Abstract
Osteosarcoma (OS) is the most common primary malignant bone tumor. Recent evidence suggests that the novel long noncoding RNA DHRS4 antisense RNA 1 (DHRS4-AS1) serves an important role in cancer progression and metastasis. However, its function and molecular mechanism in OS remain largely unknown. In the present study, DHRS4-AS1 expression was detected in OS cells by quantitative PCR. Gain- and loss-of-function experiments were conducted to study the effects of DHRS4-AS1 on the proliferation and apoptosis of OS cells. The potential mechanism of DHRS4-AS1 was examined through bioinformatics analysis and rescue experiments. DHRS4-AS1 was downregulated in OS cell lines. DHRS4-AS1 depletion promoted proliferation and inhibited apoptosis in OS cells, whereas DHRS4-AS1 overexpression had the opposite effects. Further research suggested that DHRS4-AS1 inhibited OS progression by regulating the microRNA-362-5p/aminopeptidase puromycin sensitive axis. The present findings suggested that DHRS4-AS1 may serve as a potential therapeutic target for OS.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-87246-7.
Keywords: Osteosarcoma, DHRS4-AS1, Long noncoding RNAs, MiR-362-5p, NPEPPS
Subject terms: Bone cancer, Tumour biomarkers
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor in children and adolescents, and is associated with high invasion and early metastasis1. Furthermore, 10–25% of patients will have lung metastasis, and lung injury is the main cause of death of OS2,3. For patients with metastatic OS, the survival rate is < 30%4. The high heterogeneity of OS makes it difficult to understand its molecular mechanism5. The effective molecular mechanism of OS occurrence and development has not been fully revealed. Identification of key functional molecules in OS is urgently required to develop effective preventive measures and reasonable treatment methods for OS.
Long noncoding RNAs (lncRNAs) are a class of RNA polymerase II transcripts with > 200 nucleotides6. lncRNAs can regulate the biological processes of numerous diseases, including cancer7. One of their mechanisms is to ‘adsorb’ microRNAs (miRNAs/miRs) and competitively inhibit their biological functions8. DHRS4-AS1 is a newly identified lncRNA and belongs to a natural antisense transcript with an enhancer located 13.8 kb downstream of the transcription initiation site of the dehydrogenase/reductase 4 (DHRS4) gene9. The human DHRS4 gene can encode an NADPH-dependent enzyme belonging to the short chain dehydrogenase/reductase family10,11. With the identification of this novel lncRNA, several studies have demonstrated that DHRS4-AS1 serves a regulatory role in tumor development12–15; however, there is insufficient research on the related mechanism and, to the best of our knowledge, there is no research on DHRS4-AS1 in OS.
Therefore, the purpose of the present study was to examine the expression levels and mechanism of DHRS4-AS1 in OS and to provide evidence for further research on OS. The present study revealed that the expression levels of DHRS4-AS1 were downregulated in OS. Overexpression of DHRS4-AS1 inhibited the proliferation of OS cells and promoted their apoptosis. Subsequently, using bioinformatics methods and multiple databases, it was predicted that DHRS4-AS1 may have a function similar to that of a competing endogenous RNA (ceRNA). Through experiments, the present study revealed that it can regulate the proliferation and apoptosis of OS cells via the miR-362-5p/aminopeptidase puromycin sensitive (NPEPPS) axis.
Materials and methods
Cell culture. The hFOB1.19 human osteoblast cell line (CL-0353), and the U2-OS (CL-0236) and MG63 (CL-0157) human OS cell lines were purchased from Procell Life Science & Technology Co., Ltd. The 143B human OS cell line was purchased from iCell Bioscience Inc, Shanghai iCell-h231). hFOB1.19 cells were cultured in DMEM/F12 (SH30126.01; HyClone; Cytiva) containing 10% FBS (10099; Thermo Fisher Scientific, Inc.) and 0.3 mg/ml G418 (PB180125; Procell Life Science & Technology Co., Ltd.) with 5% CO2 at 37˚C. The U2-OS, MG63 and 143B OS cell lines were cultured in RPMI 1640 medium (SH30255.01; HyClone; Cytiva) containing 10% FBS (Thermo Fisher Scientific, Inc.) with 5% CO2 at 37˚C.
Cell transfection and infection. A short hairpin RNA (shRNA/sh) sequence specific for human DHRS4-AS1 and the negative sequence were purchased from GeneChem Co., Ltd (Shanghai, China). The full length of DHRS4-AS1 (ID: ENSG00000215256) was constructed into virus DNA by GeneChem Co., Ltd, control cells were infected with virus with empty vector. Both shRNAs were transfected into OS cell lines with an MOI of 20. Subsequently, Transfection at room temperature for 72 h, the cells were continuously cultured for 1 week using complete medium containing puromycin (ST551; 10 µg/ml; Beyotime Institute of Biotechnology) to construct cell lines with stable DHRS4-AS1 knockdown or overexpression. The miR-362-5p mimic and mimic negative control (NC) were synthesized and provided by Sangon Biotech Co., Ltd. The pCMV-NPEPPS vector for upregulation of the NPEPPS gene was obtained from Sangon Biotech Co., Ltd. OS cells were transfected with the aforementioned oligonucleotides and plasmids using Lipofectamine 2000 reagent (11668-019; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. Briefly, the OS cells 5 × 104 were seeded in a 6-well plate and infected when the cells reach a confluence of 70%. Lipofectamine 2000 and overexpression plasmid/empty plasmid were diluted in the culture medium respectively and then mixed in a 1:1 ratio, 300ul of mixed solution was added to the 6-well plate. After incubation for 8 h, fresh culture medium was replaced to culture the cells for another 48 h. qPCR wsa used to verify the success or failure of gene modification. During the in vitro experiment, the following groups were established: DHRS4-AS1 group, in which DHRS4-AS1 was overexpressed through lentiviral transfection; shDHRS4-AS1 group, in which DHRS4-AS1 was silenced through lentiviral transfection. Negative control (NC) group, which only underwent lentiviral transfection with negative sequence/empty vector, and blank control (BC) group, which received no treatment. In the miR-362-5p mimic group (mm), miR-362-5p was overexpressed through miR-362-5p mimic transfection. Cells in the mimic NC group (mNC) underwent the same treatment without miR-362-5p overexpression. In the DHRS4-AS1 + miR-362-5p mimic group (D + mm), both DHRS4-AS1 and miR-362-5p were overexpressed. In the DHRS4-AS1 + mimic NC group (D + mNC), DHRS4-AS1 was overexpressed but not miR-362-5p. In the pCMV NPEPPS group (pN), NPEPPS was overexpressed using the pCMV NPEPPS vector. In the miR-362-5p mimic + pCMV NPEPPS group (mm + pN), both miR-362-5p and NPEPPS were overexpressed using miR-362-5p mimic transfection and the pCMV NPEPPS vector. The sequences of all shRNAs and miRNA mimics are provided in Supplementary Table 1.
