted commercialization rights and, therefore, have entered clinical practice: the multitarget stool DNA test (mt-sDNA), later commercialized as Cologuard by Exact Sciences; the multitarget stool RNA test (mt-sRNA), commercialized as ColoSense; and the multi-target FIT (mtFIT), which was recently tested in a large Dutch population, represent the most popular of such products.
The multi-target stool DNA test improved FIT’s performance in detecting advanced adenomas
The Cologuard multi-target stool DNA test (mt-sDNA) was the first to exemplify how a simple test like FIT could be enhanced by incorporating additional analytes. In its multiple iterations, the Cologuard mt-sDNA test has investigated various substances, consistently retaining hemoglobin as a cornerstone [9, 10]. In its first generation, it assessed the presence of 21 mutations in 3 genes (APC, KRAS, and P53), the presence of BAT-26 microsatellite instability marker, and a marker of long DNA thought to represent anoikis (epithelial cell apoptosis). The 2004 trial enrolled 2507 study participants and demonstrated a CRC sensitivity of 51.6% vs. 12.9%, and it detected 18.2% of AA, compared to 10.8% with the guaiac fecal occult blood test (FOBT) [67]. However, compared to the subsequent generations of FOBT tests, a multicenter, triple-blinded prospective trial revealed that the mt-sDNA test offered a similar sensitivity for detecting screening-relevant neoplasms (20% vs. 21%) [81]. A decade later, the second generation of the mt-sDNA test retained KRAS mutations and hemoglobin only, and incorporated the promoter methylation status of BMP3 and NDRG4 genes, along with β-actin (as a reference) [68]. In the 2014 DeeP-C study, Imperiale et al. compared this new mt-sDNA test to FIT in a population of 9989 participants and demonstrated a 92% sensitivity for CRC (vs. 73.8%) and 42% for AA (vs. 23.8%) at the cost of a lower specificity (86.6% vs. 94.9%). [68]. However, further advancements in next-generation FIT tests continuously narrowed the sensitivity gap between FIT and mt-sDNA to the point of becoming clinically insignificant [59]. Furthermore, a subsequent addendum to these results, prompted by the increasing incidence of CRC among young adults [82], revealed that the sensitivity was even lower in younger individuals, particularly for AAs (33%) [83]. In response to these concerns, the latest mt-sDNA retained fecal hemoglobin as a core element and added three methylated DNA markers (LASS4, LRRC4, PPP2R5C, and ZDHHC1 as a reference) [69]. In the 2024 BLUE-C trial, the newest mt-sDNA test was compared to FIT in 20,176 participants aged 40 or older, and it demonstrated a higher sensitivity than FIT for CRC (93.9% vs. 67.3%) and for AAs (43.4% vs. 23.3%) with a high sensitivity for screening-relevant CRCs (stages I-III: 92.7%). While the overall specificity was improved from the previous generation, it was still lower than that of FIT (90.6% vs. 94.8%) [69]. Ultimately, this implied a PPV of 33.7% for colorectal neoplasias (CRC or AAs) and an NPV of 93.0% [69, 84]. The Cologuard mt-sDNA test has been cleared by the FDA for average-risk individuals only, but preliminary results suggested 100% sensitivity and 89% specificity for IBD-associated CRC for BMP3 and NDRG4 methylation alone [85]. A recent multi-center case–control study conducted in an IBD population tested stool samples for these two molecular biomarkers, minus the hemoglobin component, and observed a 73.3% sensitivity for all-stage CRC and 76.2% for advanced colorectal neoplasias [86]. Even more interesting for the IBD population was the 100% sensitivity for IBD-associated high-grade dysplasia [86]. To date, Cologuard’s mt-sDNA remains the fecal test with the longest track record of high-quality data in support of its clinical use. Despite its merits, however, the prohibitive costs hindered its adoption into national healthcare systems [87–92]. Moreover, because it evaluates significantly more analytes than FIT, it requires almost an entire bowel movement rather than a few grams of feces, and therefore, the issue of patient compliance remains largely unaddressed [9].
The multi-target stool RNA and the multi-target FIT tests
More recently, the multitarget stool RNA (mt-sRNA) test was designed in a manner similar to mt-sDNA by combining a commercially available FIT with eight target RNA transcripts (ACY1, AREG, CDH1, EGLN2, GAPDH, KRAS, SMAD4, and TNFRSF10B) and the smoking status. The mt-sRNA test originally contained 15 mRNA transcripts and was developed for the sole detection of AAs [93], but it was later refined to only 8 transcripts and combined with FIT and the smoking status to achieve 62% sensitivity for AAs and 85% specificity at cross-validation [94]. Based on such preliminary findings, the phase III CRC-PREVENT trial was designed as a blinded and prospective clinical trial to compare the mt-sRNA test to FIT [70]. Enrolling nearly 9,000 individuals aged 45 or older via social media platforms, it demonstrated superior sensitivity for both CRC (94% vs. 78%) and AA (46% vs. 29%) and a similar specificity. Interestingly, the test’s sensitivity for AAs correlated with their size (50.0% for AAs > 20 mm but 35.5% for AAs < 10 mm). Furthermore, unlike the mt-sDNA test, the sensitivity values for CRC and AAs remained stable for study participants aged 45–49 years. One potential reason for the maintenance of these sensitivity values across all ages might be attributable to the fact that methylation patterns can change according to age and, therefore, may impact DNA-based tests more than RNA-based tests [95].
FIT has also been combined with additional protein targets. This multi-analyte test, termed Multi-Target FIT (mtFIT) was initially based on 27 additional proteins, later narrowed down to 10 proteins, and then finally to a 3-protein panel: hemoglobin, calprotectin, and SerpinF2 [96–100]. In the first assessment of the 10-protein panel, mtFIT improved AA sensitivity from 28.7% to 37.8%, with a negligible impact on CRC sensitivity (80.9% with FIT vs. 78.7% with mtFIT) [96]. These results led to a recent large Danish study (n = 35,786) that compared mtFIT to FIT in individuals aged 55–75 [98]. Of the 13,187 study participants analyzed, 29 had CRC, 241 AA, and 65 advanced serrated polyps, and mtFIT demonstrated a higher detection rate for AAs (1.64% vs. 0.86%) [98]. However, considering that this study was not colonoscopy-controlled (FIT or mt-FIT-negative individuals did not receive colonoscopy), this study could not provide sensitivity or specificity estimates [98].
Other stool-based tests for detecting advanced adenomas: DNA-Methylation, Non-Coding RNAs, and the Microbiome
Several other studies evaluated another stool tests based on the methylation status of a single gene (Vimentin), reporting a sensitivity for CRC ranging from 46 to 86% but only around 15% for AAs [101, 102]. These findings suggest that hemoglobin remains the most important biomarker for stool-based studies. While other analytes can enhance the performance of hemoglobin, such analytes cannot surpass it in head-to-head trials. Moreover, most studies on a vimentin-only fecal test had a case–control design that might have overestimated these values [103–106]. The second-generation stool-based vimentin test also incorporated KRAS and APC point mutations and was compared to FOBT [107]. In a multicenter, prospective, triple-blinded trial study that involved 4482 average-risk adults, this second-generation vimentin test had a higher sensitivity than FOBT for CRC/AAs (46% vs. 24%) but at a significantly lower specificity (84% vs. 95%) [81]. Another study evaluated the performance of a stool DNA-based SDC2 methylation test in a prospective cohort of 1,124 participants. While demonstrating high sensitivity for CRC (95.0%) and moderate sensitivity for AAs (47.9%), the test exhibited lower specificity (81.5%) compared to Cologuard [108]. Another study assessed the stool-based ColoDefense test (multiplexed methylated SEPT9 and SDC2) in a cohort of 529 stool specimens. This test demonstrated sensitivities of 55.0% and 88.2% for AAs and CRC, respectively, with a specificity of 85.9% [77]. Finally, the SpecColon test (methylated SFRP2 and SDC2) has been primarily evaluated in Asian populations. With approximately 5 g of stool, this test has shown sensitivities of 61.5% for AAs and 88.5% for early-stage CRC [109]. Combining this test with KRAS/BRAF/APC mutation analysis improved the specificity slightly [110].
Because tumor-secreted small noncoding RNAs are continuously released into the intestinal lumen, their profiles in the stool may reflect the presence of CRC and AAs [111]. This approach was first reported in 2010, where two miRNAs (miR-21 and miR-106a) were significantly overexpressed in FIT left-over samples from individuals with CRC and AA [112]. Ever since, several studies have attempted to develop stool-based miRNA markers for the early detection of CRC and AAs [112–117]. While most studies have been conducted by performing small RNA sequencing on the tissue and then assessing their presence in the stool, a recent interesting approach has been to conduct extensive miRNA profiling directly in the feces of patients with CRC. This study identified 25 differentially expressed miRNAs in two separate cohorts [118]. Five of these miRNAs, combined via machine learning approaches, achieved an area under the curve (AUC) values of 96% (CRC) and 82% (AAs). An exciting finding was the ability to validate these results in 185 individuals with FIT-positive leftover fecal samples, where the sensitivity was 61% for AAs and early-stage CRC. However, this study lacked a control group of individuals with negative FIT results. Moving forward, the key challenge lies in determining whether fecal miRNAs, mRNA, DNA, different FIT thresholds, or a combination of these approaches will drive the next breakthrough in CRC screening.
Mounting evidence suggests a role for the gut microbiome in CRC development and progression [119–124]. Fusobacterium nucleatum is one of the most studied bacteria in this context, with animal models supporting its potential causative role in CRC carcinogenesis [125–127]. These microbial imbalances (i.e., dysbiosis) have been explored as biomarkers for CRC and AAs [128–132], including attempts to combine FIT results with microbiota analysis [133, 134]. However, these studies have yielded inconsistent and sometimes contradictory findings, hindering the reproducibility of such analyses [135, 136]. More recently, Thomas et al. conducted a metagenomic analysis of several CRC datasets, combined with internal sequencing from two independent cohorts, and reported that a microbial signature diagnostic for CRC exists [137]. Using different approaches for model training and validation, they consistently demonstrated a reproducible AUC of 84% for CRC. However, the accuracy for AAs detection was lower: the authors could differentiate AAs from CRC (AUC 69%), but not AAs from non-disease controls (AUC 58%) [137]. In fact, none of the species included were significantly different between individuals with AAs and controls, a finding that undermines the excitement around the use of microbiota for screening purposes.
In summary, fecal hemoglobin (and FIT, by extension) continues to stand as the most effective single analyte for CRC screening purposes. While the performance of FIT for early-stage CRC and AA leaves space for improvement, all tests so far developed build on top of its performance, generally improving the sensitivity for CRC and AA at the cost of lower specificity values. Moreover, and perhaps more importantly, while FIT is cheap and requires minimal amounts of feces, mt-sDNA, and mt-sRNA tests have higher costs and significantly more stool volumes, two factors that might further reduce compliance with the screening recommendations because it may make stool-based screening less appealing than it already is.
Blood tests: circulating DNA methylation for detecting advanced adenomas
Approximately 59% of eligible persons 45 years of age or older are adherent to screening guidelines, well below the target of 80% set forth by the National Colorectal Cancer Roundtable [138]. What is more important is that screening options seem to select for an involuntary healthy-user bias, as screening is mostly undertaken by individuals who live healthier (and wealthier) lives, a fact underlined by the observation that 76% of CRC-related deaths occur in people who are not up to date with screening [139] and that, in general, CRC-related deaths are higher in some ethnic and racial minorities [140–143].
Over 90% of screening-eligible patients prefer a blood test over FIT and colonoscopy if given the option [39]. Therefore, the first generation of blood-based assays entered the market primarily to enhance patient compliance and centered on the analysis of the methylation status of a single gene (Septin-9) [33, 37, 48, 144]. Epi Pro Colon demonstrated higher adherence than FIT (99.5% vs. 88.1%) and increased screening uptake by + 7.5% among US Veterans who had previously declined both colonoscopy and FIT [40]. However, its diagnostic sensitivity in detecting AAs and CRC lagged significantly behind FIT and Cologuard [47]. Independent evaluation of Septin-9 as a biomarker for CRC has reported sensitivity values as low as 39% but ranging up to 88.9%, with similarly large brackets for specificity, ranging from 79–100%. In much the same way that efforts have been made to improve the performance of FIT by adding new analytes, attempts to improve Septin-9 tests have been performed by combining several analytes [145, 146]. In a retrospective case–control study, the methylation status of four additional genes (BCAT1, IKZF1, BCAN, and VAV3) was assessed in 348 participants. This approach increased the AAs sensitivity to 42% (33.1% – 51.5%) [74], which led to a large clinical trial where 105,285 individuals aged 40–80 years received this 5-gene panel as the primary screening method, and 6759 were positive. However, only 1.92% of the screen-positive individuals had CRC, and 13% had AAs, which raised questions about the specificity of the test. While trial results, to this date, have not been fully published, the effectiveness of this approach seems underwhelming [147]. Expanding on such an approach, cancer-specific CpG island methylation patterns have been developed in another non-invasive blood-based DNA test [148]. Originally developed in two consecutive sets of deep sequencing datasets, such a test employed a panel of 149 methylation markers and several machine learning approaches to eventually evaluate the test in a population cohort of 114,136 urban residents of Beijing, Hebei, and Guangzhou. Unlike other studies, this study did not include asymptomatic individuals but instead included a CRC risk-enriched cohort of individuals having either IBD, a familial or hereditary predisposition to CRC, or symptoms suggestive of CRC, and therefore included 3493 patients in the study. This approach yielded significantly better results than other blood-based tests, reaching 86.4% sensitivity for CRC. However, the study design and the machine learning approaches were specifically targeted at CRC, while adenomas, by definition, were considered controls and, as such, were all marked as negative [148].
