Abstract
Focusing on the Yashkun population of Gilgit-Baltistan, an administrative territory in northern Pakistan, our study investigated mtDNA haplotypes as indicators of ancient gene flow and genetic diversity. Genomic DNA was extracted and evaluated for quality using agarose gel electrophoresis. The complete control region of mtDNA (nt 16024–576) was amplified via PCR, and sequencing was performed using the Big Dye Terminator Kit on an Applied Biosystems Genetic Analyzer. Sequence alignment and analysis were conducted using Geneious software and the Cambridge Reference Sequence (CRS). Haplotypes were classified into phylogenetic haplogroups with HaploGrep 2 and the Mito Tool platform, referencing PhyloTree version 17. The study identified considerable genetic diversity within the Yashkun population, with major macro-haplogroups H (29.47%), T (13.68%), and M (13.68%) representing West Eurasian, South Asian, and East & Southeast Asian lineages. The highest genetic affiliation was with West Eurasian haplogroups (73.68%), indicating significant gene flow from the West. The population’s genetic distinctiveness was highlighted by the calculation of forensic genetic characteristics, such as power of discrimination (PD = 0.9975), genetic diversity (GD = 0.9865), and random match probability (RMP = 0.01349). The knowledge of the genetic makeup of the Yashkun population was further improved by the inference of phylogenetic links made possible by comparisons with other indigenous groups in Pakistan. These results attest to the reliability of mtDNA data for forensic applications such as ancestry tracking, individual identification, and additions to national forensic databases. The inclusion of mtDNA analysis in forensic science is strongly supported by this groundbreaking study on the Yashkun community, especially in areas like Pakistan with diverse genetic histories.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-83136-6.
Keywords: Mitochondrial DNA (mtDNA), Yashkun population, Control region, Haplotypes, Genetic diversity, Haplogroups
Subject terms: Evolution, Genetics
Background
In population studies, in addition to genomic DNA, which is an individual’s blueprint and the genetic messenger that spans generations, mitochondrial DNA (mtDNA) has been recognised as a vital tool1. MtDNA is being employed globally by forensic scientists to investigate the origin of the human species and the history of migrations, and to analyze population genetic diversity and phylogenetics2,3. Exploring mitochondrial genomic variations may depict zone and population specificities for a better understanding of uniqueness in diversity4–6. Although mtDNA genomic profiles provide insight into an individual’s maternal ancestry, owing to its manifold copies within the cells, absolute maternal inheritance, extremely variant control regions, compact structure, and smaller size, mtDNA delivers valuable information in medico-legal caseworks where genomic (nuclear) markers might not be accurate. Unlike nuclear DNA, which is more consistent with a lower possibility of genetic variation, mtDNA is more diverse and has a higher mutation rate7–9. Since mtDNA does not undergo recombination, it remains highly stable across generations, making it a reliable genetic marker for research and analysis, alongside nuclear DNA and RNA10,11.
Anatomically modern humans are thought to have colonized South Asia as one of the earliest topographical regions in the world12,13. Today, South Asia is home to one-third of the world’s population, characterized by a diverse mix of people14. It is hypothesized that, since the onset of human, many incursions and migrations have been conducted in South Asia, leading to a very diverse population. Pakistan, situated at the T-intersection of Southeast Asia, Central Asia, and the Middle East, is the second most populated South Asian country, providing a linkage around the continent, connecting the Mediterranean Basin to Asia, and hence has historic importance in early human relocations14,15. Situated at a crossroads of various geographic regions, Pakistan portrays a diversified assemblage of multiple languages, ethnicities, civilizations, and religions. Gilgit-Baltistan is the northernmost administrative unit and the hub for the Himalaya, Karakurum, and Hindu Kush, the great mountain chains of Pakistan. Tribe of Gilgit-Baltistan resides in the valleys, isolated by huge mountains, and thus encompass diversified ethnicities, norms, cultures, values, and backgrounds16–18. A comprehensive curation of mitochondrial haplogroups in global populations is imperative to leverage the benefits of mtDNA, given its abundance of genetic data crucial for forensic and phylogeographic applications. The established mitochondrial haplotypes have been linked to various diseases and have been studied in different species, as shown by numerous studies, with ongoing research continuing to explore the importance of mitochondrial DNA haplogroups19–23. Meaningful experimentation, determining the ancestry and ethnicity of a subject, and supplementing with haplotype analyses in different populations may offer the necessary data to simplify statistical analysis, such as estimating the frequency of mtDNA variants24.
The mtDNA haplotypes of the Yashkun people in Gilgit-Baltistan, a Dardic community that is part of the larger Indo-Aryan ethnolinguistic group, are the main objective of this study25. Known locally by a variety of names, including Borish, Boristi, and Dorshak, they are thought to have migrated from Central Asia through the Pamir Mountains and currently make up a sizable section of the population in Gilgit, Chitral, Punial, Yasin, and Ishkoman26. To better understand the genetic traits of the sampled community and highlight their dominance as a major ethnic group in the region, this study conducts a thorough population analysis focusing on the entire control region of mitochondrial DNA. Additionally, the study investigates genetic ties from the Bronze Age (about 3500 to 1200 BCE) by examining DNA samples to identify the Yashkun people’s ancestral phylogenetic ancestry and connect them to West Eurasian populations. This study aims to clarify genetic exchanges and migration patterns throughout the Bronze Age, a period of substantial cultural and technological growth. By examining genetic continuity or changes over time within the population in Gilgit-Baltistan, the goal is to understand the broader historical relationships that have influenced the contemporary genetic variety of the Yashkun community and reveal their enduring genetic origins. By analysing Bronze Age genetic affinities and ancient human movements and adaptations in the region, it is feasible to comprehend the genetic connections between ancient people and the present genetic environment. Understanding the Yashkun community’s genetic composition is crucial to recognising the unique aspects of their past and significantly enhancing Gilgit-Baltistan’s overall genetic diversity.
Methods
Sample collection
This study involved three maternal generations of Yashkun participants who were not linked to their mothers. Participants were chosen from a variety of Gilgit-Baltistan towns and regions after being told of the study’s goals. Blood samples and informed consent were then obtained.
Genomic DNA extraction and quantification
The Qiagen DNA Extraction kit (Qiagen, Hilden, Germany) was used to extract genomic DNA from whole blood in accordance with the manufacturer’s instructions. Agarose gel electrophoresis was used to qualitatively examine mitochondrial DNA (mtDNA) in order to measure the size of DNA pieces. Agarose (0.5 g) was dissolved in 60 ml of a 1X TAE mixture to create a 1% agarose gel. The gel mixture was supplemented with 2 µL of ethidium bromide. Isolated DNA (2 µL) was dissolved in 2 µL of loading dye and loaded onto a gel. Electrophoresis was conducted for 40 min at 80 V and the gel was photographed under ultraviolet light using a gel documentation system. For quantitative analysis, the Thermo-Scientific Nanodrop One C utilized a spectrophotometric technique, measuring light absorption at 260 nm to quantify the isolated mtDNA. This method provided a comprehensive assessment of both the quality and quantity of nucleic acids, ensuring a reliable characterization of the mitochondrial DNA samples.
mtDNA amplification and sequencing
The complete control region of mtDNA, D-loop (16024 to 576), was amplified using PCR with specific primers (Supplementary Table 1). GeneAmp PCR System 9700 was used for the PCR process (Applied Biosystems, Foster City, CA, USA). A total of 50 L reaction mixture, comprising 25 L hot start master mix, was used for the amplification process (abm, Canada, Cat. No. G906), along with 21 L of H2O that was nuclease-free (Ambion, Thermo Fisher Scientific, USA), reverse and forward pair of primers that were 2 L (10µM) and target DNA that was also 2 L. The PCR cycling procedure included initial denaturation at 94 °C for 10 min, followed by 30 amplification cycles and a final extension at 72 °C for 5 min. Sequencing was performed using the Big Dye Terminator version 3.1 Cycle Sequencing Kit on a 3730xl Applied Biosystems Genetic Analyzer (Thermo Fisher Scientific).
