Abstract
The AML treatment landscape has significantly changed in recent years with the approval of targeted therapies in the front-line and relapsed/refractory settings, including inhibitors of FLT3 and IDH1/2 mutations. More importantly, approval of the combination of the BCl-2 inhibitor, venetoclax, and hypomethylating agents or low dose cytarabine provided unprecedented breakthrough for the frontline treatment of older, unfit AML patients. Even with all this exciting progress, more targeted therapies for AML treatment are needed. Recent development of menin inhibitors targeting AML with KMT2A rearrangements or NPM1 mutations could represent a promising new horizon of treatment for patients within these subsets of AML. Our current review will focus on a summary and updates of recent developments of menin inhibitors in the treatment of AML, on the challenges ahead arising from drug resistance, as well as on the opportunities of novel combinations with menin inhibitors.
Keywords: Acute myeloid leukemia, Menin inhibitors, Novel therapies
Background
Acute myeloid leukemia (AML) treatment paradigm is undergoing a much-needed breakthrough with dedicated research, technological advances, and clinical trials. Detection of novel molecular targets, sensitive tumor detection markers and, importantly, drug discovery, are augmenting the outcomes of AML. This transformation has been heralded by approval and availability of small molecule inhibitors, such as selective antagonists of the anti-apoptotic protein BCL-2 (B cell lymphoma-2) including Venetoclax in 2017, inhibitors of mutated FLT-3 (FMS Like Tyrosine kinase 3), and IDH 1/2 (Isocitrate Dehydrogenase), as well as antibody drug conjugates(ADC). A detailed review of these and other agents in development for AML has been published previously [1].
In recent years, one of the most exciting advancements in AML treatment has been the clinical development of menin inhibitors. Here, we review the clinical development of menin inhibitors for the treatment of AML harboring mutations in the Nucleophosmin (NPM1) gene (NPM1m) or rearrangements in the histone-lysine N-methyltransferase 2A (KMT2A) gene (KMT2Ar), and for other rare genetic aberrations. Additionally, we discuss the challenges and opportunities that lie ahead in the clinical application of these compounds.
Biological basis of menin inhibition
Menin protein biology, structure, and interaction with other proteins:
The menin protein is a one-of-its kind, scaffold nuclear protein which is encoded by the tumor suppressor MEN1 (Menin 1) gene. MEN1 mutations are well known to be implicated in the causation of sporadic or autosomal dominant hereditary cancer syndromes which affect the endocrine system, known as the MEN1 syndrome. This demonstrates the tumor suppressor activity of the gene. [2, 3] While the biological basis of this phenomenon is being actively studied, loss of cell cycle regulation, and disruption of inhibition of transcription factors such as JunD resulting from MEN1 mutation and a truncated menin protein are a few proposed mechanisms [4, 5] On the contrary, the menin protein exerts a paradoxically different function in the hematopoietic pathological states, where it serves as an essential oncogenic cofactor for the maintenance and propagation of leukemogenesis initiated by the oncoprotein KMT2A, as well as the KMT2A fusion protein complexes [6–8].
Crystallographic studies and subsequent functional investigations revealed the detailed structure of menin. Its essential biological functions are diverse in nature, likely in a mutually exclusive manner, carried out through its interactions with various proteins—transcription factors, chromatin modification proteins, and other regulators of cell function [9]. [4, 5] Menin protein mediates and regulates several physiological cellular functions in a tissue or cell-specific manner by varying expression levels, influencing gene transcription or repression, cell signaling, DNA repair, chromatin modulation, and other vitals interactions. [5, 9] In turn, it is itself regulated by several factors such as prolactin, transforming growth factor beta (TGFB), etc [5, 9].
Menin has been likened to a “curved left-hand like structure”, in which a centrally located deep pocket or groove (formed by the palm) forms an essential binding site for different proteins and by which physiological and pathological functions are mediated. For example, the N-terminal portion of the KMT2A wild type (WT) protein, and the oncogenic KMT2A fusion protein (FP) bind to this location. [4, 5] The groove also forms a hub for binding other essential proteins and regulators such as JunD, or (Lens epithelium derived growth factor (LEDGF) which plays a key role in KMT2A-AF9 rearranged leukemogenesis [8].
Menin-KMT2A complex formation and interaction
Role of LEDGF/HOX/MEIS1
KMT2A is a complex chromatin modifying protein with histone-methyltransferase activity (HMT), and it is ubiquitous in cells and is necessary for normal development of neural, skeletal and hematopoietic elements [10]. Importantly, in vivo mouse studies showed that KMT2A is important for the maintenance of expression of homebox (HOX family cluster genes) in tissues. [10] Alteration of KMT2A function during chromosomal rearrangements or duplications, which results in the loss of essential Su(var)3–9, Enhancer-of-zeste, Trithorax (SET) domain and the HMT activity, has been associated with leukemogenesis and other malignancies through deregulation of normal cellular processes. [11] The KMT2A WT and KMT2A FP contain short, conserved sequences on the N-terminal called high-affinity menin binding motifs (MBM 1 and MBM 2) which bind to the central groove of menin [7, 12]. The consequence of these fusions is the upregulation of HOX family cluster genes and their cofactor, Meis homebox 1 (MEIS1), also denoted as HOX/MEIS1 complex, leading to differentiation blockade, and transformation of hematopoietic progenitors to leukemic stem cell state.
Homebox gene family (HOX) are vital determinants of structural development of tissues and organs in embryogenesis and are also involved in hematopoietic progenitor propagation, development and differentiation. Consequently, aberrant activation and expression of these genes has been found to be associated with transformation of leukemic cells. While HOX cluster genes are capable or leukemic initiation, MEIS1 and other cofactors such as PBX Homebox3 (PBX3) are critically necessary to potentiate and accelerate the transformation to leukemic cells [13, 14], Together, the upregulation and maintenance of these complexes underlie the converging pathway for leukemogenesis in various genomic alterations.
Menin is crucial for recruitment, activation and maintenance of HOX expression, as demonstrated by in vitro studies where HOX/MEIS1 gene expression is dramatically reduced upon deletion of MEN1 or with the introduction of dominant negative KMT2A mutations which interfere with KMT2A -menin interaction in transformed cells [12, 15].
The KMT2A binding sites on menin are the targets of menin inhibitors, which block the interaction with the KMT2A fusion proteins [16, 17].
LEDGF is a chromatin protein which serves a vital anchoring function for the oncoprotein KMT2A, and menin complex. Like MBM, the N-terminal of KMT2A binds to LEDGF using a LEDGF binding motifs (LBM). This KMT2A- menin- LEDGF complex formation is essential for the leukemogenesis which distinguishes it from the other protein KMT2D which does not contain LBM and therefore lacks leukemogenic properties [8].
Menin-NPM1m interaction
Early, global gene expression studies of de novo AML samples from pediatric and adult populations shed light on the association between NPM1m and concurrent overexpression of HOXA cluster and MES1, showing similarities with KMT2A driven leukemias [18, 19]. NPM1m protein is translocated to the cytoplasm through nuclear export signals (NES) to exert the oncogenic functions [20–22]. Relocation of NPM1m to the nucleus through abrogation of NES does not only reverse the differentiation blockade and but also turns off HOXA expression indicating the direct upstream effect of NPM1m on HOXA genes. [23, 24], Additionally, KMT2A-WT and menin interaction remains critical for the NPM1m leukemic states. [25, 26] This was demonstrated by in vivo gene expression studies of NPM1m leukemia models in which rapid reduction of the MEIS1/PBX3 cofactors' expression and concurrent loss of self-renewal capacity of leukemic stem cells, and upregulation of differentiation markers was observed upon treatment with menin inhibitor VTP-50469 (a precursor of revumenib) [25, 26].
The collective insights gained from these and numerous other important studies led to the investigation of inhibition of menin—KMT2A protein interaction, and the downstream repressive effect on HOX/MEIS1 gene expression as an important therapeutic strategy in AML involving KMT2Ar or NPM1m, and multiple other novel rearrangements [25, 27, 28]. Of note, the menin biding sites are also vulnerable to the emergence of mutations that confer resistance to the inhibitors, as elaborated in a later Section [16, 17].
KMT2A re-arranged AML
The KMT2A gene (previously known as mixed lineage leukemia 1 or MLL1) is located on chromosome 11q23, and translocations involving the KMT2A locus are present in approximately 5% of adult AML cases. However, the KMT2Ar incidence is higher in other contexts, for example, in up to 70% of infantile leukemias and approximately 50% of therapy related AML (t-AML) following etoposide exposure [29, 30].The translocation process is complex, with over 80 different partner genes identified, and research into the mechanisms and partner genes involved in KMT2r AML is ongoing [31]. KMT2A-r AML is typically associated with decreased sensitivity to chemotherapy, leading to high rates of relapse, and an overall poor prognosis [32, 33]. The 2022 European Leukemia Network (ELN) guidelines on AML classify KMT2Ar AML in the adverse risk disease category except for AML harboring t (9;11)(p21.3;q23.3) which is classified as an intermediate risk [34]. The fusion proteins created by translocations involving the KMT2A locus yield an aberrant transcription program characterized by increased expression of developmental genes including HOX and MEIS1, thereby promoting leukemogenesis. [35, 36] Menin protein binds to the KMT2A-fusion partner protein complexes leading to nuclear localization of the fusion product and thereby facilitating an aberrant transcription program. Specifically, menin forms a hub and links KMT2A and its fusion protein with a chromatin protein LDGF which is essential for leukemogenesis. [8] The interaction between the menin and KMT2A proteins is therefore central to the pathogenesis of KMT2Ar AML and forms a therapeutic target [12, 37].
NPM1 mutated AML
The NPM1 gene is located on chromosome 5q35 and codes for a protein involved in many basic cellular processes. Mutations in the NPM1 gene (NPM1m) are seen in approximately 30% of AML cases. [38] NPM1 mutations are considered favorable and are often associated with a better prognosis. The 2022 ELN guidelines on AML classify NPM1m AML as favorable risk when not associated with internal tandem duplications in the FMS-like tyrosine kinase gene (FLT3 ITD mutations) or other adverse risk cytogenetic abnormalities. [34] Approximately 80% of patients with NPM1m AML treated with intensive chemotherapy achieve remission. [39] However, approximately 50% eventually experience relapse, and prognosis is worse for patients over age 60 compared to younger patients. [40, 41] In NPM1m AML, the mutant NPM1 protein binds to chromatin targets that are shared by the KMT2A protein. The expression of HOX and other developmental genes is also upregulated in NPM1m AML. [25, 27, 38] The interaction between menin and wild type KMT2A therefore plays an important role in the pathogenesis of NPM1m AML [25, 42].