Blood tests: circulating DNA fragmentomics for detecting advanced adenomas
Liquid biopsies have recently seen a rise in the use of "fragmentomics" rather than epigenetic profiling alone. Fragementomics analyzes circulating cfDNA by focusing on its size patterns, end motifs, preferred ends, single-stranded jagged ends, and nucleosomal footprints [149]. Fragmentomics goes beyond just size, as it also considers how DNA is packaged within nucleosomes and machine learning-powered technologies are constantly being developed to further refine the analytical power of fragmentomics. The latest iteration of such approaches, Shield from Guardant Health, also increased participation rates by + 17%, engaging patients who had actively declined screening for over a decade [37, 150]. The ECLIPSE study, recently published, evaluated such blood-based screening technology in an average-risk and asymptomatic population [76]. While this study design could not make definitive claims in terms of superior compliance rates compared to other screening options (all participants had a colonoscopy, and none were offered feces-based testing, again potentially determining a healthy-user bias in the population enrolled), it achieved an 83.1% sensitivity for CRC, but with significantly lower sensitivity values for the clinical stage I CRC (55%, CI95% 35–73%) and AAs (13.2%, CI95% 11.3–15.3%). Moreover, the specificity was also suboptimal (89.6%) and resulted in low positive predictive values for both CRC (3.2%) and AAs (12.9%). To put it into perspective, the current recommendation to offer colonoscopy to all individuals has PPV values that are equal to the prevalence of the disease. Therefore, Shield offered an advantage for CRC, with a prevalence of 0.42% (vs. 3.2%), but not for AAs, with a prevalence of 10.8% (vs. 12.9%). In other words, the test could slightly enrich the population with neoplasia in the test-positive group but not significantly more than a test-all approach. Moreover, it should be noted that a blood-based test proves efficient only when coupled with a diagnostic colonoscopy in those with positive screening tests [151, 152], an endpoint not yet analyzed by this approach. Furthermore, it is still unclear if Shield will prove effective on a broader scale: if such tests become more popular among those already prone to testing (those who are willing to take the stool- or endoscopy-based screening), such low sensitivity values might revert back decades of screening efforts. Therefore, it remains unclear if Shield will improve CRC screening, whether at the population or individual levels.
Other non-invasive tests for detecting advanced adenomas
Several other methodologies and technologies have been developed for the non-invasive early detection of CRC and AA, including miRNA-based approaches [153–161], urinary metabolite profile [162, 163], and volatile compounds in the feces and urine [164, 165]. With the exception of miRNA-based approaches, these have not yet reached large-scale population-level validation. One interesting approach, undertaken by Freenome, has been to combine these multiple analytes (RNA, DNA, and proteins) into a single blood test [166]. This multi-omics blood test, combined with machine learning approaches, was recently evaluated in the PREEMPT CRC clinical trial, the largest to evaluate a blood-based CRC screening test (N = 48,995). Although trial results have not been published in full, the recent press release announced underwhelming results, with a 79.2% sensitivity for CRC, 57.1% for stage I CRC, and 12.5% for AAs [79].
Health economics of colorectal cancer screening tests and final remarks
With various screening options available—colonoscopy-first, stool-based tests, or blood-based tests—the optimal approach hinges on four key factors: screening compliance and participation, mortality reduction, incidence reduction, and cost-effectiveness. For example, a colonoscopy-based approach offers the highest potential for reducing CRC mortality and incidence, but its costs and low population participation, as demonstrated in the NordICC trial, may limit its widespread adoption [27]. While the U.S. population may be more receptive to colonoscopy-based screening, it remains a significant challenge to ensure adequate uptake [91, 167]. FIT, on the other hand, offers a significant CRC mortality reduction for a very low cost, but its reliance on patient adherence can be a challenge. Finally, blood-based tests promise to maximize compliance and reduce CRC mortality, but for a higher cost [168]. Naturally, there has been a growing interest in understanding the most cost-effective screening option. While studies often point to FIT, this is largely due to its disproportionately lower cost and it is important to note that this cost advantage may not always outweigh the potential benefits of other, more sensitive tests [169]. One of the first cost-effectiveness analyses compared mt-sDNA every three years to FIT every year to colonoscopy every ten years. This study found that FIT and colonoscopy were both more effective and less costly than mt-sDNA. Two more recent cost-effectiveness studies compared blood-based testing every three years to Cologuard mt-sDNA every three years to annual FIT to colonoscopy every ten years [170, 171]. Both studies concluded that FIT remains the most cost-effective option due to its lower cost. While blood-based testing is cost-effective compared to no screening, it is less cost-effective than other options, leading to a decrease in quality-adjusted life-years and increased costs. However, a blood-based test could potentially match FIT's clinical outcomes under one of two conditions: a significant increase in participation rates (40–80%), especially among reluctant populations, or a substantial improvement in sensitivity to advanced adenomas (closer to 80%). If one of these conditions is met, blood-based testing could revolutionize CRC screening [171, 172].
Another crucial aspect of screening tests is the appropriate interval between tests after a negative result. Generally, tests with higher specificity and lower sensitivity require more frequent testing to compensate for missed cases [173]. In fact, studies have shown that multiple rounds of FIT testing, often up to six, are necessary to achieve sustained reductions in CRC mortality [174, 175]. Therefore, not only is participation in screening important, but so is compliance with recommended follow-up testing after multiple negative results. Both modeling and observational studies have demonstrated that a single negative FIT test, without subsequent testing, is not sufficient to protect against CRC. In fact, a recent model comparing decennial colonoscopy to annual FOBT found colonoscopy to be more cost-effective, not due to initial participation rates, but rather because of lower compliance with repeated FOBT testing [167]. To address this issue, researchers are exploring strategies such as extending the interval between FIT tests [176] or adjusting positivity thresholds to improve sensitivity and reduce the need for frequent testing [8, 177]. Survey studies already reported that patients, particularly younger ones, would prefer testing every three years to once a year [178]. In one study, in fact, FIT testing every two years has already shown improved compliance and cost-effectiveness compared to colonoscopy every ten years [179]. Alternatively, lowering the positivity threshold of FIT to improve sensitivity could be a more effective strategy than simply increasing the testing interval [180]. For example, the Scottish Bowel Screening Program chose a threshold of 80 μg/g with an annual frequency of testing, and reported an uptake of FIT of 64% [181]. On the other hand, the Netherlands FIT program determined testing once every 2 years with a cutoff value of 47 μg/g and observed a higher participation rate of 72% [176]. Unsurprisingly, further lowering the threshold to 20 or 10 even μg/g improved the sensitivity for both early-stage CRC and even AAs, but also increased significantly the proportion of patients requiring a diagnostic colonoscopy [182]. Interestingly, a recent study compared the performance of FIT with a lower positivity threshold to mt-sRNA testing and found that a similar level of sensitivity and specificity, as reported in the CRC-PREVENT study, could be achieved by adjusting the FIT threshold without the need for additional mt-sRNA testing. These findings align with previous observations for multi-target stool DNA testing [183]
One problem that may be common to all blood-based tests is that they may pick up non-CRC cancers or not adenomas because methylation and expression level differences may be common to other cancers and, therefore, not very specific [184]. These considerations have driven forward a new generation of tests that assess the risk of cancer, in general, not only CRC, which forms the basis of multi-cancer early detection tests. Although not the primary interest of this review, Galleri (Grail) and Cancer-Guard (Exact Sciences) embody such an approach. Galleri relies on 5mC-modified cfDNA, while CancerGuard combines ctDNA mutations with protein immunoassays. However, trial sub-analyses indicated that their sensitivity for early stages CRC and AAs is either suboptimal or not yet tested [52, 54, 57].
In conclusion, blood-based tests currently appear primarily attractive because they appeal to the large number of individuals who are currently non-compliant with screening recommendations, potentially increasing the participation rate to screening. However, their performance metrics are currently not on par with those demonstrated by stool-based tests. Therefore, if they were to replace the stool- or endoscopy-based screening, the consequences may be less than ideal, particularly among those individuals who are already compliant with screening and may perceive these tests as alternatives.
The grand revolution of minimal residual disease
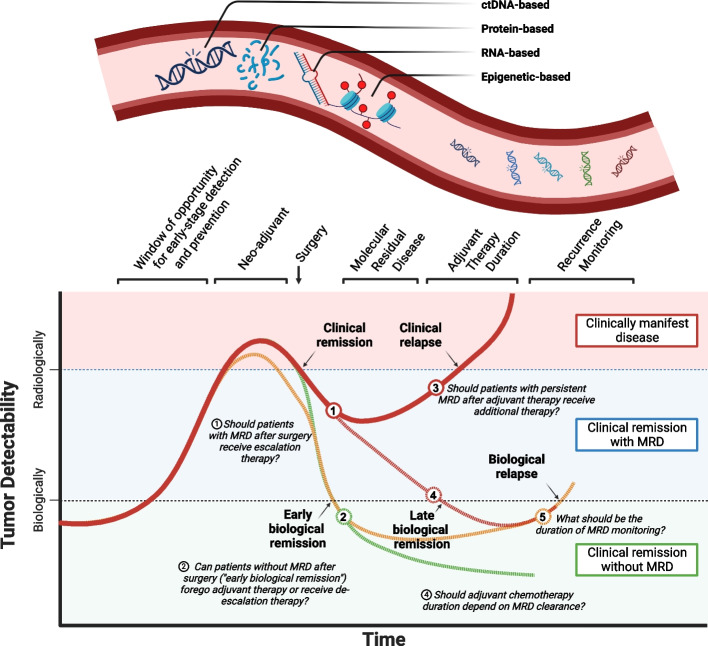
With its origin in hematology, minimal or molecular residual disease (MRD) refers to the presence of any tumor-derived molecule that represents the disease. While MRD is a conceptual framework rather than a directly measurable entity, in the context of CRC, it is commonly measured through circulating cell-free DNA (cfDNA). As a result, the two terms have become nearly synonymous within oncology, particularly within the field of CRC. However, cfDNA represents just one of many potential MRD measurement methods. Due to cfDNA's transformative impact on cancer management, particularly in challenging concepts like cancer staging, discussions of MRD in CRC often implicitly refer to cfDNA. In the context of CRC, MRD is typically assessed after surgery or adjuvant chemotherapy, while “molecular relapse” refers specifically to a subset of patients who develop MRD after initially testing negative, either after treatment completion or during monitoring. Unlike prognostic biomarkers that indicate a higher risk of recurrence, MRD directly signals persistent disease (or occult metastases) following curative-intent surgery, with a 2-year recurrence-free survival of < 5% [185–187]. The accumulating evidence on ctDNA-based MRD is transforming therapeutic algorithms and post-operative treatment decisions (Fig. 3). The revolutionary potential of cfDNA lies in its promise to redefine colon cancer management by challenging the traditional concept of stage-based treatment. Currently, post-surgical chemotherapy is administered to mitigate the risk of recurrence, treating all patients whose colon cancer has a certain risk profile (stage III and high-risk stage II). However, cfDNA will offer a more precise approach by accurately detecting MRD after surgery. This paradigm shift would enable us to transition from treating risk to targeting the actual disease that persists, potentially leading to more personalized and effective treatment strategies. Recognizing this, the National Cancer Institute (NCI) convened a task force that further fueled the development of a new wave of clinical trials exploring ctDNA's dual potential: as a biomarker for recurrence and as a surrogate endpoint of treatment success [188].
Fig. 3.