Data analysis
Mitochondrial DNA (mtDNA) analysis involves a multi-step approach using various bioinformatics tools and software. The results of sequencing of PCR-amplified mtDNA products from all subjects were aligned using Geneious27,28. Additionally, Chromas was also employed for extracting the FASTA format, while MtDNA Profiler aligns the FASTA format with the revised reference Cambridge System, enabling SNP analysis and haplotype acquisition. Sequence variations were meticulously identified, and mtDNA haplotypes of the Yashkun community were established. Haplogroups of samples are determined using MitoTool, Mitomap, and Haplogrep29–31, following PhyloTree version 17. HaploGrep 2 employs various dissimilarity metrics including Kulczynski distance, Jaccard coefficient, Kimura 2 factor, and Hamming distance to classify mtDNA haplogroups based on calculated phylogenetic measures of variants in Phylotree depending upon occurrence per position. Essential parameters of population genetics and forensics, including the total count and diversity of haplotypes, genetic diversity, discrimination power and random match probability, were computed. DnaSP is utilized to derive forensically important statistical parameters, such as nucleotide diversity and Tajima’s D. ). FST values were also computed using DnaSP v532. MEGA 11, a comprehensive software package, was applied for evolutionary distance estimation, phylogenetic tree reconstruction, and computation of basic statistical quantities from molecular data33. Graphs were plotted in using R programming language using the package ggplot2 and a heatmap was generated using R software to visualize the distinctions34.
Ethical approval
The experimental data collection followed all the rules and regulations at The University of Punjab, Lahore, Pakistan. Permissions from the authorities were obtained as required and Bioethical clearance certificate was obtained by Bioethics committee. It has been confirmed that the samples were collected and used while complying with relevant institutional, national, and international guidelines and legislation with appropriate permissions from authorities of The University of Punjab, Lahore, Pakistan. All procedures performed in current study involving human participants were in accordance with the ethical standards of the1964 Helsinki declaration and its later amendments or comparable ethical standards.
Results
In the current study, a GenBank database of Yashkun individuals (MK118774-MK118868), encompassing the mitochondrial DNA control region spanning from nucleotide positions 16,024–576. Sequence analysis identified 167 variable sites within the 1122 nucleotide regions examined. The investigation revealed 14 macrogroups constituting 86 distinct haplotypes, with six haplotypes shared among two or more participants, accounting for approximately 7% of the overall count. The remaining 80 haplotypes were unique and represented approximately 93% of the total population (Fig. 1) (Supplementary Fig. 1). Among the 86 haplotypes, four were recurrent, collectively comprising 3.16% of the selected population (Table 1).
Fig. 1.
Distribution of macrogroups among Yashkun Participants.
Table 1.
Mitochondrial DNA control region sequence polymorphism of Yashkun population of Pakistan.
| Sample ID | Haplogroup | Haplotype | Ancestral linkage |
|---|---|---|---|
| YSK-001 | H32 | 73G 152 C 263G 315.1 C | West Eurasian |
| YSK-003 | T2c1 | 16,126 C 16248T 16292T 16294T 16296T 16400T 16,519 C 16527T 73G 263G 309.1 C 315.1 C | West Eurasian |
| YSK-004 | U7a | 16309G 16318T 16,519 C 73G 151T 152 C 263G 315.1C523d 524d | West Eurasian |
| YSK-006 | U5a1d2b | 16,145 A 16192T 16256T 16270T 16,304 C 16399G 73G 263G | West Eurasian |
| YSK-014 | H7h | 16,213 A 16,519 C 146 C 263G 315.1 C | West Eurasian |
| YSK-019 | U2b2 | 16,209 C 16239T 16,352 C 16353T 73G 146 C 152 C 234G 263G | South Asian |
| YSK-021 | J1b1a1+146 | 16069T 16,092 C 16,126 C 16,145 A 16,172 C 16222T 16261T 16,271 C 73G 146 C 242T 263G 295T 309.1 C 315.1C462T 489 C | West Eurasian |
| YSK-022 | M3a1+204 | 16,126 C 16,188 A 16223T 16287T 16,519 C 73G 204 C 263G 309.1 C 315.1 C 482 C 489 C 573.1 C | South Asian |
| YSK-023 | T1a1’3 | 16,126 C 16163G 16186T 16,189 C 16294T 16,519 C 73G 152 C 195 C 263G 309.1 C 309.2 C 315.1 C | West Eurasian |
| YSK-024 | M65a+@16,311 | 16223T 16289G 16,519 C 73G 263G 315.1 C 489 C 511T | South Asian |
| YSK-026 | P+16,176 | 16176T 16309G 16,318 C 16,368 C 16,519 C 73G 152 C 200G 263G 309.1 C 315.1 C 523d 524d | South East Asia |
| YSK-028 | H32 | 73G 146 C 152 C 153G 234G 263G 309.1 C 315.1 C | West Eurasian |
| YSK-030 | H2a2a | 263G 309.1 C 315.1 C | West Eurasian |
| YSK-032 | M30+16,234 | 16223T 16234T 16,519 C 73G 195 A 263G 315.1 C | South Asian |
| YSK-034 | A8a | 16242T 16289.1T 16,293 C 16,319 A 64T 73G 146 C 235G 263G 309.1 C 315.1 C 523d 524d | East Asian |
| YSK-037 | H6 | 16111T 16239T 16,362 C 16482G 239 C 263G | West Eurasian |
| YSK-038 | U2b2 | 16,209 C 16239T 16,311 C 16,352 C 16353T 73G 146 C 152 C 153G 195 C 234G 263G 309.1 C 315.1 C | South Asian |
| YSK-039 | M30+16,234 | 16223T 16234T 16,519 C 73G 195 A 263G 315.1 C 489 C 523d 524d | South Asian |
| YSK-040 | A+152+16,362 | 16,125 A 16223T 16290T 16,311 C 16,319 A 16,362 C 73G 152 C 235G 263G 309.1 C 315.1 C 523d 524d | East Asian |
| YSK-041 | R31a | 73G 146 C 234G 263G 309.1 C 315.1 C 338T | West Eurasian |
| YSK-045 | K1b2 | 16,224 C 16,311 C 16320T 16,519 C 73G 146 C 195 C 198T 263G | West Eurasian |
| YSK-046 | U2b2 | 16,209 C 16239T 16,311 C 16,352 C 16353T 73G 146 C 152 C 153G 234G 263G 309.1 C 315.1 C | South Asian |
| YSK-048 | K1b2 | 16,224 C 16,311 C 16320T 16,519 C 73G 146 C 195 C 263G 315.1 C 524.1 A 524.2 C 524.3 A 524.4 C | West Eurasian |
| YSK-051 | H1e1a1 | 16,178 C 93G 152 C 263G 309.1 C 309.2C315.1 C 524.1 A 524.2 C | West Eurasian |