Menin inhibitors
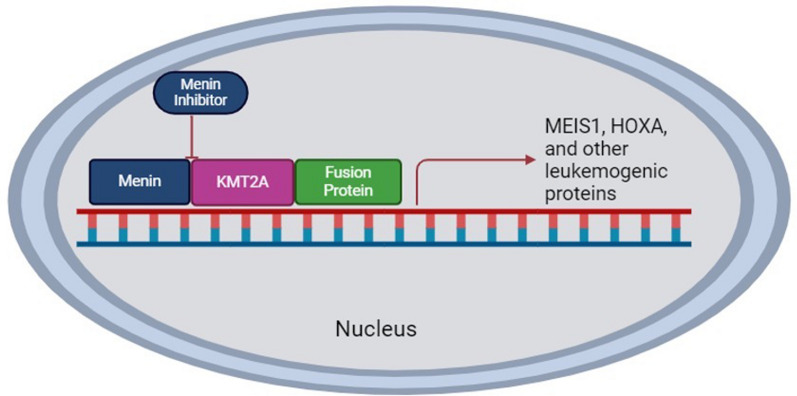
Menin inhibitors are small molecule inhibitors that bind with high affinity to the KMT2A binding pocket of menin. These agents inhibit the interaction between menin and KMT2A which inhibits KMT2A-dependent transcription of downstream target genes. These agents thereby inhibit an important component of the pathogenesis of KMT2Ar and NPM1m AML. Similarly, disruption and inhibition of the menin and KMT2A interaction is also being studied as a therapeutic strategy in leukemic states driven by other molecular rearrangements which result in increased HOXA gene transcription. [43–45] The mechanism of action of menin inhibitors is depicted in Fig. 1. The menin protein structure, and common mutation hotspots contributing to resistance to menin inhibitors are depicted in Fig. 2.
Fig. 1.
Mechanism of action of Menin Inhibitors
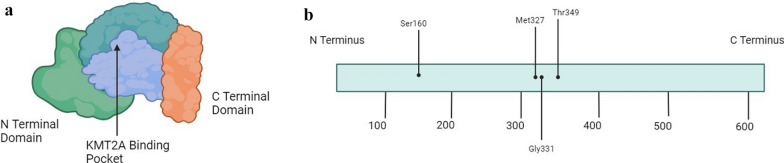
Fig. 2.
The Menin Protein Amino Acid Map
The binding of menin to KMT2A allows the KMT2A/fusion protein to promote downstream transcription of MEIS1, HOXA, and other leukemogenic proteins. Menin inhibitors block the interaction between menin and KMT2A and thereby inhibit this downstream transcription.
Physical depiction of the menin protein structure including the N terminal domain, C terminal domain, and middle region. The crystal structure of the middle region is often likened to a palm, and the KMT2A binding pocket is in this region.
Amino acid map of the menin protein which is comprised of 610 amino acids. There are various mutational hotspots that contribute to acquired resistance to menin inhibitor therapy. These hotspots are depicted here and include Ser160, Met327, Gly331, and Thr349. These mutations interfere with the interaction between menin and menin inhibitors but do not affect the interaction between menin and KMT2A.
Preclinical and clinical data for menin inhibitors in development
There are currently six different menin inhibitors being evaluated in clinical trials that are at various stages of development. These menin inhibitors include revumenib (SNDX-5613), ziftomenib (KO-539), DSP-5336, Bleximenib (JNJ-7526617), DS-1594, and BMF-219, which will be reviewed in the following sections.
Revumenib
Revumenib, also known as SNDX-5613, is the first menin inhibitor to be studied in humans. VTP-5046 is a close analogue of revumenib that demonstrated promising results in pre-clinical studies. VTP-50469 was found to be active in vitro in KMT2Ar AML cell lines. VTP-50469 was found to displace menin from KMT2A which impaired KMT2A binding to chromatin at select genes. This led to changes in gene expression with associated cellular differentiation and apoptosis. Similarly, xenograft models from patients with KMT2Ar AML or KMT2Ar acute lymphoblastic leukemia (ALL) treated with VTP-50469 demonstrated significant reduction in disease burden. [28] VTP-50469 was also found to induce loss of MEIS1 gene expression and increased differentiation of NPM1m leukemic cells in vitro. This agent also inhibited growth and stimulated differentiation of leukemic blasts in patient-derived xenograft (PDX) models containing NPM1m cells [26].
These promising pre-clinical studies led to the development of a phase I/II trial of revumenib in patients with relapsed/refractory (R/R) KMT2Ar or NPM1m acute leukemias including AML, ALL, and mixed phenotype acute leukemia (MPAL) (AUGMENT-101). This study included adult and pediatric patients aged 30 days or older. The phase 1, dose-escalation portion of the study enrolled patients not taking (arm A) or taking (arm B) strong CYP3A4 inhibitors. Revumenib was administered orally every 12 h (q12h) on days 1–28 per 28-day cycle. For arm A, four different dose levels were studied including 113 mg q12h, 226 mg q12h, 276 mg q12h, and 339 mg q12h. For arm B, three dose levels were studied including 113 mg q12h, 163 mg q12h, and 226 mg q12h. Overall, revumenib was well tolerated. Dose limiting toxicities in both arms of the phase 1 dose escalation portion included asymptomatic grade 3 QTc prolongation without any ventricular arrhythmias observed. Treatment with revumenib was found to downregulate key target genes involved in leukemogenesis and to upregulate the expression of genes involved in cellular differentiation. The revumenib doses of 226 mg q12h and 276 mg q12h in arm A and 113 mg q12 h and 163 mg q12h in arm B met the specified criteria for Recommended Phase 2 Dosing (RP2D) based on safety, tolerability and pharmacokinetic (PK) parameters. PK analysis found that drug exposure was dose-proportional in both arm A and arm B. The half-life of revumenib in arm A was approximately three hours on the 276 mg q12h dose level at the cycle 1, day 8 assessment. The half-life was approximately eight hours on the 163 mg q12h dose level in Arm B at the same assessment. The overall response rate (ORR) in the phase 1 portion was 53% and included patients with complete remission (CR), complete remission with partial hematologic recovery (CRh), complete remission with incomplete platelet recovery (CRp), and complete remission with incomplete count recovery (CRi). The median overall survival for the phase 1 population was 7 months [46].
Recently, results were reported from the phase 2 portion of the study involving patients with KMT2Ar ALL, KMT2Ar MPAL, and KMT2Ar AML. Analysis is ongoing for the cohort of patients with NPM1m disease and has not yet been reported. In the phase 2 portion of the trial, patients with KMT2Ar acute leukemias were treated with revumenib 163 mg (or 95 mg/m2 for body weight < 40 kg) every 12 h with a concurrent strong CYP3A4 inhibitor. This was a recommended phase 2 dose determined for patients in Arm B of the phase 1 portion of the study. Patients continued treatment on the phase 2 study until they experienced unacceptable toxicity or failed to achieve a morphologic leukemia free state (MLFS) or better response by the end of cycle 4. A total of 94 patients with KMT2Ar acute leukemias received at least one dose of study drug and were included in the safety analysis. The patient population was overall heavily pre-treated with 43.6% of patients receiving 3 or more lines of therapy prior to enrollment, and 50% having undergone prior allogeneic hematopoietic stem cell transplantation (alloHSCT). The most frequent adverse events related to treatment were nausea, differentiation syndrome (DS) seen in 26.6% of patients and QTc prolongation seen in 23.4% of patients. The most common grade 3 or higher treatment related adverse events were DS (16%), QTc prolongation (13.8%), and febrile neutropenia (13.8%). Despite the relatively high rates of DS and QTc prolongation, none of the patients discontinued study therapy because of these two adverse events. DS is an important adverse event associated with menin inhibitors that needs to be monitored. It can present in different ways with signs and symptoms including fever, hypotension, edema, weight gain, pleural effusions, pericardial effusions, dyspnea, radiographic changes on chest imaging, acute kidney injury, rash, and a rapid increase in white blood cell count. Treatment of DS includes immediate initiation of steroids and holding the menin inhibitor in cases of severe or progressive DS.
A total of 57 patients with KMT2Ar acute leukemias were included in the interim phase 2 efficacy analysis. At a median follow up of 6.1 months, 22.8% of patients had achieved a CR + CRh which outperformed the pre-specified efficacy boundary for KMT2Ar disease. The median duration of CR + CRh was 6.4 months. Composite complete response (CRc) for the phase 2 efficacy analysis was defined as CR + CRh + CRp + CRi and was 43.9%. ORR included CRc in addition to MLFS and partial remission and was reported to be 63.2%. Notably, MRD-negativity was achieved in 68.2% of patients with CRc who had MRD reported. A total of 38.9% of patients went on to transplant. Patients were allowed to resume revumenib after transplant, and it was resumed in 50% of patients. Based on this analysis, the KMT2Ar portion of the study was terminated early for observed efficacy. [47] Notably, in the phase I/II AUGMENT-101 study of revumenib in R/R AML, 9 patients with AML, including one with detectable MRD, were able to resume revumenib following ASCT per protocol amendment. While 4 patients had progressive disease, the remaining 5 patients were able to continue therapy and maintained composite complete remission( CRc), including the patient with detectable MRD who later regained undetectable MRD status. Duration of therapy ranged between 23 and 588 days. Authors reported a tolerable safety profile with dose adjustments for cytopenia. These results sparked interest in the role of menin inhibitors in deepening responses in heavily pretreated patients. More clinical trials are underway and are being designed to study MRD eradication and maintenance strategies [48].