Opportunities for minimal residual disease assessment to complement clinical management. Numbers represent scientific question that are being explored by trials summarized in Table 2
The scope of the problem for stage II and III colon cancer
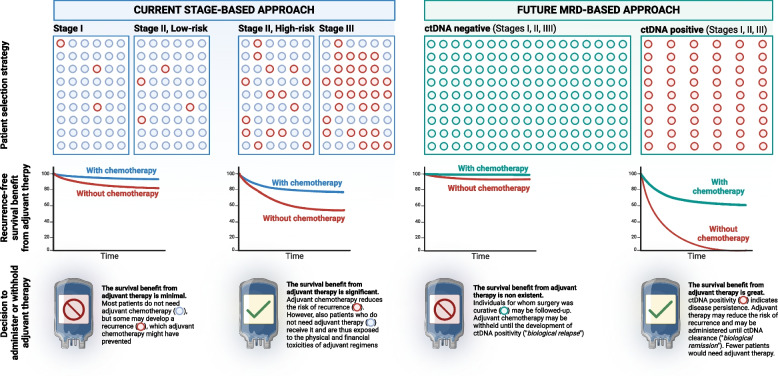
The decision to administer adjuvant chemotherapy following curative surgery for colon cancer hinges on a two-step approach based on stage and histological risk factors. All patients with stage III colon cancer are recommended for adjuvant chemotherapy. Additionally, patients with stage II colon cancer exhibiting high-risk histological features also receive it. These guidelines, although in place since 2004, have been recognized as inadequate by several societies, including ASCO, because the recurrence risk for patients with stage II/III colon cancer is only 20–30% [189–193]. On the other hand, ctDNA-positivity appears to be a better predictor of recurrence [186, 187, 194–197]. Furthermore, the IDEA trial results already supported a transition towards a shorter 3-month chemotherapy regimen, but because surgery can be curative even in stage III colon cancer, an additional de-escalation strategy might be to skip adjuvant chemotherapy entirely for ctDNA-negative patients [198, 199]. In fact, adjuvant chemotherapy reduces the recurrence risk by 5–10% at most for those with stage II colon cancer, at the significant cost of unnecessary toxicity, including peripheral neuropathy [191–193, 200–204]. Because a substantial proportion of patients with stage II and, to a lesser degree, stage III are receiving therapies they do not need, an MRD-based approach, instead, would spare them from these physical and financial toxicities (Fig. 4) [205–208]. Another slightly different application of MRD is prognosis prediction for stage IV disease [209]. In the metastatic setting, ctDNA may be used to quantify the overall tumor burden (thus gauge how well a treatment is working), tumor evolution (allowing for earlier adjustments to therapy), and predicting immunotherapy response (to choose the most appropriate checkpoint inhibitor).
Fig. 4.
The transformative changes promised by the implementation of a molecular residual disease (MRD)-based approach
Finally, another application is early recurrence monitoring, which is conceptually different from MRD because of the time component. Because CRC may recur years after their original treatment, periodic ctDNA testing may be performed over an extended time period. Unlike the previous two applications, in this case, the focus shifts from a time point to a time window, during which the development of a test’s positivity is assessed. The idea here is more practical than innovative. Patients already undergo various tests like colonoscopies, imaging, and physical exams. While CEA blood tests are used for monitoring, their sensitivity for recurrence is below 70%, so ctDNA may be integrated into existing blood draws.
A molecular background to cfDNA-Based MRD
The tumor-derived fraction of cfDNA is usually referred to as ctDNA, which comprises below 1% of cfDNA [194, 210, 211], especially in the early stage settings [194, 212]. From a design standpoint, MRD assays can be summarized in two options, either tumor-informed or tumor-agnostic; the first looks for patient-specific tumor DNA mutations in plasma after the patient’s tumor has been sequenced, while the second looks for DNA mutations without a priori knowledge of the mutational status of the patient’s tumor. Plasma-only approaches are generally less sensitive but more practical because they do not need to obtain, sequence, and analyze the tumor tissue to create a custom panel for each patient [187]. Nevertheless, recent years have demonstrated a significant shift towards applying a tumor-informed approach, because although more expensive and logistically challenging, it increases the sensitivity of the assay and reduces the false-positive rate compared to a tumor-naïve approach [213–215]. This is crucial because minimizing false positives is essential for clinical adoption, especially when positive test results are poised to lead to interventions. From a purely statistical standpoint, however, tumor-based approaches may also encounter low specificity values because, as the number of variants assessed increases, so does the risk of a false-positive result. To combat this loss in specificity, many tumor-informed methods require at least two variants to be present in both tumor and plasma and attempt to limit the number of variants that are being tracked (currently ranging from 16 to 115) [185, 214].
Clinical and observational studies on ctDNA-based MRD
In one of the first studies to explore the use of ctDNA in stage II colon cancer, 44/231 patients received adjuvant chemotherapy based on histological risk factors, and 178 did not. However, 14 of these 178 had detectable ctDNA levels after surgery, and 79% of these experienced a CRC recurrence at a median follow-up of 27 months [216]. Similarly, 4 of the 44 who received adjuvant chemotherapy had persistent ctDNA after chemotherapy completion, and they all developed a recurrence within 11 months [216]. These results provided the initial rationale to hypothesize that ctDNA detection should determine the administration of adjuvant treatment and that the inability to clear ctDNA after adjuvant chemotherapy represents a poor prognostic indicator. Similar results were later observed by others across all CRC stages. In a study of 96 stage III colon cancer patients, those with detectable ctDNA levels had a higher risk of recurrence even after adjusting for other risk factors (HR, 7.5; CI95%, 3.5–16.1). Moreover, the inability to clear ctDNA levels after chemotherapy was also associated with poorer 3-year DFS outcomes (HR, 6.8; CI95%, 11.0–157.0) [186]. Furthermore, during post-surgical surveillance, the development of ctDNA positivity precedes radiological and clinical evidence of recurrence by a median of at least 3 months [217]. Across different studies, the detectability of ctDNA after surgery could predict relapse with a median lead time of 9–12 months (range 1.8–11.5 months) [185–187, 196, 217–220]. On the other hand, the ability to clear ctDNA permanently is independently associated with prolonged recurrence-free survival, while almost all patients with a transient or persistent surge of ctDNA levels relapse [221]. Therefore, different therapeutic approaches are being studied to assess whether patients with persistent ctDNA positivity may need an intensified triplet chemotherapy, a longer treatment period (bringing it back to 6 months), or a different therapy altogether. Interestingly, several studies have demonstrated a direct relationship between the use of adjuvant chemotherapy and the conversion of ctDNA-positive patients into ctDNA negativity (about 25%). The IDEA-France study, for example, demonstrated an improvement in DFS survival by administering 6 months of chemotherapy to stage III CRC patients with detectable ctDNA post-operatively [222].
It should be sadly highlighted that much of the research on MRD has involved colon cancer, leaving rectal cancer behind, and, most importantly, locally advanced rectal cancer, a field where much improvement is needed. One such study reported that, in 159 patients with locally advanced rectal cancer, ctDNA was detectable in 8% after neoadjuvant-chemoradiotherapy and 12% after surgery [223]. Moreover, ctDNA-positivity conferred a higher risk of recurrence, both with chemotherapy (HR = 10) and without it (HR = 22) [223].
In most of the aforementioned studies, no interventional decisions were taken based on ctDNA results, which were blinded to the treating physician. As evidence accumulated about the potential of ctDNA to truly transform clinical practice, interventional clinical trials were initiated to explore several outcomes and potential avenues to transform the way we treat CRC.
The efficiency advantage of ctDNA-Based clinical trials
Because of the substantial evidence in favor of a ctDNA-based stratification of high- and low-risk, clinical trials have been designed with different endpoints to evaluate how ctDNA may be used in clinical practice (Table 2). Interestingly, ctDNA-based trials offer a more efficient study design, substantially reducing the trial sample size by increasing statistical power. Because statistical power depends on the recurrence rate, smaller sample sizes are possible for ctDNA-positive cohorts (leading to approximately an eightfold reduction compared to traditional study designs) [209]. Beyond reduced sample sizes, ctDNA offers another significant advantage – its potential as a surrogate endpoint for treatment response. Traditionally, DFS serves as a primary endpoint in clinical trials. However, ctDNA negativity could replace DFS, leading to further improvement in clinical trial design efficiency and conduct. Because ctDNA positivity indicates a higher risk of recurrence sooner than clinical or radiological evidence, ctDNA-based trials may have shorter follow-up periods, translating to significant cost reductions through several channels: less time needed for follow-up, centralized ctDNA analyses instead of geographically dispersed imaging analyses, and reduced reliance on expensive imaging expertise. Estimates suggest that a ctDNA-based approach could also lead to a 75% reduction in patient costs [209]. Moreover, in line with such advantages, the act of repeated sampling further increases the power of a study. For example, the MEDOCC-CrEATE study indicated a sample size of 60 ctDNA-positive patients (after testing 1320 patients) sufficient to analyze recurrence rates [224]. In other words, this strategy allowed a sample size reduction of almost 20 folds. For the DYNAMIC-II trial, the use of a ctDNA-based approach allowed the intervention group to be of only 30 ctDNA-positive patients stage II colon cancer patients to receive adjuvant single-agent chemotherapy (from an overall sample size of 455).
Table 2.
Clinical trial results from minimal residual disease assays based on ctDNA (numbers refer to Fig. 2)
|
Trial’s name [REF] Design |
Target(s) tested (assay name) | Population tested | Internvention(s) tested | Responds to question No | Sample size | Study design and endpoints |
|---|---|---|---|---|---|---|
|
BESPOKE [225] Observational |
Tumor-informed ctDNA (Signatera) | Stage II/III | Local preference | ① | 1000 | Non-randomized trial. the treating physician received the ctDNA results and, if positive, may decide to assign the patient to adjuvant therapy (escalation) or observation, based on the clinicians’ preference |
| Stage I-IV | Local preference | ① | 2000 | |||
|
Phase III |
WIF1 and NPY methylation by ddPCR | Stage II | mFOLFOX6 or CapOx for 3/6 months | ①② | 198 |
Escalation. ctDNA-positive patients are randomized (2:1) to chemotherapy or observation De-escalation. ctDNA-negative patients are randomized (1:4) to follow-up within trial or follow-up outside of trial |
|
CIRCULATE-US [227] Phase II/III |
Tumor-informed ctDNA (Signatera) | Stage II/III | FOLFOX6 or CAPEOX for 3–6 months | ①②③⑤ | N.A |
De-escalation. ctDNA-negative patients are randomized to adjuvant therapy (FOLFOX6 or CAPEOX) or ctDNA-based surveillance Escalation. ctDNA-positive patients are randomized to double therapy (FOLFOX6/CAPEOX) or triplet therapy (FOLFIRINOX) Cross-over design allowed. ctDNA-negative patients who develop ctDNA positivity during surveillance may enter the escalation arm and be randomized to FOLFOX6/CAPEOX or FOLFIRINOX |
|
CIRCULATE-Germany/Austria [228] Phase III |
Tumor-informed ctDNA (Signatera) | Stage II | Capecitabine ± oxaliplatin (investigator's choice) | ①② | 554 |
Escalation. ctDNA-positive patients are randomized (2:1) to chemotherapy or follow-up De-escalation. ctDNA-negative patients are randomized (1:4) to follow-up within study or outside of study |
|
COBRA Escalation [229] Phase II/III |
Reveal (Guardant health) * | Stage IIA (low-risk) | Oxaliplatin, capecitabine, leucovorin, leucovorin-calcium, fluorouracil | ② | 1408 (prematurely stopped) | Escalation. ctDNA positive patients are assigned to treatment, while ctDNA negative patients to observation. The phase II study assesses ctDNA clearance as an endpoint while the phase III part recurrence free survival |
|
CIRCULATE-Japan [230] Umbrella |
Tumor-informed ctDNA (Signatera) | Stage II, III, or IV resectable | See below | ①②③ | 1039 | Ongoing umbrella trial with three sub-parts: GALAXY, ALTAIR, and VEGA (see below) |
|
GALAXY [231] Observational |
Tumor-informed ctDNA (Signatera) | Stage II, III, or IV resectable | Observational | ① | 1039 | Observational. Part of the CIRCULATE-Japan study; ctDNA positivity at 4 weeks after surgery associated with recurrence (HR 10.0), also in stage II/III (HR 10.82), and identified patients who derived benefit from ACT (HR 6.59) |
|
ALTAIR [231] Phase III |
Tumor-informed ctDNA (Signatera) | Stage III after completion of CapOx (3 months) | Trifluridine/tipiracil or SOC | ③ | 240 |
Escalation. Part of the CIRCULATE-Japan study. Ongoing randomized phase III trial to investigate ctDNA-guided second-line adjuvant therapy management ctDNA positive patients are randomization to treatment escalation vs. surveillance. VEGA is the comparator arm |
|
VEGA [231] Phase III |
Tumor-informed ctDNA (Signatera) | Stage II, III, or IV resectable | Observation | ② | 1240 | De-escalation. Part of the CIRCULATE-Japan study. Randomized phase III trial of treatment de-escalation for ctDNA negative patients |
|
CIRCULATE-IDEA Phase III |
Tumor-informed ctDNA (Signatera) |
Stage II high-risk Stage III low-risk |
CapOx | ①②③ | 1912 |
De-escalation. Sub-analysis of the CIRCULATE-Japan study, designed to create an international repository for ctDNA studies and evaluating 3-year DFS as the endpoint Patients are randomized to receive ctDNA-based treatment vs. standard of care. In the intervention arm, ctDNA negative patients receive observation while ctDNA positive patients receive CapOx. In the standard of care arm, all patients receive CapOx, regardless of ctDNA results |