| YSK-052 | H6 | 16049.1G 16111T 16239T 16,362 C 16482G 239 C 263G 309.1 C 315.1 C | West Eurasian |
| YSK-054 | H2a2a | 16,519 C 263G 315.1 C | West Eurasian |
| YSK-056 | H2a2a | 263G 309.1 C 315.1 C | West Eurasian |
| YSK-058 | M3 | 16,126 C 16147T 16223T 16,519 C 73G 195 C 263G 309.1 C 315.1 C 482 C 489 C | South Asian |
| YSK-059 | R30b1 | 16049.1G 16183d 16189d 16194d 16,195 C 16,196 C 16,298 C 16299G 16326T 16351T 16,352 C 73G 152 C 195 C 263G 299d 309.1 C 315.1 C 373G 493T 509T 524.1 A 524.2 C | West Eurasian |
| YSK-060 | H15a1b | 16,124 C 16184T 16,311 C 44.1 C 55 C 57 C 146 C 197G 263G 309.1 C 315.1 C | West Eurasian |
| YSK-061 | T2 | 16,126 C 16294T 16296T 16,325 C 16,519 C 16527T 73G 263G 315.1 C 523d 524d | West Eurasian |
| YSK-062 | U1 | 16182d 16183d 16189d 16,194 C 16,195 C 16,196 C 16,390 A 16,519 C 73G 241 C 263G 285T 309.1 C 309.2 C 315.1 C | West Eurasian |
| YSK-063 | M30 | 16,093 C 16223T 16,519 C 16526T 73G 195 A 263G 315.1 C 489 C 523d 524d | South Asian |
| YSK-065 | T2 | 16,126 C 16201T 16294T 16296T 16,519 C 16527T 73G 263G 315.1 C 523d 524d | West Eurasian |
| YSK-066 | HV2 | 16,093 C 16,217 C 16,359 C 73G 152 C 195 C 263G 315.1 C 523d 524d | West Eurasian |
| YSK-067 | U7a | 16309G 16,318 C 16,519 C 73G 151T 152 C 263G 309.1 C 315.1 C | West Eurasian |
| YSK-070 | M28 | 16148T 16223T 16,519 C 73G | South Asian |
| YSK-072 | J1b1a1+146 | 16069T 16,092 C 16,126 C 16,145 A 16,172 C 16222T 16261T 16,271 C 73G 146 C 242T 263G 295T 309.1 C 315.1C462T 489 C | West Eurasian |
| YSK-073 | M65a+@16,311 | 16185T 16,189 C 16223T 16289G 16,362 C 16,519 C 73G 263G 309.1 C 309.2 C 315.1 C 489 C 511T | South Asian |
| YSK-074 | H32 | 16309G 16,318 C 16,368 C 16,519 C 73G 152 C 200G 263G 315.1 C 496T 509T 523d 524d 550.1 C | West Eurasian |
| YSK-075 | H2a2a | 263G 315.1 C | West Eurasian |
| YSK-076 | U7a | 16309G 16318T 16,519 C 73G 151T 152 C 263G 315.1 C 499 A 523d 524d | West Eurasian |
| YSK-077 | J | 16,519 C 73G 185 A 263G 271T 295T 315.1 C 458.1T 462d 463d489C 523d 524d | West Eurasian |
| YSK-078 | T | 16,126 C 16294T 16,304 C 16,519 C 73G 263G 309.1C315.1 C | West Eurasian |
| YSK-079 | A+152+16,362 | 16,125 A 16223T 16290T 16,311 C 16,319 A 16,362 C 73G 152 C 235G 263G 309.1 C 315.1 C 523d 524d | East Asian |
| YSK-081 | N | 16,126 C 16147T 16223T 16,519 C 73G 195 C 263G 315.1 C | West Eurasian |
| YSK-085 | J1b1a1 | 16069T 16,126 C 16,145 A 16,172 C 16222T 16261T 16,519 C 73G 242T 263G 295T 315.1 C 462T 489 C | West Eurasian |
| YSK-088 | R5a2 | 16266T 16,304 C 16,311 C 16,356 C 16524G 73G 146 C 152 C 263G 315.1 C 523d 524d | West Eurasian |
| YSK-089 | U2b2 | 16051G 16,209 C 16239T 16,352 C 16353T 73G 146 C 152 C 234G 263G 309.1 C 315.1 C | South Asian |
| YSK-090 | H2a2a1 | 315.1 C | West Eurasian |
| YSK-091 | J1b1b | 16069T 16,126 C 16,145 A 16185T 16261T 16,519 C 73G 207 A 263G 271T 295T 309.1 C 315.1 C 459d 462T 489 C 523d 524d | West Eurasian |
| YSK-092 | H1+152 | 16,519 C 152 C 263G 309.1 C 315.1 C 523d 524d | West Eurasian |
| YSK-093 | M65a+@16,311 | 16223T 16289G 16,519 C 73G 263G 315.1 C 489 C 511T | South Asian |
| YSK-094 | U2a1a | 16051G 16,154 C 16,206 C 16230G 16,311 C 16,519 C 73G 263G 315.1 C | South Asian |
| YSK-095 | M3a1+204 | 16,126 C 16,188 A 16223T 16287T 16,519 C 73G 204 C 263G 309.1C315.1 C 482 C 489 C | South Asian |
| YSK-096 | T2 | 16,126 C 16,213 A 16294T 16296T 16,519 C 73G 263G 315.1 C | West Eurasian |
| YSK-098 | A+152+16,362 | 16,125 A 16223T 16290T 16,311 C 16,319 A 16,362 C 73G 152 C 235G 263G 309.1 C 315.1 C 523d 524d | East Asian |
| YSK-100 | T1a1’3 | 16,126 C 16163G 16186T 16,189 C 16294T 16,519 C 73G 152 C 195 C 263G 309.1 C 315.1 C | West Eurasian |
| YSK-102 | T | 16,092 C 16,126 C 16,129 C 16,183 C 16193.1 C 16294T 16,362 C 16,391 A 16,519 C 73G 152 C 199 C 263G 309.1 C 309.2 C 315.1 C | West Eurasian |
| YSK-104 | P2 | 16,519 C 73G 194T 200G 263G 309.1 C 315.1 C | South East Asia |
| YSK-105 | M3a1+204 | 16,519 C 73G 204 C 263G 309.1 C 315.1 C 482 C 489 C | South Asian |
| YSK-106 | H4a1a+195 | 152 C 195 C 263G 309.1 C 309.2 C 315.1 C | West Eurasian |
| YSK-107 | U5a1f1 | 16192T 16256T 16270T 16,304 C 16,311 C 16399G 73G 195 C 263G 315.1 C | West Eurasian |
| YSK-108 | H2a2a1 | 16,093 C 16182d 16183d 16189d 16,194 C 16,195 C 16,196 C 285T 297 C 309.1 C 309.2 C 309.3 C 315.1 C | West Eurasian |
| YSK-109 | N1a1b1 | 16223T 16,311 C 16,391 A 16,519 C 73G 143 A 199 C 204 C 250 C 263G | West Eurasian |
| YSK-110 | H14a | 16256T 16,352 C 16,422 C 263G 315.1 C | West Eurasian |
| YSK-112 | J1b1b | 73G 263G 271T 295T 309.1 C 315.1 C 459d 462T 489 C 523d524d | West Eurasian |
| YSK-115 | T | 16,126 C 16,145 A 16146T 16147T 16149d 16150T 16155d 16156d 16157.1 C 16157.2T 16157.3T 16158T 16294T 16326T 16,519 C 73G 152 C 195 C 263G 309.1 C 315.1 C | West Eurasian |
| YSK-116 | T2 | 16,126 C 16294T 16296T 16,325 C 16,519 C 16527T 73G 263G 315.1 C | West Eurasian |
| YSK-117 | H1e1a4 | 16,311 C 16,519 C 263G 309.1 C 315.1 C | West Eurasian |
| YSK-119 | J1b1a1 | 16069T 16,126 C 16,145 A 16,172 C 16222T 16261T 16,519 C 73G 242T 263G 295T 315.1 C 356.1 C 462T 489 C | West Eurasian |
| YSK-120 | C4a2b2 | 16,129 A 16223T 16,298 C 16327T 16,519 C 73G 249d263G 309T 310 C 315.1 C 489 C | East Asian |
| YSK-121 | M30c1 | 16166d 16223T 16,519 C 73G 146 C 195 A 263G 315.1 C 489 C 505G 523d 524d | South Asian |
| YSK-123 | X2m1 | 16223T 16292T 16,519 C 73G 143 A | West Eurasian |
| YSK-127 | T2 | 16049.1G 16,126 C 16,213 A 16294T 16296T 16,519 C 73G 263G 309.1 C 315.1 C | West Eurasian |
| YSK-128 | J1b1a1 | 16069T 16,126 C 16,145 A 16,172 C 16222T 16261T 16,519 C 73G 242T 263G 295T 315.1 C 462T 489 C | West Eurasian |
| YSK-129 | I | 16,129 A 16223T 16,391 A 16,519 C 73G 199 C 204 C 250 C 263G | West Eurasian |
| YSK-130 | H15a1b | 16,124 C 16184T 16,311 C 44.1 C 55 C 57 C 146 C 263G 309.1 C 315.1 C 494T | West Eurasian |