Ziftomenib
Ziftomenib, also known as KO-539, is another small molecular inhibitor of the menin-KMT2A interaction. Preclinical studies found that ziftomenib induced differentiation and inhibited proliferation of leukemic cell lines containing a KMT2A rearrangement or NPM1 mutation. [49] Ziftomenib was also found to lead to clearance of leukemia cells in mouse xenograft models derived from KMT2Ar and NPM1m cells with 40% of surviving mice remaining free of detectable leukemia after four weeks [50].
These encouraging pre-clinical results led to the creation of a phase I/II study of ziftomenib in patients with R/R AML with a dose escalation phase followed by a dose expansion phase which specifically included patients with KMT2Ar or NPM1m leukemias (KOMET-001). The phase 1a portion of this trial studied varying doses of ziftomenib ranging from 50 to 1000 mg daily in 30 patients with R/R AML regardless of cytogenetic or molecular profile. Clinical benefit with decreasing blast counts or decreasing hydroxyurea requirement were seen at all dose levels. The phase 1b portion of this study was a dose-validation study exploring two of the phase 1a doses (200 mg and 600 mg) in 24 patients with R/R KMT2Ar or NPM1m AML to determine the optimal biologically active dose. The patient population in the phase 1b study was heavily pre-treated with a median of 2.5 prior lines of treatment in the patients receiving the 200 mg dose and a median of 4 prior lines of treatment in patients receiving the 600 mg dose. Treatment emergent adverse events of grade 3 or higher included neutropenic fever, neutropenia, anemia, and thrombocytopenia at 25% each, DS and leukocytosis at 17% each, sepsis and leukopenia at 13% each. The incidence and severity of DS initially prompted the FDA to place a hold on enrollment, but this was removed after the implementation of mitigation strategies. The severity of DS in this trial subsequently decreased. No QTc prolongation or other significant cardiac toxicity has been reported. The 200 mg dose notably yielded stable or decreasing blast counts but no overall responses. The 600 mg dose yielded a CR/CRh rate of 25%, a composite CR(CRc) of 33%, and an ORR of 41.7%. Among patients achieving CRc, 75% were MRD-negative. [51] Overall, these results are quite encouraging. PK studies of ziftomenib have determined that peak drug concentrations are seen between two and three hours after daily oral dosing and that the half-life is greater than 24 h. [52] Additional pharmacology studies involving ziftomenib have demonstrated a dose proportional exposure increase up to the RP2D of 600 mg with saturation occurring at this dose and higher doses. Ziftomenib is also not associated with clinically significant interactions with strong CYP3A4 inhibitors [53].
The phase 2 portion of this trial is ongoing, and patients with NPM1m R/R AML are being treated with ziftomenib 600 mg daily (NCT04067336). Ziftomenib is also being studied in newly diagnosed as well as relapsed AML patients in combination with conventional cytotoxic induction regimens as well with azacitidine, venetoclax and gilteritinib. These trials are elaborated in the subsequent sections.
DSP-5336
DSP-5336 is a small molecule inhibitor of the menin-KMT2a interaction. In pre-clinical studies, this agent has demonstrated inhibition of growth in human KMT2Ar and NPM1m cell lines. [25, 54] This agent has also demonstrated decreased expression of HOXA9 and MEIS1 genes along with upregulated expression of the differentiation marker CD11b in KMT2Ar AML cell lines. Treatment with DSP-5336 also yielded decreased leukemic cell burden in PDX models of KMT2Ar and NPM1m AML [55].
A first in human phase I/II study of DSP-5336 is currently ongoing in patients with R/R AML, ALL, or acute leukemia of ambiguous lineage (ALAL) with a focus on patients with KMT2Ar and NPM1m disease. Preliminary data was presented at the 2023 meeting of the American Society of Hematology (ASH) on the first 24 patients enrolled. This agent was well tolerated with no dose-limiting toxicities. One possible grade 4 differentiation syndrome was observed, but concurrent pneumonia and sepsis in this patient made attribution difficult. No other DS events have been reported, and no cardiac toxicities including QTc prolongation have been reported. Pharmacodynamic (PD) studies revealed rapid downregulation of menin-KMT2A target genes including HOXA9 and MEIS1 and an increase in the differentiation marker CD11b. The mean apparent half-life of this agent ranged from 2–5 h in patients not taking a concurrent strong CYP3A4 inhibitor and 3–7 h in patients taking a concurrent strong CYP3A4 inhibitor. The maximum serum concentration was observed within two hours of dosing this agent. A subset of patients achieved complete remission with incomplete count recovery (CRi), morphologic leukemia free state (MLFS), or stable disease. The study is ongoing to determine a recommended phase 2 dose along with additional efficacy and safety data. [56] Updated results from the study were presented at the recent EHA 2024 meeting. Of the 58 patients accrued and treated, doses ranged between 40 mg BID and 300 mg BID, with more responses noted at 140 mg BID doses. In the 140 mg BID cohort, ORR (CR + CRi + MLFS) was 45% (10/22) and the CR + CRh rate was 23% (5/22), while across all the dose levels, the ORR and CR + CRh were 32% (12/38) and 16% (6/38), respectively. No DLT were observed, and DS was reported in 3 patients, and grade 3 QTc in 1 patient [57].
Bleximenib
Bleximenib (JNJ-75276617) is a potent, highly selective inhibitor of menin-KMT2A binding. In KMT2Ar and NPM1m AML cells, this agent has been demonstrated to inhibit the binding of menin to KMT2A, thereby downregulating the expression of menin-KMT2A target genes including MEIS1. Treatment of these cell lines with this agent was also shown to increase expression of differentiation markers and to induce apoptosis. Exposure of pre-clinical animal AML models to Bleximenib also resulted in decreased leukemic cell burden and a dose-dependent survival benefit [58].
This promising pre-clinical data led to the development of a phase 1, open-label, dose-finding study in patients with R/R KMT2Ar or NPM1m AML or ALL. Preliminary results from 58 patients were presented at the 2023 ASH annual meeting. Treatment emergent adverse events (TRAEs) were reported in 52% of patients with 29% experiencing a grade ≥ 3 adverse event. DS was notably seen in 14% of patients, with 5% of patients experiencing grade ≥ 3 DS. PD studies demonstrated down-regulation of menin-KMT2A target genes along with increased expression of markers of differentiation. Of the 41 patients with disease evaluation data, there were 12 responders across all dosing cohorts. The recommended phase 2 dose remains undetermined, and the dose escalation phase is ongoing. [59] There is another active phase 1b study assessing the efficacy and safety of Bleximenib in combination with venetoclax and azacitidine in patients with R/R KMT2Ar or NPM1m AML, and preliminary results have been reported recently. The safety data set for these preliminary results included 45 patients. Grade ≥ 3 TRAEs occurred in 60% of patients with the most common being thrombocytopenia, leukopenia, and neutropenia. Notably, no DLTs were reported and cases of DS or QTc prolongation were seen. The efficacy dataset for these preliminary results included 21 patients who received doses of ≥ 50 mg bid of the drug. Composite CR rate (CR/CRh/CRi) was 48% with CR/CRh of 24% [60].
DS-1594
DS-1594 is another small molecule inhibitor of the menin-KMT2A interaction with promising pre-clinical results. Human AML and ALL cell lines with KMT2Ar or NPM1m are highly sensitive to this agent. This agent was also found to be superior to cytarabine at eradicating leukemia-initiating KMT2Ar cells in vitro. DS-1594 also demonstrated significant antileukemic activity in patient-derived xenograft models of KMT2Ar or NPM1m acute leukemias in vivo [61].
This pre-clinical data led to the creation of a phase I/II trial of DS-1594 with or without mini-HCVD, azacitidine, or venetoclax in patients with R/R AML or ALL. This study is complete, but results are not yet reported (NCT04752163).
BMF-219
BMF-219 is a highly selective, covalent, irreversible inhibitor of the menin-KMT2A interaction which has demonstrated encouraging pre-clinical anti-leukemic activity in vitro and in vivo. Pre-clinical studies have also demonstrated activity of this agent in multiple myeloma (MM), chronic lymphocytic leukemia (CLL), and diffuse large B cell lymphomas (DLBCL). This is likely in part because the activity of the MYC oncoprotein is dependent on its interaction with menin, and BMF-219 has also been designed to inhibit MYC. [62] There is an ongoing phase 1 dose escalation and expansion study of this agent in patients with KMT2Ar and NPM1m AML or ALL. This study also includes patients with DLBCL, MM, and CLL (NCT05153330). Preliminary results for the cohort of patients with acute leukemias have been reported. Among 26 patients, this agent was well tolerated overall with no DLTs, and no treatment discontinuations due to adverse events. Common TRAEs occurring in 10% or more of patients included DS in 13% and vomiting in 13%. The efficacy evaluable patient population for this analysis included patients receiving doses at or near those predicted to demonstrate efficacy and who had completed at least one response assessment or seven doses of therapy. This included five patients, of whom 1 experienced a CR and 1 experienced a CRi. [63] This agent has also interestingly demonstrated the ability to regenerate insulin-producing beta cells and is currently being studied in the treatment of type 1 diabetes mellitus.
A summary of characteristics and preliminary results associated with menin inhibitors in the treatment of R/R Acute Leukemias is presented in Table 1.
Table 1.