|
DYNAMIC-III [196] Phase II/III |
Tumor-informed ddPCR | Stage III | Fluoropyrimidine, or fluoropyrimidine + oxaliplatin, or FOLFOXIRI | ①② | 1000 | De-escalation and escalation. Patients are randomized to receive ctDNA-based treatment vs. standard of care. In the intervention arm, ctDNA positive patients receive escalation therapy and ctDNA negative receive de-escalation. In the comparator standard of care arm, ctDNA results are blinded |
|
DYNAMIC-II [196] Phase II |
Tumor-informed ddPCR | Stage II | Single-agent oxaliplatin or fluoropyrimidine | ①② | 455 |
De-escalation. Patients are randomized to receive ctDNA-based treatment vs. standard of care. In the intervention arm, ctDNA positive patients received adjuvant therapy and the ctDNA negative patients received observation. All patients in the standard of care arm received adjuvant therapy A lower percentage of patients in the ctDNA-guided group than in the standard-management group received adjuvant chemotherapy (15% vs. 28%; relative risk, 1.82; 95% confidence interval [CI], 1.25 to 2.65). In the evaluation of 2-year recurrence-free survival, ctDNA-guided management was noninferior to standard management (93.5% and 92.4%, respectively; absolute difference, 1.1 percentage points; 95% CI, − 4.1 to 6.2 [noninferiority margin, − 8.5 percentage points]). Three-year recurrence-free survival was 86.4% among ctDNA-positive patients who received adjuvant chemotherapy and 92.5% among ctDNA-negative patients who did not |
| MEDOCC-CrEATE [224] | Tumor-informed ddPCR | Stage II | FOLFOX or CapOx for 6 months | ①② | 1320 | Escalation. Patients are randomized (1:1) to receive ctDNA-based treatment vs. standard of care. ctDNA-positive patients will receive escalation adjuvant therapy |
|
PEGASUS De-escalation [232] Phase II |
Reveal (Guardant health) * | Stage III and high-risk stage II | CapOx, Capecitabile, Folgiri | ④ | 135 | De-escalation.This study evaluates the conversion from ctDNA negative to ctDNA positive during adjuvant therapy. ctDNA positive patients receive CapOx while ctDNA negative patients receive capecitabine alone. However, if ctDNA negative patients who convert to ctDNA positive after 1 cycle switch to CapOx |
| TRACC Part C [233] | Tumor-informed ctDNA (Guardant Reveal) | Stage III and high-risk stage II | CapOx | ①② | 1620 | De-escalation. Patients are randomized to receive ctDNA-based treatment vs. standard of care. In the intervention arm, ctDNA positive patients wll receive CapOx, while ctDNA negative patients receive de-escalation therapy. For the comparator standard of care arm, patients receive CapOx |
*Formerly known as “Lunar-1”
Recent and ongoing clinical trials on ctDNA
CIRCULATE-Japan is a large, nationwide, multicenter (92 institutions) umbrella study that is investigating the role of ctDNA-based MRD in stage II/III colon and rectal cancer as well as resectable stage IV CRC [230]. It represents the largest and likely the most important endeavor toward the characterization of ctDNA in clinical settings. Its design allows for several sub-studies (GALAXY, observational; VEGA, de-escalation; ALTAIR, escalation) with different purposes and endpoints, partly completed and partly ongoing [230, 231]. The GALAXY study represents the prospective registry designed to monitor longitudinally the ctDNA status of patients with stage II-IV CRC who can undergo complete surgical resection, and it directs patients to one of two randomized ctDNA-guided interventional phase III trials (VEGA and ALTAIR). In this study, the liquid biopsy was tumor-informed, utilizing up to 16 patient-specific clonal, somatic single nucleotide variants that informed the design of personalized multiplex polymerase chain reaction-based NGS assays (detection of 2/16 variants defined positivity). The study initially tested 1039 patients 4 weeks after radical surgery with a median follow-up of 16.74 months and assessed for DFS as the primary endpoint and ctDNA clearance as a secondary endpoint [185]. Interestingly, the MRD status at 4 weeks after surgery was highly prognostic (18-month DFS 38.4 vs. 90.5% for MRD positive vs negative, HR 10.0, CI95%: 7.7–14.0). Furthermore, in a multivariate analysis that included all clinicopathological risk factors, ctDNA positivity at 4 weeks was the only factor independently associated with DFS after the MRD status was incorporated. Surprisingly, ctDNA-positive stage I CRC had higher rates of recurrence than oligometastatic CRC with ctDNA-negative results, a finding that corroborated previous studies [234]. Among the ctDNA-positive, those receiving adjuvant chemotherapy had an improved 18-month DFS, while among the ctDNA-negative, there was no benefit. Another outstanding finding of this study is that ctDNA-positive patients could become ctDNA-negative after chemotherapy. In fact, 3.8% converted from ctDNA-negative at week 4 to ctDNA-positive at week 12, but 7.4% converted back to negative, and their 18-month DFS was identical to the ctDNA-negative group they converted to. In essence, this supports the use of ctDNA to monitoring both the response to treatment and the disease recurrence [235]. More recently, with an additional two years of follow-up, the GALAXY study confirmed previous findings and further demonstrated that cfDNA status is not only predictive of disease recurrence but also an independent prognostic factor for overall survival [235]. While the GALAXY study is limited by its observational nature, the randomized phase III trials VEGA and ALTAIR will randomize ctDNA-negative patients to de-escalation (VEGA) or ctDNA-positive patients to treatment escalation or intensification (ALTAIR) [236]. The ALTAIR trial is a phase III study to investigate the safety and efficacy of preemptive treatment with trifluridine/tipiracil vs. placebo on DFS for CRC, in patients with ctDNA-positivity and no evidence of clinical recurrence [230]. The VEGA trial instead is a phase III non-inferiority trial randomizing ctDNA-negative stage III and high-risk stage II CRC patients to observation vs adjuvant capecitabine plus oxaliplatin. Interestingly both trials incorporate a cross-over design such that VEGA participants who become ctDNA-positive can enter the ALTAIR trial. In another recently published study that included 839 patients with stage II-III CRC, the risk of recurrence for those cfDNA-positive was significantly higher than for those cfDNA negative (HR 11.3). Interestingly, cfDNA positivity preceded radiological recurrence in 94% of cases and the inability to clear cfDNA during chemotherapy was associated with a significant risk of recurrence (HR 13.1) [237]. Several other trials are also trying to develop de-escalation strategies, like the BESPOKE and COBRA trials [225, 229]. However, in light of the old technology that the COBRA trial was built upon, Guardant Health decided to prematurely interrupt the trial. On the other hand, the BESPOKE trial demonstrated that among 350 stage II-III CRC patients, those with detectable cfDNA had a significantly higher risk of recurrence (HR 20.8). Notably, the use of adjuvant chemotherapy was beneficial only in cfDNA-positive patients, while no benefit was observed in cfDNA-negative patients [238]. These findings support the hypothesis that adjuvant chemotherapy may be withheld in low-risk, cfDNA-negative patients, while therapeutic efforts should be focused on the high-risk, cfDNA-positive population [238].
Other ongoing trials, instead, use a marker-based strategy, where patients are randomly assigned to MRD testing vs standard of care and then to receive care based on test results: ctDNA positive patients receive treatment escalation, ctDNA negative receive treatment de-escalation, and, as an active comparator arm, patients with no information on ctDNA receive standard of care. The DYNAMIC trial was a prospective randomized clinical trial of 455 stage II CRC patients who were randomized to standard treatment vs. ctDNA-based treatment in a 1:2 ratio, with the intervention being fluoropyrimidine- or oxaliplatin-based adjuvant chemotherapy. During a median follow-up of 37 months, 147 received standard management (adjuvant chemotherapy based on clinicopathological risk factors), and 294 were treated according to ctDNA results (only ctDNA positive receiving chemo). This comparison resulted in a lower proportion of patients receiving adjuvant chemotherapy (28% vs. 15%, RR = 1.82) without compromising the 2-year DFS (93.5% vs. 92.4%). Importantly, given the observation that most patients with stage II ctDNA-positive colon cancer recur within 2 years, the 3-year DFS rate for ctDNA-positive patients receiving single agent chemotherapy was particularly encouraging at 86.4% [196]. However, it should be emphasized that the time to chemotherapy initiation was almost a month longer than the current recommendations in the ctDNA-based intervention arm. Additionally, being the study designed before the IDEA meta-analysis [185, 186, 196, 197, 221, 239], most patients completed 6 months of adjuvant chemotherapy, rather than three. Other trials with a similar design include the DYNAMIC II, DYNAMIC III, TRACC, and ADNCirc. More specifically, the TRACC is a prospective multicenter, UK-based study, divided in parts B (observational) and C (treatment escalation to ctDNA-positive, treatment de-escalation in ctDNA-negative vs. standard of care) [233]. The recently completed TRACC-B study evaluated 143 stage I-III CRC patients. Notably, 14.7% of patients were cfDNA-positive and had a significantly higher risk of disease recurrence compared to cfDNA-negative patients (HR: 6.5). Interestingly, ctDNA positivity often preceded radiological evidence of recurrence in only 41.2% of cases, but unlike the GALAXY and BESPOKE studies, TRACC-B employed a tumor-agnostic testing approach [240].
In conclusion, as clinical trials are ongoing and will provide further answers in this area of active research, the duration, intensity, and composition of adjuvant chemotherapy regimens may be optimized based on ctDNA results rather than the TNM stage.
ctDNA vs. methylated-cfDNA and RNA expression
Although most of the studies on MRD concern somatic mutations, aberrant changes in DNA methylation and RNA expression are also widespread in tumor tissue and reflected in plasma [241–244]. Several studies have indicated that DNA methylation holds promise as an MRD marker [245, 246]. Some studies have begun to investigate the effect of well-known biomarkers already discussed in the AAs section of this manuscript, including SEPT-9, BCAT1, and IKZF1 [241, 245, 246]. Unfortunately, though, for methylation-based and RNA-based approaches, the sensitivity has been generally below 65% [217–220, 246, 247]. Most studies based on methylation status or the RNA expression levels have suffered from underpowered sample sizes, variability in the timing of sampling, and lack of uniformity in the timing and frequency of testing; therefore, the utility of methylation-based MRD is less firmly established than mutation-based MRD. Nevertheless, methylation-based testing may be used in tandem with mutation-based approaches to increase the sensitivity of tumor-naïve cfDNA approaches for detecting recurrences [187]. For example, the PRODIGE-GERCOR trial assessed the methylation status of WIF1 and NPY in plasma from 1,345 patients with stage III colon cancer, and 13.8% tested positive. After a median follow-up of over 6 years, this tumor-agnostic assay demonstrated that cfDNA-positive subjects had a significantly higher risk of recurrence at three years (33% vs. 24%) [222, 226].
Biological, technical, and technical limitations of the MRD approach
One challenge with ctDNA testing is a condition called clonal hematopoiesis of indeterminate potential, where hematopoietic stem cells develop cancer-mimicking mutations. These mutations become more common with age, and up to 25% of patients with advanced cancer may have them [248–251]. However, this risk can be mitigated by sequencing paired peripheral blood mononuclear cells for in silico filtering of variants common to both tumors and peripheral blood cells. Another limitation of ctDNA concerns the human anatomy and the presence of blood barriers at the peritoneum and the brain, which could sequester DNA originating from metastases at these two sites. Therefore, ctDNA detection may reach a plateau not because of the technological limits but rather by the fact that some CRCs do not shed DNA and patients whose tumors have reached the brain or the peritoneum will not be detectable [252].
The limitations of traditional, pathology- and stage-based approaches to adjuvant chemotherapy are increasingly evident. The wealth of evidence emerging in 2024 strongly supports a paradigm shift towards cfDNA-based strategies. However, critical questions remain to be answered through ongoing clinical trials. Firstly, given that cfDNA-positive patients represent a high-risk group, the optimal duration of chemotherapy may need to be extended beyond the standard 3 months, possibly back to 6 months or even until complete cfDNA clearance. Secondly, the intensity of chemotherapy regimens may require escalation, potentially necessitating triplet therapy rather than doublet therapy. Thirdly, the composition of chemotherapy regimens may need to be adjusted to target this high-risk population more effectively. Conversely, for cfDNA-negative patients with a very low risk of recurrence, the potential to reduce or even eliminate adjuvant chemotherapy should be explored. Moreover, the role of ctDNA as a primary clinical endpoint, potentially replacing traditional radiological assessments, is a promising area of research. By using MRD clearance as a treatment endpoint, we could revolutionize the definition of remission and cure, reducing reliance on costly and time-consuming imaging studies. Finally, while significant progress has been made in colon cancer, further research is needed to establish the role of ctDNA in rectal cancer. Addressing these knowledge gaps will be essential to optimize cfDNA-guided treatment strategies and improve patient outcomes.