| YSK-131 | H14a | 16256T 16,352 C 16,422 C 263G 315.1 C | West Eurasian |
| YSK-132 | H1+152 | 152 C 189G 263G 309.1 C 315.1 C 509T | West Eurasian |
| YSK-133 | H1e1a1 | 16,178 C 93G 152 C 263G 297 C 309.1 C 309.2 C 315.1 C | West Eurasian |
| YSK-134 | J1b1a1+146 | 16069T 16,092 C 16,126 C 16,145 A 16,172 C 16222T 16261T 16,271 C 73G 146 C 242T 263G 295T 309.1 C 315.1C462T 489 C | West Eurasian |
| YSK-136 | U7a | 16,274 A 16309G 16318T 16,519 C 73G 151T 152 C 263G 315.1 C 523d 524d | West Eurasian |
| YSK-137 | H1e1a6 | 16183d 16189d 16194d 16,195 C 16,196 C 16258d 150T 263G 315.1 C | West Eurasian |
| YSK-138 | H2a2a | 263G 309.1 C 315.1 C | West Eurasian |
| YSK-139 | H2a2a1g | 16,182 C 16,183 C 16,189 C 309.1 C 309.2 C 315.1 C | West Eurasian |
| YSK-144 | H14a | 16256T 16,352 C 16,422 C 263G 315.1 C | West Eurasian |
| YSK-145 | N5 | 16111T 16223T 16292T 16,519 C 73G 143 A | West Eurasian |
| YSK-146 | M30d | 16223T 16302G 16,519 C 73G 143 A 195 A 263G 315.1C489C | South Asian |
| YSK-147 | J2b1a | 16069T 16,126 C 16193T 16278T 16,519 C 73G 150T 152 C 235G 263G 295T 315.1 C 489 C | West Eurasian |
| YSK-148 | T2 | 16,126 C 16201T 16294T 16296T 16,519 C 16527T 73G 263G 315.1 C | West Eurasian |
| YSK-149 | T2d1b | 16,126 C 16294T 16,519 C 73G 194T 200G 263G 309.1C315.1 C | West Eurasian |
| YSK-151 | H2a2a1g | 16,189 C 309.1 C 315.1 C | West Eurasian |
| YSK-152 | HV14 | 16,519 C 263G 294 C 309.1 C 315.1 C 480 C 523d 524d | West Eurasian |
| YSK-153 | H2a1 | 16354T 263G 315.1 C | West Eurasian |
In this study, 95 sequences were partitioned into two datasets using DnaSP v5DnaSP to analyze different evolutionary lineages facilitating more targeted genetic analyses, with one dataset comprising 41 sequences and the other containing 54 sequences. The calculation of principal forensic and population genetics parameters revealed a high genetic diversity of 0.9975 among the Yashkun individuals. The Random Match Probability (RMP) estimated between unrelated individuals was 0.01349, indicating a low probability of random matches. Furthermore, the Power of Discrimination was 0.9865, underscoring the efficacy of the genetic markers in distinguishing between individuals within the Yashkun population. FST values were computed for various populations using DnaSP, and a heatmap was generated using R software to visualize these distinctions34 The heatmap displays a color gradient where where lower FST values (depicted in blue) indicate greater genetic similarity, and higher values (depicted in red) suggest greater genetic differentiation. A visual portrayal is used to better comprehend the genetic structure and interactions within and between the populations under study. Yashkun’s closest genetic relationships appear with the neighboring populations such as the Kalash and Shina Gilgit, indicated by the blue or light-colored blocks, suggesting high levels of genetic similarity. In contrast, the Yashkun population is more genetically distinct from distant populations like the Franco Cantabrian, Hungarians, and Spanish, as evidenced by the darker red blocks, indicating higher FST values and thus greater differentiation. Using forensic and population parameters, the study provides an inter-comparison (Table 2) among the Yashkun community and other indigenous groups in Pakistan. Hierarchical clustering of the FST values helps understand the genetic landscape and possible evolutionary histories of these groups by revealing how closely linked the populations are. Additionally, the study examines the genetic diversity and forensic applicability of the mtDNA haplotypes found in various ethnic groups in Pakistan. Significant genetic diversity (0.9975) and strong discrimination power (0.9865) were demonstrated by the Yashkun community, suggesting that mtDNA analysis is a useful method for internal group identification. The Yashkuns’ low random match probability (0.01349) adds credence to the usefulness of mtDNA analysis in forensics. Additionally, the genetic profiles of other ethnic groups—Shin, Saraiki, Sindhi, Makran, Pathan, Kashmiri, Hazara, Kalash, Wakhi, and Balti—were investigated. The distinct genomic profiles of each group varied in terms of genetic variation and forensic significance. Due to their strong discrimination strength and extremely high genetic diversity (0.9996 and 0.957, respectively), Shin and Saraiki may be useful in forensic investigations. The Balti population, while slightly lower in genetic diversity (0.9939), also showed effective discrimination power (Table 2). A comparative analysis of the phylogenetic structure of the Yashkun population with that of other indigenous groups in Pakistan is shown in Figure 2, providing further insights into the genetic relationships and evolutionary history of these populations.
Table 2.
Inter-comparison of forensic and genetic diversity indices of mtDNA control region of main ethnic groups of Pakistan.
| Parameters | Yashkun | Shin | Saraiki | Sindhi | Makran | Pathan | Kashmiri | Hazara | Kalash | Wakhi | Balti | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of samples | 95 | 79 | 85 | 88 | 99 | 230 | 317 | 319 | 76 | 40 | 52 | ||
| No. of haplotypes | 86 | 75 | 63 | 66 | 71 | 192 | 251 | 189 | 31 | 40 | 43 | ||
| No. of unique haplotypes | 80 | 72 | 58 | 50 | 54 | 128 | 201 | 124 | 23 | 39 | 37 | ||
| Genetic diversity | 0.9975 | 0.9996 | 0.957 | 0.9924 | 0.9905 | 0.9978 | 0.9977 | 0.9945 | 0.9012 | 0.998 | 0.9939 | ||
| Power of discrimination | 0.9865 | 0.9871 | 0.9458 | 0.9811 | 0.7172 | 0.8348 | 0.7918 | 0.5925 | 0.8894 | 0.974 | 0.9748 | ||
| Random match probability | 0.0135 | 0.0129 | 0.0542 | 0.0188 | 0.0195 | 0.0066 | 0.0054 | 0.0085 | 0.1106 | 0.026 | 0.025 | ||
Fig. 2.
Pairwise FST matrix, arranging populations based on the hierarchical clustering of their FST values.