Key Features of Menin Inhibitors with Reported Clinical Trial Results in R/R Acute Leukemias
| Significant CYP3A4 Inhibition | Differentiation Syndrome | QTc Prolongation | ORR (CR + CRh + CRp + CRi) | |
|---|---|---|---|---|
| Revumenib [46, 47](SNDX-5613) | Yes | 26.6% in phase 2 | 23.4% in phase 2 | 53% in phase 1 (KMT2Ar and NPM1m) 43.9% in phase 2 (KMT2Ar) |
| Ziftomenib [51](KO-539) | No | 29% in phase 1b | 0% phase 1a or 1b | 33% at 600 mg dose of phase 1b (KMT2Ar and NPM1m) |
| DSP-5336[56] | Yes | Possibly 1/24 (4%) in phase 1, but attribution difficult | 0% in phase 1 | 33% among 6 patients treated on phase 1 at doses projected to be effective (KMT2Ar and NPM1m) |
| Bleximenib (JNJ-75276617) [59] | No | 14% in phase 1 | 1% reported in phase 1 | 35% in patients treated at higher dose levels in phase 1 (n = 20) (KMT2Ar and NPM1m) |
| BMF-219[63] | Yes | 13% in phase 1 | 0% reported in phase 1 | 2 out of 5 efficacy evaluable patients in phase 1 (40%) (KMT2Ar and NPM1m) |
CYP3A4, cytochrome P450 3A4; QTc, QT interval corrected for heart rate; ORR, overall response rate; CR, complete remission; CRh, complete remission with partial hematologic recovery; CRp, complete remission with incomplete platelet recovery, Cri, complete Remission with incomplete count recovery; KMT2A, histone-lysine n-methyltransferase 2A; NPM1 Nucleophosmin 1
Clinical development of menin inhibitors in combinations
Preclinical studies indicate plausible synergy between menin blocking agents and various other approved and investigational cytotoxic and small molecule inhibitors, resulting in enhancement of antileukemic activity. Unlike the other targeted agents such as FLT3 inhibitors, IDH-1 and 2 inhibitors, which are specific to the presence of the mutated protein for their activity, menin inhibitor activity converges on modifying HOXA/MES1 gene expression which appears to be the common pathway of leukemogenesis and maintenance. The function of menin as a hub for chromatin regulators and gene transcription factors provides a potential for broader application and an essential target for menin inhibition, and combination therapies in the treatment of AML.
Menin inhibitors with chemotherapeutic agents
Conventional intensive induction therapy for fit, eligible patients includes a combination of cytarabine infusion and an anthracycline (7 + 3 or DA) which has been the standard of care (SOC) for several decades. Though historically CR (complete remission) rates have ranged around 80%, long term survival and cure without transplant have remained poor. [64] Persistent efforts to optimize SOC with the introduction of newer agents such as Gemtuzumab ozagomycin, or the FLT3 inhibitors (midostaurin and quizartinib) in combination with the standard induction and consolidation regimens led to improved PFS and OS for specific molecular sub-groups, such as core binding factor AML, or FLT3-ITD mutated AML, respectively [65–67].
As previously discussed, though NPM1m AML is considered as favorable risk by conventional classifications, and high CR rates and potential cures are achievable with intensive induction therapies, relapses are still quite common, approaching around 50%, particularly in older patients. [40, 68, 69] Similarly, long-term overall survival outcomes remain poor with KMT2Ar AML even with intensive chemotherapy, due to early relapses, and there remains a huge unmet need to improve survival in these cohorts. With the encouraging results of clinical efficacy of menin inhibitors demonstrated in the phase I trials of relapsed/refractory AML with NPM1m or KMT2Ar, it is imperative that these agents are tested in combination with the intensive and non-intensive induction regimens for newly diagnosed AML to improve long term survival and cures. Phase I dose escalation and expansion study KOMET-007 is actively investigating the safety and efficacy of ziftomenib in combination with 7 + 3 in newly diagnosed AML harboring NPM1m or KMT2Ar. Concurrently, another phase I dose escalation and expansion study KOMET-008 is investigating the combination of ziftomenib and alternative intensive and non-tensive chemotherapy regimens Fludarabine, cytarabine and Idarubicin(FLAG-Ida), or low dose cytarabine(LDAC), respectively, for R/R AML.
It is inevitable that all the other menin inhibitor compounds will be tested in combinations with intensive and non-tensive SOC regimens in frontline as well as for R/R AML, and such trials are underway.
Combining menin-KMT2A Inhibition with BCL2 Inhibition:
Combination of menin inhibitors with Bcl-2 inhibitor in newly diagnosed or relapsed refractory AML is also an active area of investigation.
BCL-2 family proteins are anti-apoptotic proteins that inhibit cancer cell death by binding to the BH3 domain of pro-apoptotic proteins, thereby sequestering them. Venetoclax is a BH3 mimetic which blocks this interaction thereby allowing the pro-apoptotic proteins to stimulate apoptosis through cytochrome C activation in the mitochondria. [70] Venetoclax is currently approved in combination with azacitidine or low dose cytarabine for the treatment of newly diagnosed AML in patients who are ineligible for intensive chemotherapy. [71, 72] Venetoclax is also under active investigation in combination with multiple other agents in the treatment of AML. In pre-clinical studies, revumenib was found to decrease the level of BCL-2 proteins in AML cell lines, and combined exposure to revumenib and venetoclax yielded synergistic lethality in these cells. [44] Similarly, apoptotic priming using ziftomenib also induced more potent activity of venetoclax in preclinical studies. [73] Another important observation from the study is that menin inhibitor ziftomenib could potentially re-sensitize leukemic cells to BCL2 inhibition. [73] This latter observation is particularly exciting given the emerging challenge of venetoclax-refractory AML. We await validation of these study findings. Ziftomenib is being studied in combination with venetoclax + azacitidine, venetoclax, or 7 + 3 in patients with newly diagnosed or R/R AML in KOMET-007 study (NCT05735184).
Revumenib is also being investigated in combination with venetoclax and azacitidine as frontline therapy in patients aged 60 and older with newly diagnosed NPM1m or KMT2Ar AML as part of the BEAT-AML master trial. [74] Another study involving this triplet combination is being created for pediatric and young adult patients with R/R AML or acute leukemia of ambiguous lineage (ALAL) with NPM1m, KMT2Ar, or other defined cytogenetic and molecular abnormalities (NCT06177067).
Combining menin-KMT2A Inhibition with FLT3 Inhibition
Mutations in the FMS-like tyrosine kinase 3 gene (FLT3) are present in approximately 30% of newly diagnosed AML cases. Most of these mutations are internal tandem duplication mutations affecting the juxtamembrane domain of the protein (ITD mutations) or point mutations in the tyrosine kinase domain (TKD mutations). FLT3 ITD mutations are known to be an adverse prognostic factor while the prognostic significance of TKD mutations remains less certain. [75, 76] The FLT3 inhibitors midostaurin and quizartinib are approved in combination with intensive chemotherapy as frontline treatment for patients with FLT3-mutated AML. [66, 77] The FLT3 inhibitor gilteritinib is approved as monotherapy for patients with R/R FLT3-mutated AML. FLT3 and NPM1 mutations frequently co-occur in AML with one study finding concurrent NPM1 mutations in 61% of FLT3-mutated AML cases. [78] FLT3 mutations can also be seen in KMT2Ar AML with one study of KMT2Ar AML finding concurrent FLT3 TKD mutations in 8% of cases and FLT3 ITD mutations in 4% of cases. [79] The FLT3 gene is notably a putative transcription target of the MEIS1 protein. [25, 80] In the phase 1 AUGMENT-101 trial, menin inhibition with revumenib was found to downregulate FLT3 expression from screening through the end of cycle 1 of treatment. [81] Pre-clinical studies have also demonstrated synergy of combined menin and FLT3 inhibition in AML models harboring NPM1 mutations and KMT2A rearrangements. The combination of menin inhibition and FLT3 inhibition demonstrated a greater decrement in cell viability than monotherapy with menin or FLT3 inhibition alone in human AML cell lines harboring co-mutations in NPM1 and FLT3. Similarly, combination therapy with a menin inhibitor and FLT3 inhibitor was found to yield superior survival compared to monotherapy with either agent alone in mouse xenograft models of KMT2Ar AML harboring a concurrent FLT3 mutation. [80] Combined menin and FLT3 inhibition may therefore have the potential to improve survival outcomes in patients with FLT3-mutated AML harboring a concurrent NPM1 mutation or KMT2A rearrangement. Early phase combination studies are currently being developed in this patient population. A phase 1 trial of revumenib combined with gilteritinib in patients with R/R FLT3-mtuated AML harboring a concurrent NPM1 mutation or KMT2A rearrangement has recently opened to accrual (NCT06222580). Similarly, the KOMET-008, phase 1 trial also has an arm testing the combination of ziftomenib and giltertinib for FLT3-ITDm R/RAML.
Composite results and experience of all the above studies are expected to lead to yet another change in the landscape of treatment of NPM1m and KMT2Ar AML towards improved outcomes.
Potential combinations with menin inhibitors for future development
DOT1L and menin inhibitors
DOT1L (disruptor of telomeric silencing 1-like) is an enzyme that catalyzes methylation of histone H3 lysine 79 (H3K79) and performs various key functions such as telomeric silencing, cell-cycle regulation, transcription, differentiation and is necessary for normal functional hematopoiesis. [82–84] The DOT1L- H3K79 methyltransferase complex interacts with the fusion partners of KMT2A to promote and maintain leukemogenesis and increased activity levels of H3K79 were observed in correlation to gene expression because of KMT2A fusion. [82, 85, 86] This observation led to preclinical and clinical investigation of DOT1L as a therapeutic target. [87] The first-in-class compound Pinometostat (formerly EPX5676) only showed modest activity in phase 1 study with 2 out of 51 patients responding, showing proof of concept, although the maximum tolerated doses could not be reached, and further dose expansion studies were not pursued. [88] Subsequent efforts and optimization led to drug discovery of second generation, novel compounds that showed more potent activity in early preclinical and PDX model studies. [89] In these studies, the novel compounds were effective in reducing leukemic cell burden and recovery of normal hematopoiesis and exhibited good safety profile. On target efficacy was demonstrated by significant reduction of MEIS1 and HOXA gene expression, as well as the H3K79 activity levels. These compounds are now in clinical trials. Additionally, due to their activity being independent of menin displacement from the chromatin, potential combinations of DOT1L and menin inhibitors and other demethylating agents are critically necessary [89].
XPO1 inhibition with menin inhibitors
Mutant NPM1 is characterized by its aberrant translocation to the cytoplasm (NPM1c) upon acquisition of nuclear export signals (NES) at the c-terminus. [20] The nuclear exporter, or exportin-1 XPO1(also known as CRM1) assists in the nucleocytoplasmic shuttling of various molecules and plays a key role in the pathobiology of NPM1m AML whereby its interaction with XPO1 results in dislocation to cytoplasm, [38] and consequently persistent overexpression of HOXA cluster genes. [38] While menin inhibitors are actively being investigated in NPM1m leukemias, there has also been interest in interrupting the XPO1- NPM1m interaction utilizing XPO1 inhibitors. Selinexor is the first clinically tested XPO1 inhibitor, currently FDA approved for treating relapsed multiple myeloma, and diffuse large B cell lymphoma. However, results from the preliminary trials of selinexor monotherapy in AML were disappointing due to suboptimal efficacy and due to its intolerable adverse effects, such as anorexia. Based on these observations, a preclinical study of ziftomenib and selinexor was completed, which reported significant synergy and improved survival of combination compared to the individual agents alone, in the PDX models. [90] Other important observations from a recent preclinical study revealed that persistent XPO1 inhibition was essential for activity, and eltanexor, a second generation XPO1 inhibitor was found to be more tolerable and raised excitement for future clinical development [91].