Conclusions
CRC research exemplifies the continuous translation of scientific discovery into clinical practice. Biomarkers, playing a pivotal role in this evolution, have become indispensable tools for improving early detection, personalizing treatment strategies, and monitoring MRD.
Current CRC screening has witnessed a remarkable evolution, transitioning from the early days of fecal occult blood testing to FIT to the modern days of multi-target stool testing. While these advancements have undeniably improved early detection, they primarily refine the established cornerstone: the FIT. While FIT effectively detects established diseases, its sensitivity for AAs still remains limited. Blood-based tests hold promise but require further refinement to compete with stool-based tests and endoscopic screening which together remain the gold standard for CRC screening. The ideal scenario lies in leveraging a multi-modality approach to maximize both detection rates and patient participation. Blood tests could be particularly valuable for increasing screening participation among individuals hesitant about traditional methods. Combining these modalities – endoscopy for those comfortable with it, stool-based tests for the more reluctant, and blood tests for the most averse – would create a multi-pronged approach, maximizing sensitivity by utilizing the strengths of each method while maintaining high patient compliance by catering to individual preferences. Future research should focus on identifying complementary biomarkers that can significantly enhance FIT’s adenoma detection capabilities.
The management of stage II and III colon cancer is undergoing a radical transformation driven by the transformative power of biomarkers. The advent of MRD testing, particularly through ctDNA analysis, represents an unprecedented shift. By analyzing these tumor markers in a patient’s blood after surgery, MRD can guide treatment decisions. This personalized approach has the potential to not only spare patients from unnecessary adjuvant chemotherapy but also redefine how we classify disease severity. In the future, ctDNA positivity may hold a more ominous connotation than traditional cancer stages, prompting closer monitoring and potentially more aggressive treatment regimens.
The future of CRC management is inextricably linked to further biomarker exploration. Research efforts, highly sensitive and specific blood-based tests for adenoma detection, novel stool-based biomarkers, and the implementation of MRD testing for personalized adjuvant therapy decisions have made CRC a less deadly disease. By addressing these crucial areas, biomarkers are continuously improving CRC management, leading to earlier detection, more personalized treatment, and ultimately, improved patient outcomes.
Acknowledgements
Not applicable.
Abbreviations
- AA
Advanced adenoma
- cfDNA
Cell-free DNA
- CRC
Colorectal cancer
- ctDNA
Circulating tumor DNA
- DNA
Deoxyribonucleic acid
- FIT
Fecal immunochemical test
- FOBT
Fecal occult blood test
- MRD
Minimal residual disease
- mRNA
Messenger ribonucleic acid
- mtFIT
Multi-target FIT
- mt-sDNA
Multi-target stool DNA
- mt-sRNA
Multi-target stool RNA
- RNA
Ribonucleic acid
Authors’ contributions
A.G and A.M, both conceptulaized the article and wrote this together.
Funding
This work was supported by CA72851, CA181572, CA184792, CA187956, CA202797, CA214254, and CA271443 grants from the National Cancer Institute, National Institute of Health, and by Fight Colorectal Cancer and the Collaborative Group of the Americas on Inherited Gastrointestinal Cancer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to participate
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hol L, van Leerdam ME, van Ballegooijen M, et al. Screening for colorectal cancer: randomised trial comparing guaiac-based and immunochemical faecal occult blood testing and flexible sigmoidoscopy. Gut. 2010;59(1):62–8. [DOI] [PubMed] [Google Scholar]
- 2.Shaukat A, Mongin SJ, Geisser MS, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369(12):1106–14. [DOI] [PubMed] [Google Scholar]
- 3.Kubisch CH, Crispin A, Mansmann U, et al. Screening for Colorectal Cancer Is Associated With Lower Disease Stage: A Population-Based Study. Clin Gastroenterol Hepatol. 2016;14(11):1612-8 e3. [DOI] [PubMed] [Google Scholar]
- 4.Ricciardiello L, Ferrari C, Cameletti M, et al. Impact of SARS-CoV-2 Pandemic on Colorectal Cancer Screening Delay: Effect on Stage Shift and Increased Mortality. Clin Gastroenterol Hepatol. 2021;19(7):1410-7 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baglioni P. The IARC Perspective on Colorectal Cancer Screening. N Engl J Med. 2018;379(3):301. [DOI] [PubMed] [Google Scholar]
- 6.Lauby-Secretan B, Vilahur N, Bianchini F, et al. The IARC Perspective on Colorectal Cancer Screening. N Engl J Med. 2018;378(18):1734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinsky P, Rabeneck L, Lauby-Secretan B. The IARC Perspective on Colorectal Cancer Screening. N Engl J Med. 2018;379(3):301–2. [DOI] [PubMed] [Google Scholar]
- 8.Toes-Zoutendijk E, Kooyker AI, Elferink MA, et al. Stage distribution of screen-detected colorectal cancers in the Netherlands. Gut. 2018;67(9):1745–6. [DOI] [PubMed] [Google Scholar]
- 9.The Medical Letter. A stool DNA test (Cologuard) for colorectal cancer screening. JAMA. 2014;312(23):2566. [DOI] [PubMed]
- 10.Clebak KT, Nickolich S, Mendez-Miller M. Multitarget Stool DNA Testing (Cologuard) for Colorectal Cancer Screening. Am Fam Physician. 2022;105(2):198–200. [PubMed] [Google Scholar]
- 11.Chen H, Lu M, Liu C, et al. Comparative Evaluation of Participation and Diagnostic Yield of Colonoscopy vs Fecal Immunochemical Test vs Risk-Adapted Screening in Colorectal Cancer Screening: Interim Analysis of a Multicenter Randomized Controlled Trial (TARGET-C). Am J Gastroenterol. 2020;115(8):1264–74. [DOI] [PubMed] [Google Scholar]
- 12.Dominitz JA, Robertson DJ, Ahnen DJ, et al. Colonoscopy vs. Fecal Immunochemical Test in Reducing Mortality From Colorectal Cancer (CONFIRM): Rationale for Study Design. Am J Gastroenterol. 2017;112(11):1736–46. [DOI] [PubMed] [Google Scholar]
- 13.Forsberg A, Westerberg M, Metcalfe C, et al. Once-only colonoscopy or two rounds of faecal immunochemical testing 2 years apart for colorectal cancer screening (SCREESCO): preliminary report of a randomised controlled trial. Lancet Gastroenterol Hepatol. 2022;7(6):513–21. [DOI] [PubMed] [Google Scholar]
- 14.Holme O, Loberg M, Kalager M, et al. Effect of flexible sigmoidoscopy screening on colorectal cancer incidence and mortality: a randomized clinical trial. JAMA. 2014;312(6):606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller EA, Pinsky PF, Schoen RE, et al. Effect of flexible sigmoidoscopy screening on colorectal cancer incidence and mortality: long-term follow-up of the randomised US PLCO cancer screening trial. Lancet Gastroenterol Hepatol. 2019;4(2):101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilonis ND, Bugajski M, Wieszczy P, et al. Long-Term Colorectal Cancer Incidence and Mortality After a Single Negative Screening Colonoscopy. Ann Intern Med. 2020;173(2):81–91. [DOI] [PubMed] [Google Scholar]
- 17.Quintero E, Castells A, Bujanda L, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med. 2012;366(8):697–706. [DOI] [PubMed] [Google Scholar]
- 18.Schoen RE, Pinsky PF, Weissfeld JL, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012;366(25):2345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Schootbrugge-Vandermeer HJ, Kooyker AI, Wisse PHA, et al. Interval post-colonoscopy colorectal cancer following a negative colonoscopy in a fecal immunochemical test-based screening program. Endoscopy. 2023;55(12):1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Ma W, Wu K, et al. Long-Term Colorectal Cancer Incidence and Mortality After Colonoscopy Screening According to Individuals’ Risk Profiles. J Natl Cancer Inst. 2021;113(9):1177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C, Liu L, Li J, et al. Effect of flexible sigmoidoscopy-based screening on colorectal cancer incidence and mortality: an updated systematic review and meta-analysis of randomized controlled trials. Expert Rev Anticancer Ther. 2023;23(11):1217–27. [DOI] [PubMed] [Google Scholar]
- 22.Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370(14):1298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X, Hang D, Wu K, et al. Long-term Risk of Colorectal Cancer After Removal of Conventional Adenomas and Serrated Polyps. Gastroenterology. 2020;158(4):852-61 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminski MF, Wieszczy P, Rupinski M, et al. Increased Rate of Adenoma Detection Associates With Reduced Risk of Colorectal Cancer and Death. Gastroenterology. 2017;153(1):98–105. [DOI] [PubMed] [Google Scholar]
- 25.Song M, Emilsson L, Bozorg SR, et al. Risk of colorectal cancer incidence and mortality after polypectomy: a Swedish record-linkage study. Lancet Gastroenterol Hepatol. 2020;5(6):537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Toledo D, Breekveldt ECH, Jeg IJ, et al. Advanced serrated polyps as a target of screening: detection rate and positive predictive value within a fecal immunochemical test-based colorectal cancer screening population. Endoscopy. 2023;55(6):526–34. [DOI] [PubMed] [Google Scholar]
- 27.Bretthauer M, Loberg M, Wieszczy P, et al. Effect of Colonoscopy Screening on Risks of Colorectal Cancer and Related Death. N Engl J Med. 2022;387(17):1547–56. [DOI] [PubMed] [Google Scholar]
- 28.Bernardo WM, Averbach M, Moura EGH. Critical Appraisal of the Clinical Trial: Effect of Colonoscopy Screening on Risks of Colorectal Cancer and Related Death. Arq Bras Cir Dig. 2023;36: e1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner H, Heisser T, Cardoso R, et al. Which results would the NordICC trial have found if screening colonoscopy had prevented all incident colorectal cancers? Gastrointest Endosc. 2023;98(5):878–9. [DOI] [PubMed] [Google Scholar]
- 30.Bretthauer M, Kaminski MF, Loberg M, et al. Population-Based Colonoscopy Screening for Colorectal Cancer: A Randomized Clinical Trial. JAMA Intern Med. 2016;176(7):894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminski MF, Bretthauer M, Zauber AG, et al. The NordICC Study: rationale and design of a randomized trial on colonoscopy screening for colorectal cancer. Endoscopy. 2012;44(7):695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladabaum U, Schoen RE, Meester R. NordICC’s 10-Year Interim Results Are Unexpected and Inconsistent With Modeling Predictions. Gastroenterology. 2024;166(3):543–4. [DOI] [PubMed] [Google Scholar]
- 33.Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19(8):521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel RL, Wagle NS, Cercek A, et al. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73(3):233–54. [DOI] [PubMed] [Google Scholar]
- 35.van den Berg DMN, de Nascimento Lima P, Knudsen AB, et al. NordICC Trial Results in Line With Expected Colorectal Cancer Mortality Reduction After Colonoscopy: A Modeling Study. Gastroenterology. 2023;165(4):1077-9 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong MCS, Huang J, Liang PS. Is the practice of colorectal cancer screening questionable after the NordICC trial was published? Clin Transl Med. 2023;13(10): e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coronado GD, Jenkins CL, Shuster E, et al. Blood-based colorectal cancer screening in an integrated health system: a randomised trial of patient adherence. Gut. 2024;73(4):622–8. [DOI] [PubMed] [Google Scholar]
- 38.Dekker E, Spaander MCW. The Ideal Screening Test Is the Test That Is Done. Gastroenterology. 2023;165(1):23–5. [DOI] [PubMed] [Google Scholar]
- 39.Ioannou S, Sutherland K, Sussman DA, et al. Increasing uptake of colon cancer screening in a medically underserved population with the addition of blood-based testing. BMC Cancer. 2021;21(1):966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang PS, Zaman A, Kaminsky A, et al. Blood Test Increases Colorectal Cancer Screening in Persons Who Declined Colonoscopy and Fecal Immunochemical Test: A Randomized Controlled Trial. Clin Gastroenterol Hepatol. 2023;21(11):2951-7 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–38. [DOI] [PubMed] [Google Scholar]
- 42.Botteri E, Hoff G, Randel KR, et al. Characteristics of nonparticipants in a randomised colorectal cancer screening trial comparing sigmoidoscopy and faecal immunochemical testing. Int J Cancer. 2022;151(3):361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das TS, Rauch J, Shaukat A. Colorectal cancer screening-what does the recent NordICC trial mean for the U.S. population? Transl Gastroenterol Hepatol. 2023;8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaukat A, Church T. Colorectal cancer screening in the USA in the wake of COVID-19. Lancet Gastroenterol Hepatol. 2020;5(8):726–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malla M, Loree JM, Kasi PM, et al. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J Clin Oncol. 2022;40(24):2846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gyparaki MT, Basdra EK, Papavassiliou AG. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J Mol Med (Berl). 2013;91(11):1249–56. [DOI] [PubMed] [Google Scholar]