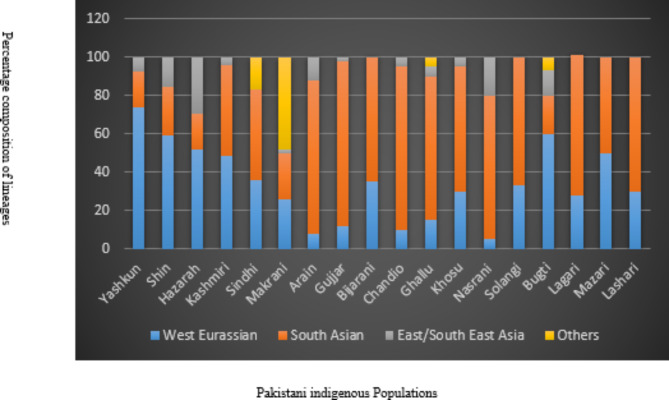
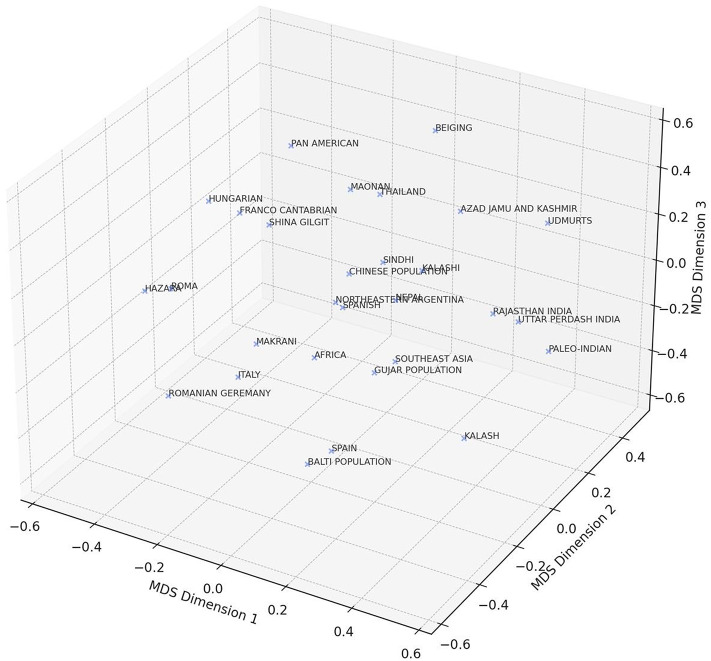
The haplotypes identified within the Yashkun population were systematically categorized into phylogenetic haplogroups. These haplogroups were broadly classified into three main classes: West Eurasian, East and Southeast Asian, and South Asian, comprising 73.68%, 18.95%, and 7.37% of the observed haplotypes, respectively (Figure 3). Notably, a substantial genetic lineage linkage was observed in West Eurasians compared to the Southeast Asian and South Asian lineages. The genetic makeup of the Yashkun population revealed a noteworthy influence from West Eurasia, implying substantial genetic divergence originating from the western region. T2, the predominant haplogroup was observed in 6.3% of Yashkun participants (n=6). Additional haplogroups observed among Yashkun individuals included H2a2a (5.26%, n=5), U2b2 (4.21%, n=4), U7a (4.21%, n=4), A+152+16362 (3.16%, n=3), H14a (3.16%, n=3), H32 (3.16%, n=3), J1b1a1+146 (3.16%, n=3), J1b1a1 (3.16%, n=3), M3a1+204 (3.16%, n=3), T (3.16%, n=3), M65a+@16311 (3.16%, n=3), H1+152 (2.11%, n=2), H15a1b (2.11%, n=2), H1e1a1 (2.11%, n=2), H2a2a1 (2.11%, n=2), H2a2a1g (2.11%, n=2), H6 (2.11%, n=2), J1b1b (2.11%, n=2), K1b2 (2.11%, n=2), M30+16234 (2.11%, n=2), T1a1’3 (2.11%, n=2), A8a (1.05%, n=1), C4a2b2 (1.05%, n=1), H1e1a4 (1.05%, n=1), H1e1a6 (1.05%, n=1), H2a1 (1.05%, n=1), H4a1a+195 (1.05%, n=1), H7h (1.05%, n=1), HV14 (1.05%, n=1), HV2 (1.05%, n=1), I (1.05%, n=1), J (1.05%, n=1), J2b1a (1.05%, n=1), M28 (1.05%, n=1), M3 (1.05%, n=1), M30 (1.05%, n=1), M30c1 (1.05%, n=1), M30d (1.05%, n=1), N (1.05%, n=1), N1a1b1 (1.05%, n=1), N5 (1.05%, n=1), P+16176 (1.05%, n=1), P2 (1.05%, n=1), R30b1 (1.05%, n=1), R31a (1.05%, n=1), R5a2 (1.05%, n=1), T2c1 (1.05%, n=1), T2d1b (1.05%, n=1), U1 (1.05%, n=1), U2a1a (1.05%, n=1), U5a1d2b (1.05%, n=1), U5a1f1 (1.05%, n=1), and X2m1 (1.05%, n=1) (Table 3). Based on FST results, a 3D Multidimensional Scaling (MDS) graphic was created using Python to measure genetic divergence among various global groups. An extensive visual depiction of the genetic links across several groups is offered by this MDS plot, which aids in spotting patterns of genetic similarity and divergence. With the designations MDS Dimension 1, MDS Dimension 2, and MDS Dimension 3, the genetic linkages are shown in three dimensions. With more genetically dissimilar populations positioned farther apart and more genetically similar populations located closer together, each point on the plot represents a distinct population or ethnic group. A shared ancestry or gene flow within this region is indicated by the plot’s genetic proximity to other South Asian populations, including the Sindhi, Kalash, and Balti. Due to geographic and evolutionary separation, populations from farther away, such as those from Africa, Southeast Asia, and Pan America, are found farther from Yashkun. This indicates notable genetic differences. The population of Yashkun exhibits a considerable genetic relationship to Franco Cantabrian, Hungarian, and other West Eurasian populations, including German and Romanian. The intricate genetic makeup of the Yashkun population, which is influenced by both regional South Asian and West Eurasian factors, is further supported by this visualisation. This information is crucial for understanding the complex genetic structure and historical connections among human populations (Figure 4).
Fig. 3.
Percentage composition of phylogeographical lineages in Pakistani populations.
Table 3.
Haplogroup frequencies of 95 Yashkuns from Gilgit-Baltistan, Pakistan.