Other potential combinations and indications
In addition to the above, drug compound screening studies in leukemia cell lines revealed that various other small molecular inhibitors are likely to have a synergistic antileukemic effect with menin inhibitors in NPM1m and KMT2Ar AML. [44, 73] Some of the notable prospective compounds include inhibitors of epigenetic regulation (LSD1 inhibitors), cell-cycle regulation (CDK4/6 inhibitors). Interestingly, all trans retinoic acid (ATRA) was also found to have a synergistic differentiating effect. [73] More validating preclinical and clinical studies are needed to test these combinations.
There is currently a clinical trial in development of revumenib in combination with venetoclax in patients with NPM1m, KMT2Ar, or NUP98r AML in morphologic remission but with MRD ≥ 0.1% by MFC. This trial allows for patients in first remission following intensive chemotherapy or at least 2 cycles of non-intensive therapy. It will also include patients in second remission following any therapy (NCT06284486).
A summary of ongoing or upcoming trials combining menin inhibition with other therapies is presented in Table 2.
Table 2.
Summary of select ongoing or planned trials evaluating menin inhibitors combined with standard of care therapies
| Age | Phase | Agent | Indication and line of therapy | Combination regimen | Results |
|---|---|---|---|---|---|
| Adults > 18 years | I | Ziftomenib (KO-539) KOMET-007 (NCT05735184) | Upfront and R/R AML with KMT2Ar and NPM1m | 7 + 3; (Daunorubicin cytarabine); Venetoclax. Azacitidine, and Venetoclax | In accrual |
| Adults > 18 years | I | Ziftomenib (KO-539) KOMET-008 (NCT06001788) | R/R AML with NPM1m and KMT2Ar, FLT3m | FLAG-IDA; LDAC; Gilteritinib | In accrual |
| Adults > 18 years | Ib | Revumenib (SNDX-5613) (NCT05886049) | Upfront AML with NPM1m and KMT2Ar | 7 + 3 (Daunorubicin cytarabine) | In accrual |
| Adults > 18 years | I/II | Revumenib (SNDX-5613) SAVE trial (NCT05360160) | Upfront AML with NPM1m and KMT2Ar | ASTX (oral Decitabine-Cedazuridine) | In accrual |
| Pediatric and young adults 1–30 years | I | Revumenib (SNDX-5613) (NCT06177067) | Upfront AML, ALAL KMT2Ar NUP98r, NPM1m or fusion, PICALM::MLLT10, DEK::NUP214, UBTF-TD, KAT6A::CREBBP, or SET::NUP214 | Azacitidine and Venetoclax | In accrual |
| Children > 12 years, and adults | I/II | Revumenib (SNDX-5613) (NCT06284486) | CR1 or CR2 with detectable MRD NPM1m, or KMT2Ar, or NUP98r AML | Venetoclax | Not yet open |
| Adults > 18 years | Ib | Revumenib (SNDX-5613) BEAT-AML[74] (NCT03013998) | Upfront AML with NPM1m and KMT2Ar | Composite CR(cCR) of 100% (95% CI: 75.3–100) | |
| Adults > 18 years | I | Revumenib (SNDX-5613) (NCT06222580) | R/R AML with NPM1m or KMT2Ar, and FLT3m | Gilteritinib | In accrual |
| Adults > 18 years | I | Bleximenib (JNJ-75276617(60) (NCT05453903) | R/R AML with NPM1m or KMT2Ar | Azacitidine and Venetoclax | Composite CR(CR/CRh/CRi) of 48% and CR/CRh of 24% |
R/R, relapsed/refractory; AML, acute myeloid leukemia; ALL, Acute Lymphoblastic Leukemia; KMT2A, histone-lysine N-methyltransferase 2A; NPM1, nucleophosmin 1; ALAL, acute leukemia of ambiguous lineage; MRD, measurable residual disease; HMA, hypomethylating agent; FLAG-Ida, fludarabine, cytarabine, granulocyte colony stimulating factor-idarubicin; LDAC, low dose cytarabine; ORR, overall response rate; CR, complete remission; Ccr, composite complete remission; CRh, complete remission with partial hematologic recovery; Cri, complete remission with incomplete count recovery; FLT3m, FLT3-ITD and TKD gene mutations
Opportunities for menin inhibitors in future clinical development
In addition to the activity of menin inhibition in the NPM1m and KMT2Ar cohorts as depicted above, multiple preclinical studies and a few clinical observations point towards a much wider application of menin inhibitors in rare molecular alterations where dysregulation of HOXA genes and transcription is the essential component of leukemic development and progression.
NUP98 rearranged AML
Recurrent translocations resulting in the fusion of nucleoporin NUP98, present on chromosome 11p15, with histone proteins such as histone methyltransferase nuclear receptor-binding SET domain protein 1 (NSD1), are found to be sufficient to induce AML in vivo, through over expression of HOX genes. [92] Several other fusion partners including HOXA9 ( NUP98-HOXA9), HOXA 13( NUP98-HOXA13) have also been identified. [93] These leukemias are overrepresented in pediatric populations, frequently co-occur with FLT3-ITD or WT1(Wilms tumor 1) mutations and are associated with poor survival. [93] Preclinical studies demonstrate that these leukemias are dependent on KMT2A and could be interrupted by inhibition and inactivation of KMT2A through menin inhibitors. [93–96] Similarly, other rare translocations involving NUP98 have been identified which are associated with poor prognosis and are likely susceptible to menin inhibition [97–103].
Clinical trials are now actively incorporating leukemias with NUP98r, along with the NPM1m and KMT2Ar ( NCT05326516; NCT06376162, testing revumenib and ziftomenib, respectively).
DEK-NUP214 AML
DEK is a nucleophosphoprotein which binds to the DNA and chromatin. It has been identified as an oncogene, particularly with its fusion partner NUP214, in t(6;9) AML. [104, 105] The fusion protein modulates aberrant HOX gene expression and is implicated in leukemogenesis. Similarly, SET-NUP214 fusion product also influences HOX cluster region and results in aberrant transcription. Menin inhibitors have potential therapeutic role in these rare leukemias with recurrent translocations through its negative modulation of the HOX pathway as aforementioned [106].
UBTF-TD AML
UBTF-TD (tandem duplication of upstream binding transcription factor) located on exon 13, has been found to be the third most common recurrent molecular aberration in relapsed and/or refractory pediatric AML, accounting for about 9% of the relapsed cohorts in one study, which is associated with very poor prognosis. [107]UBTF-TD shares similar transcriptional changes with those of NPM1m and NUP98-R AML sub-types. Importantly, HOX cluster genes are overexpressed across all these subtypes [108, 109]. Due to frequent associations with FLT3-ITD or WT1 mutations, responses to conventional chemotherapy are sub-optimal. In a study by Barajas et al., UBTF-TD was found to colocalize on HOXA/B cluster along with the KMT2A/menin complex and contributed to the transcriptional dysregulation and leukemogenesis. In this model, primary leukemia cells were shown to be susceptible to menin inhibition (SNDX-5613). [110] This provides the proof of concept for further clinical study of menin inhibitors in these rare, yet molecularly defined subgroups of refractory adult and pediatric leukemias.
Challenges and resistance mechanisms to menin inhibitors
Limitations to the long-term durability and efficacy of most targeted therapies is the inevitable emergence of resistance mechanisms and pathways making the drugs or molecules ineffective, and disease relapses clinically.
Somatic mutations arising in the MEN1 gene, contributing to resistance to the menin inhibitors have been identified and reported.
Important insights were obtained in a study which demonstrated for the first time in a comprehensive manner the nature of acquired, somatic MEN1 mutations and their functional consequences. Using CRISPR-Cas9 base editing, authors identified hotspot mutations at the T349, G331, and s160 residues as the most common candidate drivers of development of resistance [16].
Targeted next generation sequencing analysis of pre- and post-treatment samples indicated development of MEN1 mutations as the leading cause. Assessment of archival bone marrow samples from the FIH phase I revumenib AUGMENT-101 trial revealed that resistance causing mutations were noted at an average of 2 cycles of treatment exposure [16] Mutations were found in 12 of 31 samples analyzed (38.7%), following revumenib exposure but none in the pre-exposure group. [16] In preclinical studies invitro and on PDX models, disruption of the binding affinity of revumenib is the most likely mechanism observed with the mutations at the residues. Of note, this change did not disrupt the interaction with the normal ligand of the MLL1 protein and only affected the interactions with revumenib. This led to abrogation of displacement of menin from the chromatin that was induced by the inhibitors. They observed that the target genes for menin inhibition were not adequately repressed in the mutant cases, when compared to the MEN1-WT cells. Also, these hotspot mutations were thought to be a class effect as they demonstrated similar effect of decreased binding affinity to other compounds, KO-539, and DS-25, etc. Continuous exposure to revumenib may lead to an increased selection of the fitter clones carrying the mutations, further driving resistance and leukemic progression.
Interestingly, an analysis of a relapsed AML patient sample without detectable MEN1 mutation revealed distinct transcriptional programming, with attenuated expression of the KMT2A target genes MES1 and HOXA at baseline, indicating existence of alternate mechanisms of resistance to menin inhibition. These mutations had similar effect on both the KMT2Ar and NPM1m leukemias [16].
Second generation inhibitors that could overcome the effects of mutations and binding mechanisms, need to be developed to circumvent this emergent challenge. Combination strategies using other small molecule inhibitors, and conventional cytotoxic therapies in upfront treatment, as shown in Table 2. may likely delay or overcome some resistance pathways. [17] Clinical trials are also underway using menin inhibitors in R/R AML patients who were previously exposed to other menin inhibitor compounds and relapsed. These trial observations should answer the important question of efficacy of re-exposure to menin blockade.