- 47.Ahlquist DA, Taylor WR, Mahoney DW, et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10(3):272-7 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55(7):1337–46. [DOI] [PubMed] [Google Scholar]
- 50.Grobbee EJ, Wisse PHA, Schreuders EH, et al. Guaiac-based faecal occult blood tests versus faecal immunochemical tests for colorectal cancer screening in average-risk individuals. Cochrane Database Syst Rev. 2022;6(6):CD009276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin S, Zhu D, Shao F, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci U S A. 2021;118(5):e2017421118. [DOI] [PMC free article] [PubMed]
- 52.Killock D. Diagnosis: CancerSEEK and destroy - a blood test for early cancer detection. Nat Rev Clin Oncol. 2018;15(3):133. [DOI] [PubMed] [Google Scholar]
- 53.Marlow LAV, Schmeising-Barnes N, Warwick J, et al. Psychological Impact of the Galleri test (sIG(n)al): protocol for a longitudinal evaluation of the psychological impact of receiving a cancer signal in the NHS-Galleri trial. BMJ Open. 2023;13(7): e072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neal RD, Johnson P, Clarke CA, et al. Cell-Free DNABased Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers (Basel). 2022;14(19):4818. [DOI] [PMC free article] [PubMed]
- 55.Nicholson BD, Oke J, Virdee PS, et al. Multi-cancer early detection test in symptomatic patients referred for cancer investigation in England and Wales (SYMPLIFY): a large-scale, observational cohort study. Lancet Oncol. 2023;24(7):733–43. [DOI] [PubMed] [Google Scholar]
- 56.Pyzocha NJ. Galleri Test for the Detection of Cancer. Am Fam Physician. 2022;106(4):459–60. [PubMed] [Google Scholar]
- 57.Schrag D, Beer TM, McDonnell CH 3rd, et al. Blood-based tests for multicancer early detection (PATHFINDER): a prospective cohort study. Lancet. 2023;402(10409):1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta S, Coronado GD, Argenbright K, et al. Mailed fecal immunochemical test outreach for colorectal cancer screening: Summary of a Centers for Disease Control and Prevention-sponsored Summit. CA Cancer J Clin. 2020;70(4):283–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imperiale TF, Gruber RN, Stump TE, et al. Performance Characteristics of Fecal Immunochemical Tests for Colorectal Cancer and Advanced Adenomatous Polyps: A Systematic Review and Meta-analysis. Ann Intern Med. 2019;170(5):319–29. [DOI] [PubMed] [Google Scholar]
- 60.Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med. 2014;160(3):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson DJ, Dominitz JA, Beed A, et al. Baseline Features and Reasons for Nonparticipation in the Colonoscopy Versus Fecal Immunochemical Test in Reducing Mortality From Colorectal Cancer (CONFIRM) Study, a Colorectal Cancer Screening Trial. JAMA Netw Open. 2023;6(7): e2321730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levin TR, Corley DA, Jensen CD, et al. Effects of Organized Colorectal Cancer Screening on Cancer Incidence and Mortality in a Large Community-Based Population. Gastroenterology. 2018;155(5):1383-91 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60(9):1183–91. [DOI] [PubMed] [Google Scholar]
- 64.Ravegnini G, Zolezzi Moraga JM, Maffei F, et al. Simultaneous Analysis of SEPT9 Promoter Methylation Status, Micronuclei Frequency, and Folate-Related Gene Polymorphisms: The Potential for a Novel Blood-Based Colorectal Cancer Biomarker. Int J Mol Sci. 2015;16(12):28486–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selby K, Levine EH, Doan C, et al. Effect of Sex, Age, and Positivity Threshold on Fecal Immunochemical Test Accuracy: A Systematic Review and Meta-analysis. Gastroenterology. 2019;157(6):1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niedermaier T, Weigl K, Hoffmeister M, et al. Diagnostic performance of flexible sigmoidoscopy combined with fecal immunochemical test in colorectal cancer screening: meta-analysis and modeling. Eur J Epidemiol. 2017;32(6):481–93. [DOI] [PubMed] [Google Scholar]
- 67.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351(26):2704–14. [DOI] [PubMed] [Google Scholar]
- 68.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–97. [DOI] [PubMed] [Google Scholar]
- 69.Imperiale TF, Porter K, Zella J, et al. Next-Generation Multitarget Stool DNA Test for Colorectal Cancer Screening. N Engl J Med. 2024;390(11):984–93. [DOI] [PubMed] [Google Scholar]
- 70.Barnell EK, Wurtzler EM, La Rocca J, et al. Multitarget Stool RNA Test for Colorectal Cancer Screening. JAMA. 2023;330(18):1760–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nian J, Sun X, Ming S, et al. Diagnostic Accuracy of Methylated SEPT9 for Blood-based Colorectal Cancer Detection: A Systematic Review and Meta-Analysis. Clin Transl Gastroenterol. 2017;8(1): e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niu F, Wen J, Fu X, et al. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev. 2017;26(9):1411–9. [DOI] [PubMed] [Google Scholar]
- 73.Han YD, Oh TJ, Chung TH, et al. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenetics. 2019;11(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai G, Cai M, Feng Z, et al. A Multilocus Blood-Based Assay Targeting Circulating Tumor DNA Methylation Enables Early Detection and Early Relapse Prediction of Colorectal Cancer. Gastroenterology. 2021;161(6):2053-6 e2. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka Hiromi, Shen Shuo, Scimia Mauro, et al. ColoScape test: a molecular assay to detect early-stage colorectal cancer in plasma cell-free DNA. Cancer Research. 2023;83:6505. [Google Scholar]
- 76.Chung DC, Gray DM 2nd, Singh H, et al. A Cell-free DNA Blood-Based Test for Colorectal Cancer Screening. N Engl J Med. 2024;390(11):973–83. [DOI] [PubMed] [Google Scholar]
- 77.Dai Y, Zhao G, Yang J, et al. A simplified multiplex methylated DNA testing for early detection of colorectal cancer in stool DNA. BMC Gastroenterol. 2022;22(1):428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu YT, Chen XF, Zhai CB, et al. Clinical evaluation of a multitarget fecal immunochemical test-sDNA test for colorectal cancer screening in a high-risk population: a prospective, multicenter clinical study. MedComm (2020). 2023;4(4):e345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Freenome. Freenome Announces Topline Results for PREEMPT CRC® to Validate the First Version of its Blood-Based Test for the Early Detection of Colorectal Cancer 2024 [Available from: https://www.freenome.com/newsroom/freenome-announces-topline-results-for-preempt-crc-to-validate-the-first-version-of-its-blood-based-test-for-the-early-detection-of-colorectal-cancer/.
- 80.Niedermaier T, Balavarca Y, Brenner H. Stage-Specific Sensitivity of Fecal Immunochemical Tests for Detecting Colorectal Cancer: Systematic Review and Meta-Analysis. Am J Gastroenterol. 2020;115(1):56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008;149(7):441–50, W81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cavestro GM, Mannucci A, Balaguer F, et al. Delphi Initiative for Early-Onset Colorectal Cancer (DIRECt) International Management Guidelines. Clin Gastroenterol Hepatol. 2023;21(3):581-603 e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Imperiale TF, Kisiel JB, Itzkowitz SH, et al. Specificity of the Multi-Target Stool DNA Test for Colorectal Cancer Screening in Average-Risk 45–49 Year-Olds: A Cross-Sectional Study. Cancer Prev Res (Phila). 2021;14(4):489–96. [DOI] [PubMed] [Google Scholar]
- 84.Carethers JM. Improving Noninvasive Colorectal Cancer Screening. N Engl J Med. 2024;390(11):1045–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kisiel JB, Klepp P, Allawi HT, et al. Analysis of DNA Methylation at Specific Loci in Stool Samples Detects Colorectal Cancer and High-Grade Dysplasia in Patients With Inflammatory Bowel Disease. Clin Gastroenterol Hepatol. 2019;17(5):914-21 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Itzkowitz S, Farraye FA, Limburg PJ, et al. Assessment of Stool DNA Markers to Detect Colorectal Neoplasia in Patients with Inflammatory Bowel Disease: A Multi-site Case-control Study. J Crohns Colitis. 2023;17(9):1436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deibel A, Deng L, Cheng CY, et al. Evaluating key characteristics of ideal colorectal cancer screening modalities: the microsimulation approach. Gastrointest Endosc. 2021;94(2):379-90 e7. [DOI] [PubMed] [Google Scholar]
- 88.Itzkowitz SH, Ahlquist DA. The Case for a Multitarget Stool DNA Test: A Closer Look at the Cost Effectiveness Model. Gastroenterology. 2017;152(6):1620–1. [DOI] [PubMed] [Google Scholar]
- 89.Ladabaum U, Mannalithara A. Comparative Effectiveness and Cost Effectiveness of a Multitarget Stool DNA Test to Screen for Colorectal Neoplasia. Gastroenterology. 2016;151(3):427-39 e6. [DOI] [PubMed] [Google Scholar]
- 90.Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, et al. Stool DNA testing to screen for colorectal cancer in the Medicare population: a cost-effectiveness analysis. Ann Intern Med. 2010;153(6):368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peterse EFP, Meester RGS, de Jonge L, et al. Comparing the Cost-Effectiveness of Innovative Colorectal Cancer Screening Tests. J Natl Cancer Inst. 2021;113(2):154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Redwood DG, Dinh TA, Kisiel JB, et al. Cost-Effectiveness of Multitarget Stool DNA Testing vs Colonoscopy or Fecal Immunochemical Testing for Colorectal Cancer Screening in Alaska Native People. Mayo Clin Proc. 2021;96(5):1203–17. [DOI] [PubMed] [Google Scholar]
- 93.Barnell EK, Kang Y, Wurtzler EM, et al. Noninvasive Detection of High-Risk Adenomas Using Stool-Derived Eukaryotic RNA Sequences as Biomarkers. Gastroenterology. 2019;157(3):884-7 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barnell EK, Kang Y, Barnell AR, et al. Multitarget Stool RNA Test for Noninvasive Detection of Colorectal Neoplasias in a Multicenter, Prospective, and Retrospective Cohort. Clin Transl Gastroenterol. 2021;12(5): e00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu AT, Fei Z, Haghani A, et al. Universal DNA methylation age across mammalian tissues. Nat Aging. 2023;3(9):1144–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Klaver W, Wisse PHA, van Wifferen F, et al. Clinical Validation of a Multitarget Fecal Immunochemical Test for Colorectal Cancer Screening : A Diagnostic Test Accuracy Study. Ann Intern Med. 2021;174(9):1224–31. [DOI] [PubMed] [Google Scholar]
- 97.de Klerk CM, Wieten E, Lansdorp-Vogelaar I, et al. Performance of two faecal immunochemical tests for the detection of advanced neoplasia at different positivity thresholds: a cross-sectional study of the Dutch national colorectal cancer screening programme. Lancet Gastroenterol Hepatol. 2019;4(2):111–8. [DOI] [PubMed] [Google Scholar]
- 98.Wisse PHA, de Klaver W, van Wifferen F, et al. The multitarget faecal immunochemical test for improving stool-based colorectal cancer screening programmes: a Dutch population-based, paired-design, intervention study. Lancet Oncol. 2024;25(3):326–37. [DOI] [PubMed] [Google Scholar]
- 99.Bosch LJW, de Wit M, Pham TV, et al. Novel Stool-Based Protein Biomarkers for Improved Colorectal Cancer Screening: A Case-Control Study. Ann Intern Med. 2017;167(12):855–66. [DOI] [PubMed] [Google Scholar]
- 100.Komor MA, Bosch LJ, Coupe VM, et al. Proteins in stool as biomarkers for non-invasive detection of colorectal adenomas with high risk of progression. J Pathol. 2020;250(3):288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97(15):1124–32. [DOI] [PubMed] [Google Scholar]
- 102.Zou H, Harrington JJ, Shire AM, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2686–96. [DOI] [PubMed] [Google Scholar]
- 103.Itzkowitz SH, Jandorf L, Brand R, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5(1):111–7. [DOI] [PubMed] [Google Scholar]
- 104.Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103(11):2862–70. [DOI] [PubMed] [Google Scholar]
- 105.Baek YH, Chang E, Kim YJ, et al. Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Dis Colon Rectum. 2009;52(8):1452–9 discussion 9–63. [DOI] [PubMed] [Google Scholar]
- 106.Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27(9):858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ahlquist DA, Skoletsky JE, Boynton KA, et al. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology. 2000;119(5):1219–27. [DOI] [PubMed] [Google Scholar]
- 108.Kim CW, Kim H, Kim HR, et al. A Stool DNA-Based SDC2 Methylation Test for the Early Detection of Colorectal Cancer in an Asymptomatic, High-Risk Population: A Multicenter Prospective Randomized Trial. Am J Gastroenterol. 2024. Online ahead of print. [DOI] [PubMed]