| Broad Haplogroup | Number | Proportions % |
Haplogroup | Number | Proportions % |
|---|---|---|---|---|---|
| A | 4 | 4.21 | A+152+16,362 | 3 | 3.16 |
| A8a | 1 | 1.05 | |||
| C | 1 | 1.05 | C4a2b2 | 1 | 1.05 |
| H | 28 | 29.47 | H1+152 | 2 | 2.11 |
| H14a | 3 | 3.16 | |||
| H15a1b | 2 | 2.11 | |||
| H1e1a1 | 2 | 2.11 | |||
| H1e1a4 | 1 | 1.05 | |||
| H1e1a6 | 1 | 1.05 | |||
| H2a1 | 1 | 1.05 | |||
| H2a2a | 5 | 5.26 | |||
| H2a2a1 | 2 | 2.11 | |||
| H2a2a1g | 2 | 2.11 | |||
| H32 | 3 | 3.16 | |||
| H4a1a+195 | 1 | 1.05 | |||
| H6 | 2 | 2.11 | |||
| H7h | 1 | 1.05 | |||
| HV | 2 | 2.11 | HV14 | 1 | 1.05 |
| HV2 | 1 | 1.05 | |||
| I | 1 | 1.05 | I | 1 | 1.05 |
| J | 10 | 10.53 | J | 1 | 1.05 |
| J1b1a1 | 3 | 3.16 | |||
| J1b1a1+146 | 3 | 3.16 | |||
| J1b1b | 2 | 2.11 | |||
| J2b1a | 1 | 1.05 | |||
| K | 02 | 2.11 | K1b2 | 2 | 2.11 |
| M | 13 | 13.68 | M28 | 1 | 1.05 |
| M3 | 1 | 1.05 | |||
| M30 | 1 | 1.05 | |||
| M30+16,234 | 2 | 2.11 | |||
| M30c1 | 1 | 1.05 | |||
| M30d | 1 | 1.05 | |||
| M3a1+204 | 3 | 3.16 | |||
| M65a+@16,311 | 3 | 3.16 | |||
| N | 3 | 3.16 | N | 1 | 1.05 |
| N1a1b1 | 1 | 1.05 | |||
| N5 | 1 | 1.05 | |||
| P | 2 | 2.11 | P+16,176 | 1 | 1.05 |
| P2 | 1 | 1.05 | |||
| R | 3 | 3.16 | R30b1 | 1 | 1.05 |
| R31a | 1 | 1.05 | |||
| R5a2 | 1 | 1.05 | |||
| T | 13 | 13.68 | T | 3 | 3.16 |
| T1a1’3 | 2 | 2.11 | |||
| T2 | 6 | 6.32 | |||
| T2c1 | 1 | 1.05 | |||
| T2d1b | 1 | 1.05 | |||
| U | 12 | 12.63 | U1 | 1 | 1.05 |
| U2a1a | 1 | 1.05 | |||
| U2b2 | 4 | 4.21 | |||
| U5a1d2b | 1 | 1.05 | |||
| U5a1f1 | 1 | 1.05 | |||
| U7a | 4 | 4.21 | |||
| X | 1 | 1.05 | X2m1 | 1 | 1.05 |
Fig. 4.
3D Multidimensional Scaling (MDS) plot based on FST values by Python.
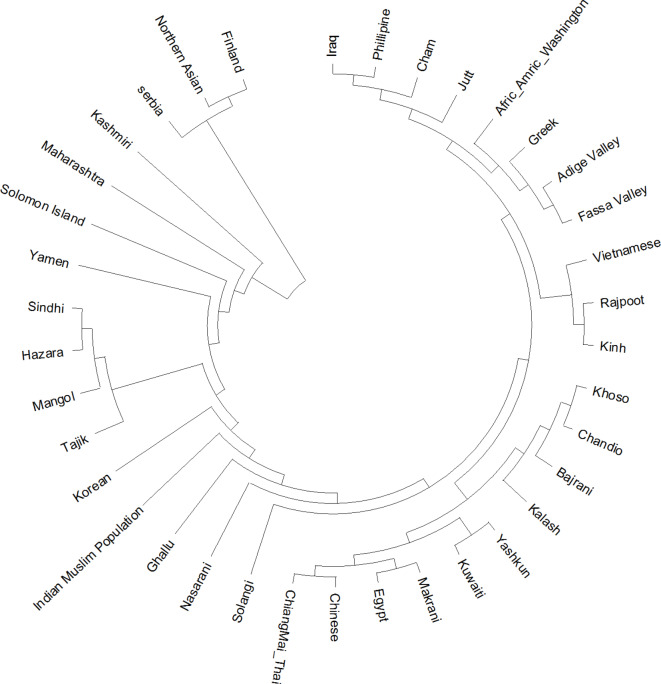
The evolutionary relationships among various populations were analyzed using MEGA11 software33 and the Neighbor-Joining method to infer lineages. The resulting phylogenetic tree is scaled to reflect branch lengths corresponding to evolutionary distances, calculated using the Maximum Composite Likelihood method and measured by the number of base substitutions per site. This analysis incorporated nucleotide sequences from different populations, producing a tree that illustrates their evolutionary connections. Each branch represents a unique evolutionary trajectory, with its length indicating the extent of genetic divergence. Populations located at the branch tips are organized so that closely related groups appear on adjacent branches, while more distantly related groups are placed on separate, longer branches, indicating greater genetic divergence. Distinct clusters on the tree highlight genetic similarities and shared evolutionary histories. For instance, populations from Northern Asia, Finland, and Serbia cluster together, suggesting close evolutionary ties, while other populations, such as those from Iraq, the Philippines, and Cham, form separate clusters, reflecting distinct ancestries. Some clusters align with regional proximity or historical connections, such as the South Asian cluster, which includes Sindhi, Hazara, Mongol, and Tajik populations, and another cluster containing Middle Eastern and African groups like Egypt, Kuwait, and Makrani (Figure 5). Additionally, a Neighbor-Joining tree was constructed among the Yashkun individuals to focus on the genetic structure and diversity of the Yashkun population (Supplementary Figure 2). A median-joining network, constructed using Network software, was also created to visualize relationships and evolutionary pathways among haplotypes (Supplementary Figure 3). This phylogenetic analysis provides insights into the genetic diversity and evolutionary patterns of the populations studied, illustrating both shared relationships and genetic variation among them.
Fig. 5.
Evolutionary analyses were conducted in MEGA11 among multiple populations. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site.
Despite being a country with tremendously diverse and heterogeneous ethnicities, diversity in population and ethnic groups in the Pakistani population has not been studied from a genetic perspective35. The present study analyzed the mtDNA hypervariable region of 95 Yashkun individuals from Gilgit-Baltistan. It revealed 167 sequence variations and 86 distinct haplotypes, among which six were found to be shared, while the remaining 80 were unique. The percentage of unique haplotypes in the study population was higher than in the other population groups in Pakistan. For instance, the Hazara population exhibited 66% unique haplotypes, the Makrani community 77%, Burusho ethnicity 72%, Saraiki individuals 74%, Parsi class 50%, Pathan 68%, Baluchi 66%, Sindhi 56%, Bangashi 60%, Khattak 52%, Brahui 58%, Mahsud 36%, and Kalash 27%36,37,38,39,40,41,42,43,44. In addition to indicating genetic variation in a population, such contrasting values, as compared to our study, reported that the occurrence of unique haplotypes in different groups of Pakistan might occur because of the smaller sample size in different studies45.
The current study revealed that the Yashkun population exhibited a genetic diversity of 0.9975. This level of genetic diversity is relatively high and comparable to that observed in other Pakistani ethnicities. For example, the Pathan community showed a genetic diversity of 0.9978, closely followed by the Sindhi population with a diversity of 0.9924. The diversity of the Hazara community was 0.9945, while that of the Kashmir residents was 0.9977. Additionally, the Makrani population exhibited a diversity of 0.9905, and the Burushos and Yousafzais populations showed diversities of 0.980 and 0.9392, respectively35,40,41,42,43,44,46. Yashkun individuals demonstrate high genetic diversity and powerful discrimination ability, resulting in a low random match probability. These findings indicate that mtDNA analysis has significant value and relevance for practical applications in forensics.