Combination with IKAROS degraders
The concern about emergence of resistance mechanisms to menin inhibitor needing immediate attention is met with concerted efforts to unravel genetic vulnerabilities that could mitigate or overcome resistance pathways. Armstrong, Bourgeoius and colleagues, [111, 112] in their extensive preclinical work, demonstrated that IKAROS occupies a key component of transcriptional complex that mediates KMT2A/menin fusion driven overexpression of HOX family along with MES1 genes, in an independent manner separate from the menin cofactor activity. Mezigdomide, the new generation cerebrelon E3 ubiquitin ligase modulator (CELmod) is an IKAROS degrader and retains more potent (nearly 1000-fold) single agent activity than the currently approved agents Lenalidomide, or Iberdomide, in the KMT2Ar or NPM1m AML PDX models. Mezigdomide synergized with menin inhibitors resulting in prolonged survival in the preclinical studies. Interestingly, the combination therapy showed apoptosis as the predominant means of anti-leukemic activity, rather than differentiation which was the prominent mechanism of action of menin inhibitors.
IKAROS degradation with Mezigdomide when combined with revumenib delayed MEN1 resistance mutation emergence and resensitized menin inhibitors to pre-existent MEN1 hotspot mutations. Aside from transient leukopenia, dominated by lymphopenia, no other safety concerns were observed. Mezigdomide is already being tested in a phase III clinical trial for multiple myeloma and the findings of the current study lay compelling groundwork for combination clinical trials of menin inhibitors and Mezigdomide in KMT2Ar and NPM1m AML patients both in upfront and relapsed setting, particularly following prior menin inhibitor exposure [112].
Conclusions
With the expansion of targeted treatment options for AML, we are entering a new era to provide better care of AML patients with improved efficacy, fewer toxicities, and more convenience for patients with oral compounds, essentially leading to a better quality of life. The clinical development of menin inhibitors is entering the mature stage, and these agents will change the treatment landscape for the patients with KMT2A re-arrangements, NPM1 mutations, and patients with other rare genetic changes as discussed in the preceding sections. Further, combination of menin inhibitors with chemotherapy and other small molecules or targeted therapies might provide new hopes for some selected AML patients. While emergence of menin resistance is inevitable, it is also encouraging to note the development of novel pathways and compounds that can overcome and/or prevent the resistance development. The ongoing clinical development of novel targeted therapies might still have long way to go, but the future of the treatment of AML remains promising.
Acknowledgements
The authors thank the editor for useful discussion and critically reviewing the first draft of the article.
Abbreviations
- Allo-SCT
Allogeneic hematopoietic stem cell transplantation
- AML
Acute Myeloid Leukemia
- BSC
Best supportive care
- CI
Confidence interval
- CR
Complete remission
- CRc
Composite CR (includes CR, CRi, and CRp)
- CRi
CR with incomplete hematologic recovery
- CRMRD
CR with absence of minimal residual disease
- CRp
CR with incomplete platelet counts
- DLT
Dose limiting toxicities
- DFS
Disease-free survival
- EFS
Event-free survival
- FDA
US Food and Drug Administration
- HR
Hazard ratio
- KM
Kaplan–Meier
- MDS
Myelodysplastic Syndrome
- MLFS
Morphological leukemia-free state
- MRD
Minimal (measurable) Residual Disease
- OS
Overall survival
- RFS
Relapse-free survival
- SCT
Stem cell transplantation
- TTE
Time-to-event
- ATRA
All-Trans Retinoic Acid
- CLL
Chronic lymphocytic leukemia
- mCR
Bone marrow CR
- HiDAC
High dose cytarabine (AraC)
- HMA
Hypomethylating agent
- LDAC
Low dose cytarabine
- ITD
Internal tandem duplication
- TKD
Tyrosine kinase domain
- ORR
Overall response rate
- RP2D
Recommended phase 2 dose
Author contributions
K.N, KS, and H.L designed and wrote the draft of the review and approved the final manuscript. KN and KS have contributed equally to this manuscript.
Funding
Not applicable.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethical approval
Not applicable.
Competing interests
KN is a site PI for the KOMET-007 study, received research support from Abbvie Inc, served at the advisory board meeting for Morphosys. KS reports no competing interests for this review. HL reports no competing interests for this review; HL served at the Advisory Board meeting for Servier and Rigel and received a consulting fee from AbbVie in the past 24 months
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kalyan VG Nadiminti and Kieran D Sahasrabudhe - These two authors have contributed equally to this work.
References
- 1.Liu H. Emerging agents and regimens for AML. J Hematol Oncol. 2021;14(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276(5311):404–7. [DOI] [PubMed] [Google Scholar]
- 3.Lemmens I, Van de Ven WJ, Kas K, Zhang CX, Giraud S, Wautot V, Buisson N, De Witte K, Salandre J, Lenoir G, Pugeat M. Identification of the multiple endocrine neoplasia type 1 (MEN1) gene. Human Molecul Gene. 1997;6(7):1177–83. [DOI] [PubMed] [Google Scholar]
- 4.Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J Biol Chem. 2011;286(36):31742–8. 10.1074/jbc.M111.258186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, Gurung B, Wan B, Matkar S, Veniaminova NA, Wan K, Merchant JL, Hua X, Lei M. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482(7386):542–6. 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiel AT, Huang J, Lei M, Hua X. Menin as a hub controlling mixed lineage leukemia. BioEssays. 2012;34(9):771–80. 10.1002/bies.201200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama A. Transcriptional activation by MLL fusion proteins in leukemogenesis. Exp Hematol. 2017;1(46):21–30. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matkar S, Thiel A, Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci. 2013;38(8):394–402. 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-gene, functions as a transcriptional maintenance factor in morphogenesis. Proceedings National Academy Sci. 1998;95(18):10632–6. 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27(8):396–402. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–18. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Cuellar M-P, Steger J, Fuller E, Hetzner K, Slany RK. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica. 2015;100(7):905–13. 10.3324/haematol.2015.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining Roles for HOX and MEIS1 Genes in Induction of Acute Myeloid Leukemia. Molecul Cellul Biol. 2001;21(1):224–34. 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275–83. 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perner F, Stein EM, Wenge DV, Singh S, Kim J, Apazidis A, Rahnamoun H, Anand D, Marinaccio C, Hatton C, Wen Y. MEN1 mutations mediate clinical resistance to menin inhibition. Nature. 2023;615(7954):913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenge DV, Armstrong SA. The future of HOXA-expressing leukemias: menin inhibitor response and resistance. Current Opin Hematol. 2024;31(2):64–70. [DOI] [PubMed] [Google Scholar]
- 18.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106(3):899–902. [DOI] [PubMed] [Google Scholar]
- 19.Mullighan CG, Kennedy A, Zhou X, Radtke I, Phillips LA, Shurtleff SA, Downing JR. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21(9):2000–9. [DOI] [PubMed] [Google Scholar]
- 20.Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, Bigerna B, Pasqualucci L, Mannucci R, Rosati R, Gorello P. Both carboxy-terminus NES motif and mutated tryptophan (s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107(11):4514–23. [DOI] [PubMed] [Google Scholar]
- 21.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–66. [DOI] [PubMed] [Google Scholar]
- 22.Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, Martelli MP, Liso A, Mecucci C, Fabbiano F, Martelli MF. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67(13):6230–7. [DOI] [PubMed] [Google Scholar]
- 23.Dovey OM, Cooper JL, Mupo A, Grove CS, Lynn C, Conte N, Andrews RM, Pacharne S, Tzelepis K, Vijayabaskar MS, Green P. Molecular synergy underlies the co-occurrence patterns and phenotype of NPM1-mutant acute myeloid leukemia. Blood J Am Soc Hematol. 2017;130(17):1911–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, Gionfriddo I, Mezzasoma F, Milano F, Nabet B, Buckley DL. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell. 2018;34(3):499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kühn MWM, Song E, Feng Z, Sinha A, Chen CW, Deshpande AJ, et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov. 2016;6(10):1166–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uckelmann HJ, Kim SM, Wong EM, Hatton C, Giovinazzo H, Gadrey JY, Krivtsov V, Rücker FG, Döhner K, McGeehan GM, Levine RL, Bullinger L, Vassiliou GS, Armstrong SA. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science. 2020;367(6477):586–90. 10.1126/science.aax5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34(3):499-512.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krivtsov AV, Evans K, Gadrey JY, Eschle BK, Hatton C, Uckelmann HJ, et al. A menin-MLL inhibitor induces specific chromatin changes and eradicates disease in models of MLL-rearranged leukemia. Cancer Cell. 2019;36(6):660-673.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Górecki M, Kozioł I, Kopystecka A, Budzyńska J, Zawitkowska J, Lejman M. Updates in KMT2A Gene Rearrangement in pediatric acute lymphoblastic leukemia. Biomedicines. 2023;11(3):821. 10.3390/biomedicines11030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.HJ Super NMMTRLMBJPB. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood. 1993;82(12):3705–11. [PubMed] [Google Scholar]