- 109.Zhao G, Liu X, Liu Y, et al. Methylated SFRP2 and SDC2 in stool specimens for Colorectal Cancer early detection: A cost-effective strategy for Chinese population. J Cancer. 2021;12(9):2665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin J, Zhang L, Chen M, et al. Evaluation of combined detection of multigene mutation and SDC2/SFRP2 methylation in stool specimens for colorectal cancer early diagnosis. Int J Colorectal Dis. 2022;37(6):1231–8. [DOI] [PubMed] [Google Scholar]
- 111.Dragomir MP, Kopetz S, Ajani JA, et al. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut. 2020;69(4):748–63. [DOI] [PubMed] [Google Scholar]
- 112.Link A, Balaguer F, Shen Y, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19(7):1766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ahmed FE, Jeffries CD, Vos PW, et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6(5):281–95. [PubMed] [Google Scholar]
- 114.Weng M, Wu D, Yang C, et al. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2017;181:108–20. [DOI] [PubMed] [Google Scholar]
- 115.Duran-Sanchon S, Moreno L, Auge JM, et al. Identification and Validation of MicroRNA Profiles in Fecal Samples for Detection of Colorectal Cancer. Gastroenterology. 2020;158(4):947-57 e4. [DOI] [PubMed] [Google Scholar]
- 116.Tarallo S, Ferrero G, De Filippis F, et al. Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut. 2022;71(7):1302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao Z, Zhu A, Bhardwaj M, et al. Fecal microRNAs, Fecal microRNA Panels, or Combinations of Fecal microRNAs with Fecal Hemoglobin for Early Detection of Colorectal Cancer and Its Precursors: A Systematic Review. Cancers (Basel). 2021;14(1):65. [DOI] [PMC free article] [PubMed]
- 118.Pardini B, Ferrero G, Tarallo S, et al. A Fecal MicroRNA Signature by Small RNA Sequencing Accurately Distinguishes Colorectal Cancers: Results From a Multicenter Study. Gastroenterology. 2023;165(3):582-99 e8. [DOI] [PubMed] [Google Scholar]
- 119.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chung L, Orberg ET, Geis AL, et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23(3):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chung L, Thiele Orberg E, Geis AL, et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23(2):203-14 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152(4):851-66 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Udayasuryan B, Zhou Z, Ahmad RN, et al. Fusobacterium nucleatum infection modulates the transcriptome and epigenome of HCT116 colorectal cancer cells in an oxygen-dependent manner. Commun Biol. 2024;7(1):551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yamamura K, Izumi D, Kandimalla R, et al. Intratumoral Fusobacterium Nucleatum Levels Predict Therapeutic Response to Neoadjuvant Chemotherapy in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2019;25(20):6170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63(12):1932–42. [DOI] [PubMed] [Google Scholar]
- 129.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–8. [DOI] [PubMed] [Google Scholar]
- 130.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 131.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vogtmann E, Hua X, Zeller G, et al. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS ONE. 2016;11(5): e0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Baxter NT, Ruffin MTT, Rogers MA, et al. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zackular JP, Rogers MA, Ruffin MTT, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7(11):1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Y, Misra BB, Liang L, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9(14):4101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao Y, Wang C, Goel A. Role of gut microbiota in epigenetic regulation of colorectal Cancer. Biochim Biophys Acta Rev Cancer. 2021;1875(1): 188490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thomas AM, Manghi P, Asnicar F, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wender R, Brooks D, Sharpe K, et al. The National Colorectal Cancer Roundtable: Past Performance, Current and Future Goals. Gastrointest Endosc Clin N Am. 2020;30(3):499–509. [DOI] [PubMed] [Google Scholar]
- 139.Doubeni CA, Fedewa SA, Levin TR, et al. Modifiable Failures in the Colorectal Cancer Screening Process and Their Association With Risk of Death. Gastroenterology. 2019;156(1):63-74 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Augustus GJ, Ellis NA. Colorectal Cancer Disparity in African Americans: Risk Factors and Carcinogenic Mechanisms. Am J Pathol. 2018;188(2):291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carethers JM. Racial and ethnic disparities in colorectal cancer incidence and mortality. Adv Cancer Res. 2021;151:197–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cunningham TJ, Croft JB, Liu Y, et al. Vital Signs: Racial Disparities in Age-Specific Mortality Among Blacks or African Americans - United States, 1999–2015. MMWR Morb Mortal Wkly Rep. 2017;66(17):444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Giaquinto AN, Miller KD, Tossas KY, et al. Cancer statistics for African American/Black People 2022. CA Cancer J Clin. 2022;72(3):202–29. [DOI] [PubMed] [Google Scholar]
- 144.Carter JV, Galbraith NJ, Yang D, et al. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116(6):762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kandimalla R, Xu J, Link A, et al. EpiPanGI Dx: A Cell-free DNA Methylation Fingerprint for the Early Detection of Gastrointestinal Cancers. Clin Cancer Res. 2021;27(22):6135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149(5):1204-25 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ding Y, Liu J, Li Y, et al. Two-year update of the prospective evaluation of ColonAiQ (PreC) study. Ann Oncol. 2023;34(2):S426. [Google Scholar]
- 148.Zhao F, Bai P, Xu J, et al. Efficacy of cell-free DNA methylation-based blood test for colorectal cancer screening in high-risk population: a prospective cohort study. Mol Cancer. 2023;22(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Thierry AR. Circulating DNA fragmentomics and cancer screening. Cell Genom. 2023;3(1): 100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Tranaltes K, Brown SR. Guardant Health Shield Screening for Colon Cancer. Am Fam Physician. 2023;107(6):655–6. [PubMed] [Google Scholar]
- 151.Gupta S, Lieberman D, Anderson JC, et al. Recommendations for Follow-Up After Colonoscopy and Polypectomy: A Consensus Update by the US Multi-Society Task Force on Colorectal Cancer. Gastrointest Endosc. 2020;91(3):463-85 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.San Miguel Y, Demb J, Martinez ME, et al. Time to Colonoscopy After Abnormal Stool-Based Screening and Risk for Colorectal Cancer Incidence and Mortality. Gastroenterology. 2021;160(6):1997-2005 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66(4):654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ozawa T, Kandimalla R, Gao F, et al. A MicroRNA Signature Associated With Metastasis of T1 Colorectal Cancers to Lymph Nodes. Gastroenterology. 2018;154(4):844-8 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wada Y, Shimada M, Murano T, et al. A Liquid Biopsy Assay for Noninvasive Identification of Lymph Node Metastases in T1 Colorectal Cancer. Gastroenterology. 2021;161(1):151-62 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 157.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–81. [DOI] [PubMed] [Google Scholar]
- 158.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–26. [DOI] [PubMed] [Google Scholar]
- 159.Zanutto S, Ciniselli CM, Belfiore A, et al. Plasma miRNA-based signatures in CRC screening programs. Int J Cancer. 2020;146(4):1164–73. [DOI] [PubMed] [Google Scholar]
- 160.Nakamura K, Hernandez G, Sharma GG, et al. A Liquid Biopsy Signature for the Detection of Patients With Early-Onset Colorectal Cancer. Gastroenterology. 2022;163(5):1242-51 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim ER, Kwon HN, Nam H, et al. Urine-NMR metabolomics for screening of advanced colorectal adenoma and early stage colorectal cancer. Sci Rep. 2019;9(1):4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang H, Tso V, Wong C, et al. Development and validation of a highly sensitive urine-based test to identify patients with colonic adenomatous polyps. Clin Transl Gastroenterol. 2014;5(3): e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Liere E, van Dijk LJ, Bosch S, et al. Urinary volatile organic compounds for colorectal cancer screening: A systematic review and meta-analysis. Eur J Cancer. 2023;186:69–82. [DOI] [PubMed] [Google Scholar]
- 165.Chandrapalan S, Bosch S, Cubiella J, et al. Systematic review with meta-analysis: volatile organic compound analysis to improve faecal immunochemical testing in the detection of colorectal cancer. Aliment Pharmacol Ther. 2021;54(1):14–23. [DOI] [PubMed] [Google Scholar]
- 166.Ulz P, Perakis S, Zhou Q, et al. Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat Commun. 2019;10(1):4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meester RGS, Lansdorp-Vogelaar I, Winawer SJ, et al. Projected Colorectal Cancer Incidence and Mortality Based on Observed Adherence to Colonoscopy and Sequential Stool-Based Screening. Am J Gastroenterol. 2024;119(7):1392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Aziz Z, Wagner S, Agyekum A, et al. Cost-Effectiveness of Liquid Biopsy for Colorectal Cancer Screening in Patients Who Are Unscreened. JAMA Netw Open. 2023;6(11): e2343392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhong GC, Sun WP, Wan L, et al. Efficacy and cost-effectiveness of fecal immunochemical test versus colonoscopy in colorectal cancer screening: a systematic review and meta-analysis. Gastrointest Endosc. 2020;91(3):684-97 e15. [DOI] [PubMed] [Google Scholar]
- 170.van den Puttelaar R, de Nascimento Lima P, Knudsen AB, et al. Effectiveness and Cost-Effectiveness of Colorectal Cancer Screening With a Blood Test That Meets the Centers for Medicare & Medicaid Services Coverage Decision. Gastroenterology. 2024;167(2):368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ladabaum U, Mannalithara A, Weng Y, et al. Comparative Effectiveness and Cost-Effectiveness of Colorectal Cancer Screening With Blood-Based Biomarkers (Liquid Biopsy) vs Fecal Tests or Colonoscopy. Gastroenterology. 2024;167(2):378–91. [DOI] [PubMed] [Google Scholar]
- 172.Mannucci A, Goel A. Stool and Blood DNA Tests for Colorectal Cancer Screening. N Engl J Med. 2024;390(23):2224. [DOI] [PubMed] [Google Scholar]
- 173.Mannucci A, Puzzono M, Danese S. Early Diagnosis of Colorectal Cancer by Biannual Fecal Immunochemical Tests: Is That Your Final Answer? Gastroenterology. 2022;162(7):2102–4. [DOI] [PubMed] [Google Scholar]
- 174.Zorzi M, Hassan C, Capodaglio G, et al. Long-term performance of colorectal cancerscreening programmes based on the faecal immunochemical test. Gut. 2018;67(12):2124–30. [DOI] [PubMed] [Google Scholar]
- 175.Giorgi Rossi P, Vicentini M, Sacchettini C, et al. Impact of Screening Program on Incidence of Colorectal Cancer: A Cohort Study in Italy. Am J Gastroenterol. 2015;110(9):1359–66. [DOI] [PubMed] [Google Scholar]
- 176.Breekveldt ECH, Lansdorp-Vogelaar I, Toes-Zoutendijk E, et al. Colorectal cancer incidence, mortality, tumour characteristics, and treatment before and after introduction of the faecal immunochemical testing-based screening programme in the Netherlands: a population-based study. Lancet Gastroenterol Hepatol. 2022;7(1):60–8. [DOI] [PubMed] [Google Scholar]
- 177.van der Vlugt M, Grobbee EJ, Bossuyt PMM, et al. Interval Colorectal Cancer Incidence Among Subjects Undergoing Multiple Rounds of Fecal Immunochemical Testing. Gastroenterology. 2017;153(2):439-47 e2. [DOI] [PubMed] [Google Scholar]
- 178.Makaroff KE, Shergill J, Lauzon M, et al. Patient Preferences for Colorectal Cancer Screening Tests in Light of Lowering the Screening Age to 45 Years. Clin Gastroenterol Hepatol. 2023;21(2):520-31 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Areia M, Fuccio L, Hassan C, et al. Cost-utility analysis of colonoscopy or faecal immunochemical test for population-based organised colorectal cancer screening. United European Gastroenterol J. 2019;7(1):105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Young GP, Woodman RJ, Ang FLI, et al. Both Sample Number and Test Positivity Threshold Determine Colonoscopy Efficiency in Detection of Colorectal Cancer With Quantitative Fecal Immunochemical Tests. Gastroenterology. 2020;159(4):1561-3 e3. [DOI] [PubMed] [Google Scholar]
- 181.Clark G, Strachan JA, Carey FA, et al. Transition to quantitative faecal immunochemical testing from guaiac faecal occult blood testing in a fully rolled-out population-based national bowel screening programme. Gut. 2021;70(1):106–13. [DOI] [PubMed] [Google Scholar]