The haplogroups of the Yashkun population, established from the current study, suggests that their genetic composition greatly contributes to West Eurasian matrilineage (73.68%), a moderate to high influence on South Asian lineages (18.95%), and a lesser proportion to East and Southeast Asian lineages (7.37%). Thus, the study revealed a strong genetic linkage with West Eurasian ancestry compared to the lineages found in South and Southeast Asia. A significant genetic association with the West Eurasian lineage has been documented in various groups of Pakistan and surrounding countries. For instance, Tarklanis and Kohistanis exhibit a 54% association. Utmankheils showed a 47% relevance, while Yousafzais exhibited a 52% association. Among Gujars, the association was reported to be 37%. The population groups in the Swat and Dir districts had a 45% association, while Pathans showed a 55% association. The Shins exhibit 60%, whereas Hazaras and Kashmiris show an association of 52% and 49 %, respectively. In contrast, Makranis displayed a 26% association, and the mitochondrial genetic configuration of the Sindhi population in Pakistan showed a 36% West Eurasian lineage21,35,37,40,41,42,43,44,46,47. Nevertheless, caste communities and tribal classes display low to modest West Eurasian percentage, for example, Gujar (12%), Arain (8%), Bijarani (35%), Ghallu (15%), Chandio (10%), while Khosu, Solangi, Lagari, Lashari, Nasrani, Bugti, and Mazari exhibit the percentages of 30%, 33%, 27.7%, 30%, 5%, 60%, and 50%, respectively36,37,48. In studies conducted on the Indian population, a significant frequency of West Eurasian mtDNA genetic ancestry has been observed. For instance, Indian Punjabis has been found to exhibit a genetic proportion that varies from 40% to 50%. Similarly, Gujaratis and Kashmiris reported a proportion of approximately 30%. However, in Indian populations following the caste approach and ethnicity in states comprising West Bengal, Tamil Nadu, Uttar Pradesh, Kerala, and Maharashtra, West Eurasian lineages were comparatively lower49,50. The Afghanistan population also exhibits a significant West Eurasian phylogenetic composition. The frequencies of this lineage have been reported to be 40%, 64%, 74%, and 89% in Hazaras, Pashtuns, Baluchis, and Tajiks, respectively51.
Amongst this evidence, the current research revealed a prominent association between West Eurasian lineages and the Yashkun population. This indicates a significant and continuous genetic flow towards Gilgit-Baltistan from West and Central Asia, which took place in various stages that can be traced back to almost 2.5 million years ago. The initial influx is believed to have occurred during the Paleolithic era when masses migrated from the Middle East towards the Indo-Gangetic plains of India and Pakistan. The gene flow was also aided by later human dispersal throughout the Neolithic and Mesolithic periods. Alexander the Great, the British government in the Indian subcontinent, and Arab and Muslim conquests were among the historical events that contributed to the introduction of West Eurasian genetic elements to the area39,52. The practice of caste marriage, which limits gene flow within particular caste groups, is responsible for the comparatively lower West Eurasian genetic structure of tribes in Pakistan and India53.
Thus, the study’s findings show that mitochondrial DNA is a crucial and effective tool for population genetics research and forensic applications in this field. In addition to improving the representation of ethnically diverse communities, incorporating this data into forensic databases also increases the accuracy of identification procedures in areas where the Yashkun population resides. This is particularly important when it comes to addressing missing person cases, unidentified remains cases, and criminal investigations. Its usefulness for worldwide forensic applications is further reinforced by the dataset’s establishment of a strong basis for comparative analyses with other populations, which facilitate cross-regional forensic investigations and population structure research. These results demonstrate the value of mtDNA analysis in forensic investigations, especially for identifying people in situations where nuclear DNA is not available. All things considered, the parameters of the study validate that the mtDNA information from the Yashkun community is thorough and trustworthy for forensic purposes, such as identifying individuals, determining ancestry, and adding information to national forensic databases. The significance of using mtDNA analysis in forensic research is shown by this evidence, particularly in areas like Pakistan where the population is genetically diverse.
Conclusion
To the best of our knowledge, the current study is the first to provide a genetic profile of the complete mtDNA control region in the Yashkun population. The results demonstrated remarkable genetic diversity, together with a low random match probability of 0.9975 and 0.0135, respectively. These findings indicate that mitochondrial DNA is a valuable and effective method for analyzing population genetics and forensic purposes in this region. The current report shows a high proportion of West Eurasia in the genetic structure of the Yashkun population, suggesting a great genetic drift from the West. The provided statistics will significantly contribute to the formation of a national mitochondrial DNA databank that can serve multiple forensic purposes.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are highly obliged to all the volunteers of Yashkun population for cooperation and provision of their blood samples. The authors would also like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2025R350), King Saud University, Riyadh, Saudi Arabia.
Author contributions
MUK wrote the initial manuscript, MIU, QA, AA, SH, and TZ provided the technical guidance regarding statistical and genetic interpretation and analysis and MFS supervised the project and helped out in the sequencing process. All authors reviewed the manuscript.
Data availability
The mtDNA control-region sequences in a fasta format of all the Yashkun participants in the present study have been made available to GenBank under MK118774-MK118868 accession number and will be available for the public for future scientific use.
Declarations
Consent for publication
Not applicable.
Conflict of interest
The authors have no conflict of interest in this publication.
Ethical approval
The study obtained a bioethical clearance approval and certificate from the Bioethics Committee of the University of the Punjab. All procedures conducted in the current research with human participants adhered to the ethical standards set by the institutional and/or national research committee, as well as the principles highlighted in the 1964 Declaration of Helsinki along with its consequent amendments and comparable ethical outlines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Muhammad Umer Khan, Email: Muhammad.umer4@mlt.uol.edu.pk.
Muhammad Farooq Sabar, Email: farooq.camb@pu.edu.pk.
Qurban Ali, Email: saim1692@gmail.com.
References
- 1.Bouchard, C. & Wackerhage, H. in Molecular Exercise Physiology 61–88Routledge, (2022).
- 2.Connell, J. et al. Mitochondrial DNA Analysis in Population Isolates: Challenges and Implications for Human Identification. Curr. Mol. Biology Rep.10, 1–8 (2024). [Google Scholar]
- 3.Syndercombe Court, D. Mitochondrial DNA in forensic use. Emerg. Top. life Sci.5, 415–426. 10.1042/etls20210204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, F. et al. Approaches and challenges in identifying, quantifying, and manipulating dynamic mitochondrial genome variations. Cell. Signal.117, 111123. 10.1016/j.cellsig.2024.111123 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Brand, J. A. et al. Mitochondrial genetic variation as a potential mediator of intraspecific behavioural diversity. Trends Ecol. Evol.39, 199–212. 10.1016/j.tree.2023.09.009 (2024). [DOI] [PubMed] [Google Scholar]
- 6.Schlebusch, C. M. & Jakobsson, M. Tales of Human Migration, Admixture, and Selection in Africa. Annu. Rev. Genom. Hum. Genet.19, 405–428. 10.1146/annurev-genom-083117-021759 (2018). [DOI] [PubMed] [Google Scholar]
- 7.DeBalsi, K. L. et al. Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res. Rev.33, 89–104. 10.1016/j.arr.2016.04.006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, H., Slone, J., Fei, L., Huang, T. & Mitochondrial, D. N. A. Variants and Common Diseases: A Mathematical Model for the Diversity of Age-Related mtDNA Mutations. Cells810.3390/cells8060608 (2019). [DOI] [PMC free article] [PubMed]
- 9.Lee, W. et al. Molecular basis for maternal inheritance of human mitochondrial DNA. Nat. Genet.55, 1632–1639. 10.1038/s41588-023-01505-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, R. et al. Nuclear genetic control of mtDNA copy number and heteroplasmy in humans. Nature620, 839–848. 10.1038/s41586-023-06426-5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleem, A. et al. Biological role and regulation of circular RNA as an emerging biomarker and potential therapeutic target for cancer. Mol. Biol. Rep.5110.1007/s11033-024-09211-3 (2024). [DOI] [PubMed]
- 12.Hublin, J. et al. How old are the oldest Homo sapiens in. Far East. Asia?118, e2101173118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blinkhorn, J. et al. Middle Palaeolithic occupation in the Thar Desert during the Upper Pleistocene: the signature of a modern human exit out of Africa? 77, 233–238 (2013).