- 31.Winters AC, Bernt KM. MLL-rearranged leukemias—an update on science and clinical approaches. Front Pediatrics. 2017;9(5):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood J Am Soc Hematol. 2017;129(4):424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102(7):2395–402. [DOI] [PubMed] [Google Scholar]
- 34.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood J Am Soc Hematol. 2010;115(3):453–74. [DOI] [PubMed] [Google Scholar]
- 35.Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MD, Barbieri BC. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32(2):273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011;6(9):1059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood J Am Soc Hematol. 2020;136(15):1707–21. [DOI] [PubMed] [Google Scholar]
- 39.Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, Whitman SP, Wu YZ, Schwind S, Paschka P, Powell BL. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene-and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clinical Oncol. 2010;28(4):596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubmann M, Köhnke T, Hoster E, Schneider S, Dufour A, Zellmeier E, et al. Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica. 2014;99(8):1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, Linch DC. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood J Am Soc Hematol. 2008;111(5):2776–84. [DOI] [PubMed] [Google Scholar]
- 42.Uckelmann HJ, Haarer EL, Takeda R, Wong EM, Hatton C, Marinaccio C, et al. Mutant NPM1 directly regulates oncogenic transcription in acute myeloid leukemia. Cancer Discov. 2023;13(3):746–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Issa GC, Ravandi F, DiNardo CD, Jabbour E, Kantarjian HM, Andreeff M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia. 2021;35(9):2482–95. [DOI] [PubMed] [Google Scholar]
- 44.Fiskus W, Boettcher S, Daver N, Mill CP, Sasaki K, Birdwell CE, Davis JA, Takahashi K, Kadia TM, DiNardo CD, Jin Q. Effective Menin inhibitor-based combinations against AML with MLL rearrangement or NPM1 mutation (NPM1c). Blood Cancer J. 2022;12(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abuhantash M, Collins EM, Thompson A. Role of the HOXA cluster in HSC emergence and blood cancer. Biochem Soc Trans. 2021;49(4):1817–27. [DOI] [PubMed] [Google Scholar]
- 46.Issa GC, Aldoss I, DiPersio JF, Cuglievan B, Stone RM, Arellano ML, Thirman MJ, Patel MR, Dickens D, Shenoy S, Shukla N. The Menin inhibitor SNDX-5613 (revumenib) leads to durable responses in patients (Pts) with KMT2A-rearranged or NPM1 mutant AML: updated results of a phase (Ph) 1 study. Blood. 2022;140(1):150–2. [Google Scholar]
- 47.Aldoss I, Issa GC, Thirman M, DiPersio J, Arellano M, Blachly JS, Mannis GN, Perl A, Dickens DS, McMahon CM, Traer E. Revumenib Monotherapy in patients with relapsed/refractory KMT2Ar Acute leukemia: topline efficacy and safety results from the pivotal augment-101 phase 2 study. Blood. 2023;21(142):5. [Google Scholar]
- 48.Zucenka A, Issa GC, Arellano M, Khazal S, Khera N, Stock W, Cuglievan B, Gu Y, Van Nguyen H, Smith AR, Stein EM. Revumenib maintenance therapy following revumenib-induced remission and transplant. Blood. 2023;28(142):4950. [Google Scholar]
- 49.Fiskus W, Daver N, Boettcher S, Mill CP, Sasaki K, Birdwell CE, Davis JA, Das K, Takahashi K, Kadia TM, DiNardo CD. Activity of menin inhibitor ziftomenib (KO-539) as monotherapy or in combinations against AML cells with MLL1 rearrangement or mutant NPM1. Leukemia. 2022;36(11):2729–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burrows F, Wu T, Kessler L, Li S, Zhang J, Zarrinkar P, Li L, Cierpicki T, Grembecka J, Ren P, Liu Y. Abstract LB-A27: a novel small molecule menin-MLL inhibitor for potential treatment of MLL-rearranged leukemias and NPM1/DNMT3A-mutant AML. Molecul Cancer Therapeut. 2018;1(17):27. [Google Scholar]
- 51.Erba HP, Fathi AT, Issa GC, Altman JK, Montesinos P, Patnaik MM, Foran JM, De Botton S, Baer MR, Schiller GJ, Walter RB. Update on a phase 1/2 first-in-human study of the menin-KMT2A (MLL) inhibitor ziftomenib (KO-539) in patients with relapsed or refractory acute myeloid leukemia. Blood. 2022;140(1):153–6. [Google Scholar]
- 52.Wang ES, Altman JK, Pettit K, De Botton S, Walter RP, Fenaux P, Burrows F, Tomkinson BE, Martell B, Fathi AT. Preliminary data on a phase 1/2A first in human study of the menin-KMT2A (MLL) inhibitor KO-539 in patients with relapsed or refractory acute myeloid leukemia. Blood. 2020;5(136):7–8. [Google Scholar]
- 53.Title: CLINICAL pharmacology and pharmacokinetic profile of ziftomenib, a menin inhibitor, in adults with relapsed/refractory acute myeloid leukemia.
- 54.Cierpicki T, Grembecka J. Challenges and opportunities in targeting the menin-MLL interaction. Future Med Chem. 2014;6(4):447–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eguchi K, Shimizu T, Kato D, Furuta Y, Kamioka S, Ban H, Ymamoto S, Yokoyama A, Kitabayashi I. Preclinical Evaluation of a novel orally bioavailable menin-MLL interaction inhibitor, DSP-5336, for the Treatment of acute leukemia patients with MLL-rearrangement or NPM1 mutation. Blood. 2021;23(138):3339. [Google Scholar]
- 56.Daver N, Zeidner JF, Yuda J, Watts JM, Levis MJ, Fukushima K, Ikezoe T, Ogawa Y, Brandwein J, Wang ES, Miyazaki Y. Phase 1/2 First-in-human study of the menin-MLL inhibitor DSP-5336 in patients with relapsed or refractory acute leukemia. Blood. 2023;28(142):2911. [Google Scholar]
- 57.EHA Library—The official digital education library of European Hematology Association (EHA) [Internet]. [cited 2024 Jun 16]. Available from: https://library.ehaweb.org/eha/2024/eha2024-congress/422236/naval.daver.first-in-human.phase.1.2.study.of.the.menin-mll.inhibitor.dsp-5336.html?f=listing%3D0%2Abrowseby%3D8%2Asortby%3D1%2Asearch%3Ds132
- 58.Kwon MC, Querolle O, Dai X, Thuring JW, Verhulst T, Marien A, Goffin D, Cai W, Keersmaekers V, Eyassu F, Verstraeten K. Pharmacological characterization of JNJ-75276617, a menin-KMT2A inhibitor, as targeted treatment for KMT2A-altered and NPM1-mutant acute leukemia. Blood. 2022;140(1):5928–9. [Google Scholar]
- 59.Jabbour E, Searle E, Abdul-Hay M, Abedin S, Aldoss I, Piérola AA, Alonso-Dominguez JM, Chevallier P, Cost C, Daskalakis N, Dillon R. A first-in-human phase 1 study of the menin-KMT2A (MLL1) inhibitor JNJ-75276617 in adult patients with relapsed/refractory acute leukemia harboring KMT2A or NPM1 alterations. Blood. 2023;28(142):57. [Google Scholar]
- 60.Wei A, et al. A phase 1b study of the menin-kmt2a inhibitor jnj-75276617 in combination with venetoclax and azacitidine in relapsed/refractory acute myeloid leukemia with alterations in kmt2a or nPM1. Europ Hematol Associat. 2024;5(14):24. [Google Scholar]
- 61.Numata M, Haginoya N, Shiroishi M, Hirata T, Sato-Otsubo A, Yoshikawa K, Takata Y, Nagase R, Kashimoto Y, Suzuki M, Schulte N. A novel Menin-MLL1 inhibitor, DS-1594a, prevents the progression of acute leukemia with rearranged MLL1 or mutated NPM1. Cancer Cell Int. 2023;23(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, Kenney TM, Wei Y, Andrews DW, Sunnerhagen M, Arrowsmith CH. MYC protein interactors in gene transcription and cancer. Nature Rev Cancer. 2021;21(9):579–91. [DOI] [PubMed] [Google Scholar]
- 63.Lancet J, Ravandi F, Montesinos P, Barrientos JC, Badar T, Alegre A, Bashey A, Burgues JM, Brunetti L, Curran EK, de Leeuw DC. Covalent menin inhibitor Bmf-219 in patients with relapsed or refractory (R/R) acute leukemia (AL): preliminary phase 1 data from the covalent-101 study. Blood. 2023;28(142):2916. [Google Scholar]
- 64.Vasu S, Kohlschmidt J, Mrózek K, Eisfeld AK, Nicolet D, Sterling LJ, Becker H, Metzeler KH, Papaioannou D, Powell BL, Kolitz JE. Ten-year outcome of patients with acute myeloid leukemia not treated with allogeneic transplantation in first complete remission. Blood Adv. 2018;2(13):1645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hills RK, Castaigne S, Appelbaum FR, Delaunay J, Petersdorf S, Othus M, Estey EH, Dombret H, Chevret S, Ifrah N, Cahn JY. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: a meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 2014;15(9):986–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erba HP, Montesinos P, Kim HJ, Patkowska E, Vrhovac R, Žák P, Wang PN, Mitov T, Hanyok J, Kamel YM, Rohrbach JE. Quizartinib plus chemotherapy in newly diagnosed patients with FLT3-internal-tandem-duplication-positive acute myeloid leukaemia (QuANTUM-First): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2023;401(10388):1571–83. [DOI] [PubMed] [Google Scholar]
- 67.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Döhner K, Marcucci G, Lo-Coco F. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. New England J Med. 2017;377(5):454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jentzsch M, Grimm J, Bill M, Goldmann K, Schulz J, Niederwieser D, Platzbecker U, Schwind S. Outcomes of older patients with NPM1 mutated and FLT3-ITD negative acute myeloid leukemia receiving allogeneic transplantation. HemaSphere. 2020;4(1): e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lachowiez CA, Loghavi S, Kadia TM, Daver N, Borthakur G, Pemmaraju N, et al. Outcomes of older patients with NPM1-mutated AML: current treatments and the promise of venetoclax-based regimens. Blood Adv. 2020;4(7):1311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute Myelogenous Leukemia. Cancer Discov. 2016;6(10):1106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei AH, Montesinos P, Ivanov V, DiNardo CD, Novak J, Laribi K, et al. Venetoclax plus LDAC for newly diagnosed AML ineligible for intensive chemotherapy: a phase 3 randomized placebo-controlled trial. Blood. 2020;135(24):2137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, Konopleva M, Döhner H, Letai A, Fenaux P, Koller E. Azacitidine and Venetoclax in previously untreated acute myeloid leukemia. New England J Med. 2020;383(7):617–29. [DOI] [PubMed] [Google Scholar]