- 182.Berry E, Miller S, Koch M, et al. Lower Abnormal Fecal Immunochemical Test Cut-Off Values Improve Detection of Colorectal Cancer in System-Level Screens. Clin Gastroenterol Hepatol. 2020;18(3):647–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Niedermaier T, Seum T, Hoffmeister M, et al. Lowering Fecal Immunochemical Test Positivity Threshold vs Multitarget Stool RNA Testing for Colorectal Cancer Screening. JAMA. 2024;332(3):251–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lo YMD. Cell-free DNA for Colorectal Cancer Screening. N Engl J Med. 2024;390(11):1047–50. [DOI] [PubMed] [Google Scholar]
- 185.Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019;5(8):1124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tie J, Cohen JD, Wang Y, et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019;5(12):1710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal Residual Disease Detection using a Plasma-only Circulating Tumor DNA Assay in Patients with Colorectal Cancer. Clin Cancer Res. 2021;27(20):5586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dasari A, Morris VK, Allegra CJ, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020;17(12):757–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.van der Stok EP, Spaander MCW, Grunhagen DJ, et al. Surveillance after curative treatment for colorectal cancer. Nat Rev Clin Oncol. 2017;14(5):297–315. [DOI] [PubMed] [Google Scholar]
- 190.O’Connell MJ, Campbell ME, Goldberg RM, et al. Survival following recurrence in stage II and III colon cancer: findings from the ACCENT data set. J Clin Oncol. 2008;26(14):2336–41. [DOI] [PubMed] [Google Scholar]
- 191.Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev. 2008;2008(3):CD005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bockelman C, Engelmann BE, Kaprio T, et al. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncol. 2015;54(1):5–16. [DOI] [PubMed] [Google Scholar]
- 193.Andre T, de Gramont A, Vernerey D, et al. Adjuvant Fluorouracil, Leucovorin, and Oxaliplatin in Stage II to III Colon Cancer: Updated 10-Year Survival and Outcomes According to BRAF Mutation and Mismatch Repair Status of the MOSAIC Study. J Clin Oncol. 2015;33(35):4176–87. [DOI] [PubMed] [Google Scholar]
- 194.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Loupakis F, Sharma S, Derouazi M, et al. Detection of Molecular Residual Disease Using Personalized Circulating Tumor DNA Assay in Patients With Colorectal Cancer Undergoing Resection of Metastases. JCO Precis Oncol. 2021;5:1166–77. [DOI] [PMC free article] [PubMed]
- 196.Tie J, Cohen JD, Lahouel K, et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med. 2022;386(24):2261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med. 2021;18(5): e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Andre T, Meyerhardt J, Iveson T, et al. Effect of duration of adjuvant chemotherapy for patients with stage III colon cancer (IDEA collaboration): final results from a prospective, pooled analysis of six randomised, phase 3 trials. Lancet Oncol. 2020;21(12):1620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sargent D, Sobrero A, Grothey A, et al. Evidence for cure by adjuvant therapy in colon cancer: observations based on individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2009;27(6):872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Boyne DJ, Cuthbert CA, O’Sullivan DE, et al. Association Between Adjuvant Chemotherapy Duration and Survival Among Patients With Stage II and III Colon Cancer: A Systematic Review and Meta-analysis. JAMA Netw Open. 2019;2(5): e194154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Andre T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350(23):2343–51. [DOI] [PubMed] [Google Scholar]
- 202.Baxter NN, Kennedy EB, Bergsland E, et al. Adjuvant Therapy for Stage II Colon Cancer: ASCO Guideline Update. J Clin Oncol. 2022;40(8):892–910. [DOI] [PubMed] [Google Scholar]
- 203.Kuebler JP, Wieand HS, O’Connell MJ, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: results from NSABP C-07. J Clin Oncol. 2007;25(16):2198–204. [DOI] [PubMed] [Google Scholar]
- 204.Land SR, Kopec JA, Cecchini RS, et al. Neurotoxicity from oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: NSABP C-07. J Clin Oncol. 2007;25(16):2205–11. [DOI] [PubMed] [Google Scholar]
- 205.Kopetz S, Tabernero J, Rosenberg R, et al. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncologist. 2015;20(2):127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pages F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–39. [DOI] [PubMed] [Google Scholar]
- 207.Venook AP, Niedzwiecki D, Lopatin M, et al. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31(14):1775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yamanaka T, Oki E, Yamazaki K, et al. 12-Gene Recurrence Score Assay Stratifies the Recurrence Risk in Stage II/III Colon Cancer With Surgery Alone: The SUNRISE Study. J Clin Oncol. 2016;34(24):2906–13. [DOI] [PubMed] [Google Scholar]
- 209.Kasi PM, Fehringer G, Taniguchi H, et al. Impact of Circulating Tumor DNA-Based Detection of Molecular Residual Disease on the Conduct and Design of Clinical Trials for Solid Tumors. JCO Precis Oncol. 2022;6: e2100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102(45):16368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Holdhoff M, Schmidt K, Donehower R, et al. Analysis of circulating tumor DNA to confirm somatic KRAS mutations. J Natl Cancer Inst. 2009;101(18):1284–5. [DOI] [PubMed] [Google Scholar]
- 212.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7(12):1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McDonald BR, Contente-Cuomo T, Sammut SJ, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11(504):eaax7392. [DOI] [PMC free article] [PubMed]
- 215.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403):eaan2415. [DOI] [PMC free article] [PubMed]
- 216.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wang Y, Li L, Cohen JD, et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol. 2019;5(8):1118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Scholer LV, Reinert T, Orntoft MW, et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res. 2017;23(18):5437–45. [DOI] [PubMed] [Google Scholar]
- 219.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625–34. [DOI] [PubMed] [Google Scholar]
- 220.Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30(11):1804–12. [DOI] [PubMed] [Google Scholar]
- 221.Henriksen TV, Tarazona N, Frydendahl A, et al. Circulating Tumor DNA in Stage III Colorectal Cancer, beyond Minimal Residual Disease Detection, toward Assessment of Adjuvant Therapy Efficacy and Clinical Behavior of Recurrences. Clin Cancer Res. 2022;28(3):507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Taieb J, Taly V, Henriques J, et al. Prognostic Value and Relation with Adjuvant Treatment Duration of ctDNA in Stage III Colon Cancer: a Post Hoc Analysis of the PRODIGE-GERCOR IDEA-France Trial. Clin Cancer Res. 2021;27(20):5638–46. [DOI] [PubMed] [Google Scholar]
- 223.Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68(4):663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Schraa SJ, van Rooijen KL, van der Kruijssen DEW, et al. Circulating tumor DNA guided adjuvant chemotherapy in stage II colon cancer (MEDOCC-CrEATE): study protocol for a trial within a cohort study. BMC Cancer. 2020;20(1):790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kasi PM, Sawyer S, Guilford J, et al. BESPOKE study protocol: a multicentre, prospective observational study to evaluate the impact of circulating tumour DNA guided therapy on patients with colorectal cancer. BMJ Open. 2021;11(9): e047831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Taieb J, Benhaim L, Laurent Puig P, et al. Decision for adjuvant treatment in stage II colon cancer based on circulating tumor DNA: The CIRCULATE-PRODIGE 70 trial. Dig Liver Dis. 2020;52(7):730–3. [DOI] [PubMed] [Google Scholar]
- 227.Sahin IH, Lin Y, Yothers G, et al. Minimal Residual Disease-Directed Adjuvant Therapy for Patients With Early-Stage Colon Cancer: CIRCULATE-US. Oncology (Williston Park). 2022;36(10):604–8. [DOI] [PubMed] [Google Scholar]
- 228.Folprecht G, Reinacher-Schick A, Weitz J, et al. The CIRCULATE Trial: Circulating Tumor DNA Based Decision for Adjuvant Treatment in Colon Cancer Stage II Evaluation (AIO-KRK-0217). Clin Colorectal Cancer. 2022;21(2):170–4. [DOI] [PubMed] [Google Scholar]
- 229.Verbus EA, Rossi AJ, Luna AJ, et al. Circulating Tumor DNA as a Predictive Biomarker in Adjuvant Chemotherapy for Patients with Stage 2A Colon Cancer (COBRA). Ann Surg Oncol. 2021;28(8):4095–7. [DOI] [PubMed] [Google Scholar]
- 230.Taniguchi H, Nakamura Y, Kotani D, et al. CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112(7):2915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kotani D, Oki E, Nakamura Y, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med. 2023;29(1):127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lonardi S, Pietrantonio F, Tarazona Llavero N, et al. The PEGASUS trial: Post-surgical liquid biopsy-guided treatment of stage III and high-risk stage II colon cancer patients. Ann Oncol. 2023;34:S1268-9. [Google Scholar]
- 233.Slater S, Bryant A, Chen HC, et al. ctDNA guided adjuvant chemotherapy versus standard of care adjuvant chemotherapy after curative surgery in patients with high risk stage II or stage III colorectal cancer: a multi-centre, prospective, randomised control trial (TRACC Part C). BMC Cancer. 2023;23(1):257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bolhuis K, van ’t Erve I, Mijnals C, et al. Postoperative circulating tumour DNA is associated with pathologic response and recurrence-free survival after resection of colorectal cancer liver metastases. EBioMedicine. 2021;70:103498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nakamura Y, Watanabe J, Akazawa N, et al. ctDNA-based molecular residual disease and survival in resectable colorectal cancer. Nat Med. 2024. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 236.Oki E, Yoshino T. Postoperative circulating tumor DNA could guide CRC adjuvant treatment. Nat Med. 2023;29(1):39–40. [DOI] [PubMed]
- 237.Henriksen TV, Demuth C, Frydendahl A, et al. Unraveling the potential clinical utility of circulating tumor DNA detection in colorectal cancer-evaluation in a nationwide Danish cohort. Ann Oncol. 2024;35(2):229–39. [DOI] [PubMed] [Google Scholar]
- 238.Kasi PM, Aushev VN, Ensor J, et al. Circulating tumor DNA (ctDNA) for informing adjuvant chemotherapy (ACT) in stage II/III colorectal cancer (CRC): Interim analysis of BESPOKE CRC study. J Clin Oncol. 2024;42;suppl.3.
- 239.Grothey A, Sobrero AF, Shields AF, et al. Duration of Adjuvant Chemotherapy for Stage III Colon Cancer. N Engl J Med. 2018;378(13):1177–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Slater S, Bryant A, Aresu M, et al. Tissue-Free Liquid Biopsies Combining Genomic and Methylation Signals for Minimal Residual Disease Detection in Patients with Early Colorectal Cancer from the UK TRACC Part B Study. Clin Cancer Res. 2024;30(16):3459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Fu B, Yan P, Zhang S, et al. Cell-Free Circulating Methylated SEPT9 for Noninvasive Diagnosis and Monitoring of Colorectal Cancer. Dis Markers. 2018;2018:6437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Jung G, Hernandez-Illan E, Moreira L, et al. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17(2):111–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Miyazaki K, Wada Y, Okuno K, et al. An exosome-based liquid biopsy signature for pre-operative identification of lymph node metastasis in patients with pathological high-risk T1 colorectal cancer. Mol Cancer. 2023;22(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kandimalla R, Gao F, Matsuyama T, et al. Genome-wide Discovery and Identification of a Novel miRNA Signature for Recurrence Prediction in Stage II and III Colorectal Cancer. Clin Cancer Res. 2018;24(16):3867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Murray DH, Symonds EL, Young GP, et al. Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival. J Cancer Res Clin Oncol. 2018;144(9):1741–50. [DOI] [PubMed] [Google Scholar]
- 246.Musher BL, Melson JE, Amato G, et al. Evaluation of Circulating Tumor DNA for Methylated BCAT1 and IKZF1 to Detect Recurrence of Stage II/Stage III Colorectal Cancer (CRC). Cancer Epidemiol Biomarkers Prev. 2020;29(12):2702–9. [DOI] [PubMed] [Google Scholar]
- 247.Tham C, Chew M, Soong R, et al. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer. 2014;120(20):3131–41. [DOI] [PubMed] [Google Scholar]
- 248.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Coombs CC, Zehir A, Devlin SM, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell. 2017;21(3):374-82 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Young AL, Challen GA, Birmann BM, et al. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Dasari A, Grothey A, Kopetz S. Circulating Tumor DNA-Defined Minimal Residual Disease in Solid Tumors: Opportunities to Accelerate the Development of Adjuvant Therapies. J Clin Oncol. 2018;36(35):JCO2018789032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.