- 14.Yeung, W. J. J., Desai, S. & Jones, G. W. Families in southeast and South Asia. Ann. Rev. Sociol.44, 469–495 (2018). [Google Scholar]
- 15.Waring, T. M., Wood, Z. T. & Szathmáry, E. Characteristic processes of human evolution caused the Anthropocene and may obstruct its global solutions. Philos. Trans. R. Soc. Lond. B Biol. Sci.379, 20220259. 10.1098/rstb.2022.0259 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marzen, C. G. & Balochistan Self-Determination, and US Foreign Policy. NCJ Int’l L. 45, 195 (2019). [Google Scholar]
- 17.Aroosa et al. Analysis to uncover relationships between morphological traits, sex and their distribution in various chicken breeds. Bull. Biol. Allied Sci. Res.2024 (1), 78. 10.54112/bbasr.v2024i1.78 (2024). [Google Scholar]
- 18.Price, M. The Politics of State Recognition: Norms, Geopolitics, and the East Pakistan Crisis. Global Stud. Q.4, ksae043 (2024). [Google Scholar]
- 19.Basu, A. & Majumder, P. P. in Evolution of the Human Genome II: Human Evolution Viewed from Genomes 153–164Springer, (2021).
- 20.Ferreira, L. L. et al. A pilot study of mitochondrial genomic ancestry in admixed Brazilian patients with type 1 diabetes. Diabetol. Metab. Syndr.1610.1186/s13098-024-01342-8 (2024). [DOI] [PMC free article] [PubMed]
- 21.Khan, M. U. et al. Forensic and genetic characterization of mtDNA lineages of Shin, a Unique Ethnic Group in Pakistan. Pakistan J. Zool.53, 133 (2021). [Google Scholar]
- 22.Terrado-Ortuño, N., May, P. & Forensic, D. N. A. Phenotyping: a review on SNP panels, genotyping techniques, and prediction models. Forensic Sci. Res., owae013 (2024).
- 23.Ahmed, M. et al. Investigating the inheritance patterns and potential associations of selected human morphogenetic traits. Bull. Biol. Allied Sci. Res.2024 (1), 77. 10.54112/bbasr.v2024i1.77 (2024). [Google Scholar]
- 24.Butler, J. M. Recent activities in the United States involving the National commission on forensic science and the organization of scientific area committees for forensic science. Australian J. Forensic Sci.49, 526–540 (2017). [Google Scholar]
- 25.Hussain, B. et al. Cross-cultural ethnobotany of the Baltis and Shinas in the Kharmang district, Trans-Himalaya India-Pakistan border. Heliyon10, e28613. 10.1016/j.heliyon.2024.e28613 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, P. Gilgit Baltistan: Between Hope and DespairVol. 14 (Institute for Defence Studies and Analyses, 2013). [Google Scholar]
- 27.Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinf. (Oxford England). 28, 1647–1649. 10.1093/bioinformatics/bts199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, D. et al. A PCR-independent approach for mtDNA enrichment and next-generation sequencing: comprehensive evaluation and clinical application. J. translational Med.2210.1186/s12967-024-05213-8 (2024). [DOI] [PMC free article] [PubMed]
- 29.Andrews, R. M. et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet.23, 147. 10.1038/13779 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Arouk, N. I. Fingerprinting of Human Genome via New Mitochondrial-Based Approach.
- 31.Holland, M. M. & Parson, W. in Forensic DNA Applications 67–86CRC Press, (2023).
- 32.Zimmermann, B. et al. Improved visibility of character conflicts in quasi-median networks with the EMPOP NETWORK software. Croatian Med. J.55, 115–120. 10.3325/cmj.2014.55.115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol.38, 3022–3027. 10.1093/molbev/msab120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu, Z. Complex heatmap visualization. iMeta1 (e43). 10.1002/imt2.43 (2022). [DOI] [PMC free article] [PubMed]
- 35.Ullah, I. et al. High Y-chromosomal Differentiation Among Ethnic Groups of Dir and Swat Districts, Pakistan. Ann. Hum. Genet.81, 234–248. 10.1111/ahg.12204 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Bhatti, S. et al. Genetic perspective of uniparental mitochondrial DNA landscape on the Punjabi population, Pakistan. Mitochondrial DNA Part. DNA Mapp. sequencing Anal.29, 714–726. 10.1080/24701394.2017.1350951 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Bhatti, S. et al. Problems in Mitochondrial DNA forensics: while interpreting length heteroplasmy conundrum of various Sindhi and Baluchi ethnic groups of Pakistan. Mitochondrial DNA Part. DNA Mapp. sequencing Anal.29, 501–510. 10.1080/24701394.2017.1310853 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Hayat, S. et al. Mitochondrial DNA control region sequences study in Saraiki population from Pakistan. Leg. Med.17, 140–144 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Quintana-Murci, L. et al. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am. J. Hum. Genet.74, 827–845. 10.1086/383236 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakha, A. et al. EMPOP-quality mtDNA control region sequences from Kashmiri of Azad Jammu & Kashmir, Pakistan. Forensic Sci.Int. Genet.25, 125–131. 10.1016/j.fsigen.2016.08.009 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Rakha, A. et al. Forensic and genetic characterization of mtDNA from Pathans of Pakistan. Int. J. Legal Med.125, 841–848. 10.1007/s00414-010-0540-7 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Siddiqi, M. H. et al. Genetic characterization of the Makrani people of Pakistan from mitochondrial DNA control-region data. Legal Med. (Tokyo Japan). 17, 134–139. 10.1016/j.legalmed.2014.09.007 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Siddiqi, M. H. et al. Current pool of ultimate collection of mitochondrial DNA from remnants of Kalash. Mitochondrial DNA Part. B Resour.6, 2410–2414. 10.1080/23802359.2021.1952119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakha, A. et al. mtDNA sequence diversity of Hazara ethnic group from Pakistan. Forensic Sci.Int. Genet.30, e1–e5. 10.1016/j.fsigen.2017.07.004 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Ullah, I. et al. Mitochondrial genetic characterization of Gujar population living in the Northwest areas of Pakistan. Advancements Life Sci.4, 84–91 (2017). [Google Scholar]
- 46.Yasmin, M., Rakha, A., Noreen, S. & Salahuddin, Z. Mitochondrial control region diversity in Sindhi ethnic group of Pakistan. Legal Med. (Tokyo Japan). 26, 11–13. 10.1016/j.legalmed.2017.02.001 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Singh, G. et al. Genetic characterisation of the North-West Indian populations: analysis of mitochondrial DNA control region variations. Ann. Hum. Biol.48, 166–172. 10.1080/03014460.2021.1879933 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Bhatti, S. et al. Mitochondrial DNA variation in the Sindh population of Pakistan. Australian J. Forensic Sci.49, 201–216 (2017). [Google Scholar]
- 49.Ahmed, M. Ancient Pakistan-an Archaeological History: Volume III: Harappan Civilization-the Material CultureAmazon,. (2014).
- 50.Kivisild, T. et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet.72, 313–332. 10.1086/346068 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whale, J. W. Mitochondrial DNA analysis of four ethnic groups of Afghanistan (University of Portsmouth Portsmouth, 2012). [Google Scholar]
- 52.McElreavey, K. & Quintana-Murci, L. A population genetics perspective of the Indus Valley through uniparentally-inherited markers. Ann. Hum. Biol.32, 154–162. 10.1080/03014460500076223 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Pemberton, T. J. et al. Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India. Ann. Hum. Genet.72, 535–546. 10.1111/j.1469-1809.2008.00457.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mtDNA control-region sequences in a fasta format of all the Yashkun participants in the present study have been made available to GenBank under MK118774-MK118868 accession number and will be available for the public for future scientific use.