- 73.Rausch J, Dzama MM, Dolgikh N, Stiller HL, Bohl SR, Lahrmann C, Kunz K, Kessler L, Echchannaoui H, Chen CW, Kindler T. Menin inhibitor ziftomenib (KO-539) synergizes with drugs targeting chromatin regulation or apoptosis and sensitizes acute myeloid leukemia with MLL rearrangement or NPM1 mutation to venetoclax. Haematologica. 2023;108(10):2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.EHA Library—The official digital education library of European Hematology Association (EHA) [Internet]. [cited 2024 Jun 16]. Available from: https://library.ehaweb.org/eha/2024/eha2024-congress/422238/joshua.zeidner.phase.1b.study.of.azacitidine.venetoclax.and.revumenib.in.newly.html?f=listing%3D0%2Abrowseby%3D8%2Asortby%3D1%2Asearch%3Ds134
- 75.Fröhling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 20020808th ed. 2002;100(13):4372–80. [DOI] [PubMed]
- 76.Mead AJ, Linch DC, Hills RK, Wheatley K, Burnett AK, Gale RE. FLT3 tyrosine kinase domain mutations are biologically distinct from and have a significantly more favorable prognosis than FLT3 internal tandem duplications in patients with acute myeloid leukemia. Blood J Am Soc Hematol. 2007;110(4):1262–70. [DOI] [PubMed] [Google Scholar]
- 77.Stone RM, Mandrekar SJ, Sanford BL, Laumann K, Geyer S, Bloomfield CD, Thiede C, Prior TW, Döhner K, Marcucci G, Lo-Coco F. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. New England J Med. 2017;377(5):454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jahn N, Jahn E, Saadati M, Bullinger L, Larson RA, Ottone T, Amadori S, Prior TW, Brandwein JM, Appelbaum FR, Medeiros BC. Genomic landscape of patients with FLT3-mutated acute myeloid leukemia (AML) treated within the CALGB 10603/RATIFY trial. Leukemia. 2022;36(9):2218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bill M, Mrózek K, Kohlschmidt J, Eisfeld AK, Walker CJ, Nicolet D, Papaioannou D, Blachly JS, Orwick S, Carroll AJ, Kolitz JE. Mutational landscape and clinical outcome of patients with de novo acute myeloid leukemia and rearrangements involving 11q23/KMT2A. Proceed National Academy Sci. 2020;117(42):26340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dzama MM, Steiner M, Rausch J, Sasca D, Schönfeld J, Kunz K, et al. Synergistic targeting of FLT3 mutations in AML via combined menin-MLL and FLT3 inhibition. Blood. 2020;136(21):2442–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Issa GC, Aldoss I, DiPersio J, Cuglievan B, Stone R, Arellano M, Thirman MJ, Patel MR, Dickens DS, Shenoy S, Shukla N. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature. 2023;615(7954):920–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dafflon C, Craig VJ, Mereau H, Gräsel J, Schacher Engstler B, Hoffman G, Nigsch F, Gaulis S, Barys L, Ito M, Aguadé-Gorgorió J. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia. 2017;31(6):1269–77. [DOI] [PubMed] [Google Scholar]
- 84.Chen CW, Armstrong SA. Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond. Experimental Hematol. 2015;43(8):673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dafflon C, Craig VJ, Mereau H, Gräsel J, Schacher Engstler B, Hoffman G, Nigsch F, Gaulis S, Barys L, Ito M, Aguadé-Gorgorió J. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia. 2017;31(6):1269–77. [DOI] [PubMed] [Google Scholar]
- 86.Riedel SS, Haladyna JN, Bezzant M, Stevens B, Pollyea DA, Sinha AU, Armstrong SA, Wei Q, Pollock RM, Daigle SR, Jordan CT. MLL1 and DOT1L cooperate with meningioma-1 to induce acute myeloid leukemia. J Clinic Investigat. 2016;126(4):1438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP, Waters NJ. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood J Am Soc Hematol. 2013;122(6):1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, Jongen-Lavrenic M, Altman JK, Thomson B, Blakemore SJ, Daigle SR. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood J Am Soc Hematol. 2018;131(24):2661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perner F, Gadrey JY, Xiong Y, Hatton C, Eschle BK, Weiss A, Stauffer F, Gaul C, Tiedt R, Perry JA, Armstrong SA. Novel inhibitors of the histone methyltransferase DOT1L show potent antileukemic activity in patient-derived xenografts. Blood J Am Soc Hematol. 2020;136(17):1983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uddin MH, Aboukameel A, Khan H, Bannoura S, Deol A, Yang J, Dyson G, Azmi A, Polin L, Maciejewski JP, Balasubramanian SK. The clinical menin inhibitor ziftomenib and the nuclear export inhibitor selinexor synergistically inhibit the growth of MLL-r AML. Blood. 2023;142(1):4168. [Google Scholar]
- 91.Pianigiani G, Gagliardi A, Mezzasoma F, Rocchio F, Tini V, Bigerna B, Sportoletti P, Caruso S, Marra A, Peruzzi S, Petito E. Prolonged XPO1 inhibition is essential for optimal antileukemic activity in NPM1-mutated AML. Blood Adv. 2022;6(22):5938–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nature Cell Biol. 2007;9(7):804–12. [DOI] [PubMed] [Google Scholar]
- 93.Heikamp EB, Henrich JA, Perner F, Wong EM, Hatton C, Wen Y, Barwe SP, Gopalakrishnapillai A, Xu H, Uckelmann HJ, Takao S. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood J Am Soc Hematol. 2022;139(6):894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, Hsieh JJ. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell. 2016;30(6):863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rasouli M, Blair H, Troester S, Szoltysek K, Cameron R, Ashtiani M, Krippner-Heidenreich A, Grebien F, McGeehan G, Zwaan CM, Heidenreich O. The MLL–Menin Interaction is a Therapeutic Vulnerability in NUP98-rearranged AML. Hemasphere. 2023;7(8): e935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michmerhuizen NL, Klco JM, Mullighan CG. Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood J Am Soc Hematol. 2020;136(20):2275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu H. NUP98 rearrangement in B lymphoblastic leukemia with hyperdiploidy. Blood J Am Soc Hematol. 2020;136(8):1011. [DOI] [PubMed] [Google Scholar]
- 98.Lauw MI, Qi Z, Eversmeyer L, Prakash S, Wen KW, Yu J, Monaghan SA, Aggarwal N, Wang L. Distinct pathologic feature of myeloid neoplasm with t (v; 11p15); NUP98 rearrangement. Human Pathol. 2022;1(123):11–9. [DOI] [PubMed] [Google Scholar]
- 99.Lim HH, An GD, Woo KS, Kim KH, Kim JM, Kim SH, Han JY. NUP98 rearrangement in acute myelomonocytic leukemia with t (11; 19)(p15; p12): the first case report worldwide. Annal Laborat Med. 2017;37(3):285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Romana SP, Radford-Weiss I, Ben Abdelali R, Schluth C, Petit A, Dastugue N, Talmant P, Bilhou-Nabera C, Mugneret F, Lafage-Pochitaloff M, Mozziconacci MJ. NUP98 rearrangements in hematopoietic malignancies: a study of the groupe francophone de cytogenetique hematologique. Leukemia. 2006;20(4):696–706. [DOI] [PubMed] [Google Scholar]
- 101.Xie W, Raess PW, Dunlap J, Hoyos CM, Li H, Li P, Swords R, Olson SB, Yang F, Anekpuritanang T, Hu S. Adult acute myeloid leukemia patients with NUP98 rearrangement have frequent cryptic translocations and unfavorable outcome. Leukemia Lymphoma. 2022;63(8):1907–16. [DOI] [PubMed] [Google Scholar]
- 102.Bertrums EJ, Smith JL, Harmon L, Ries RE, Wang YC, Alonzo TA, Menssen AJ, Chisholm KM, Leonti AR, Tarlock K, Ostronoff F. Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia. Haematologica. 2023;108(8):2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, Hsieh JJ. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell. 2016;30(6):863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Potluri S, Kellaway SG, Coleman DJ, Keane P, Imperato MR, Assi SA, Cockerill PN, Bonifer C. Gene regulation in t (6; 9) DEK: NUP214 acute myeloid leukemia resembles that of FLT3-ITD/NPM1 acute myeloid leukemia but with an altered HOX/MEIS axis. Leukemia. 2024;38(2):403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sandahl JD, Coenen EA, Forestier E, Harbott J, Johansson B, Kerndrup G, Adachi S, Auvrignon A, Beverloo HB, Cayuela JM, Chilton L. t (6; 9)(p22; q34)/DEK-NUP214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica. 2014;99(5):865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Khan I, Amin MA, Eklund EA, Gartel AL. Regulation of HOX gene expression in AML. Blood Cancer J. 2024;14(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duployez N, Vasseur L, Kim R, Largeaud L, Passet M, L’Haridon A, Lemaire P, Fenwarth L, Geffroy S, Helevaut N, et al. UBTF tandem duplications define a distinct subtype of adult de novo acute myeloid leukemia. Leukemia. 2023;37(6):1245–53. 10.1038/s41375-023-01906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Umeda M, Ma J, Huang BJ, Hagiwara K, Westover T. Integrated genomic analysis identifies UBTF tandem duplications as a recurrent lesion in pediatric acute myeloid leukemia. Blood Cancer Discov. 2022;3(3):194–207. 10.1158/2643-3230.BCD-21-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barajas J, Umeda M, Ma J, Khanlari M, Walsh MP, Westover T, Contreras L, Song G, Ma X, Thomas M III, Klco JM. UBTF tandem duplications in pediatric MDS and AML: implications for clinical screening and diagnosis. Blood. 2023;28(142):838. [Google Scholar]
- 110.Barajas JM, Rasouli M, Umeda M, Hiltenbrand R, Abdelhamed S, Mohnani R, Arthur B, Westover T, Thomas ME III, Ashtiani M, Janke LJ. Acute myeloid leukemias with UBTF tandem duplications are sensitive to menin inhibitors. Blood. 2024;143(7):619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aubrey BJ, Cutler JA, Bourgeois W, Donovan KA, Gu S, Hatton C, Perlee S, Perner F, Rahnamoun H, Theall AC, Henrich JA. IKAROS and MENIN coordinate therapeutically actionable leukemogenic gene expression in MLL-r acute myeloid leukemia. Nature Cancer. 2022;3(5):595–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bourgeois W, Cutler JA, Aubrey BJ, Wenge DV, Perner F, Martucci C, Henrich JA, Klega K, Nowak RP, Donovan KA, Boileau M. Mezigdomide is effective alone and in combination with menin inhibition in preclinical models of KMT2A-r and NPM1c AML. Blood. 2024;143(15):1513–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.