Quantitative PCR (qPCR). Total RNA was isolated from tissues and cells using TRIzol reagent (10296-028; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. The ImProm-IITM Reverse Transcription System (A3800; Promega Corporation) was then used to reverse transcribe total RNA (1 µg) into cDNA. Target primers were amplified using SYBR Green Mix (C11733046; Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of the primers are listed in Supplementary Table 2. All primers were synthesized by Sangon Biotech Co., Ltd. The miR-362-5p level was measured using the TaqMan MicroRNA Assay Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions. All results were calculated and presented as 2-ΔΔCT 16. The expression of the small noncoding RNA U6 was used as an endogenous control for miR-362-5p, and GAPDH was used as an endogenous control for DHRS4-AS1. Each experiment was conducted in triplicate.
Western blotting. Total protein was extracted by whole cell lysis (KGP701; Nanjing KeyGen Biotech Co., Ltd.). Protein samples (100 µg) were subjected to SDS-PAGE (KGP113; Nanjing KeyGen Biotech Co., Ltd.) and transferred to PVDF membranes (IPVH00010; MilliporeSigma). The membranes were blocked with 5% skim milk for 1 h and incubated with the primary antibody at 4˚C for 12 h. The membranes were then incubated with the secondary antibody (horseradish peroxidase conjugated; S598224; 1:500; Aladdin Biochemical Technology Co., Ltd) for 2 h at room temperature, and the expression of the target protein was detected using the ECL method (KGP1121; Nanjing KeyGen Biotech Co., Ltd.). The following antibodies were used: NPEPPS (GTX88877; 1:1,000; GeneTex International Corporation), caspase 3 (9662 S; 1:800; Cell Signaling Technology, Inc.), Bcl2 (15071 S; 1:800; Cell Signaling Technology, Inc.), BAX (5023 S; 1:800; Cell Signaling Technology, Inc.) and Actin (AF0003; 1:1,000; Beyotime Institute of Biotechnology).
Cell proliferation analysis via Cell Counting Kit-8 (CCK-8) assays. The cells to be tested (~ 1,000 cells/well) were inoculated in 96-well plates (three replicate wells per group), with a total medium volume of 100 µl. CCK-8 solution reagent (10 µl; Dojindo Laboratories, Inc.) was then added to each well every 24 h, and the absorbance value (optical density) was measured at 450 nm after incubation at 37˚C for 2 h.
TUNEL cell apoptosis analysis. Cells were treated with 4% paraformaldehyde (P0099; Beyotime Institute of Biotechnology) and Triton X-100 (ST797; Beyotime Institute of Biotechnology) at room temperature for 15 and 5 min, respectively. The TUNEL test solution (C1088; Beyotime Institute of Biotechnology) was prepared according to the manufacturer’s instructions. The cells were then incubated with TUNEL test solution in the dark at 37˚C for 60 min and treated with Hoechst (C1101; Beyotime Institute of Biotechnology). The nuclei and TUNEL-positive cells were observed under a fluorescence microscope (Nikon Corporation), and the positive rate (TUNEL/Hoechst) of five random fields was calculated.
Grouping and treatment of nude mice. A total of nine BALB/C nude mice (~ 4 weeks old, male, 15 g) were purchased from the Animal Center of Southern Medical University. Nude mice are kept in an environment where the temperature is maintained between 20 and 26 °C, the humidity is maintained between 40 and 70%, there is 12 h of light and 12 h of darkness, and there is sufficient and high quality food and water. The nude mice were randomly divided into three groups: DHRS4-AS1 knockdown group (shDHRS4-AS1), NC group and BC group. Cells (~ 2 × 106) were subcutaneously injected into the left front leg of nude mice. Observe the formation of subcutaneous tumors once a day. The longest and shortest diameters of transplanted tumors were measured with a Vernier caliper every 5 days, the volume of subcutaneous transplanted tumors (V = L x W2 x 0.5) was calculated, and the tumor growth curve was drawn. At 1 month after the injection, the nude mice were sacrificed. Without pre-charging the chamber, place the mice in the chamber and introduce 100% CO2 at a fill rate of 50% displacement of the chamber volume per minute with CO2, added to the existing air in the chamber. Then, tumor tissue was removed, weighed and imaged. All animal experiments were approved by the Ethics Committee of the Zhujiang Hospital of Southern Medical University (LAEC-2022-043; Guangzhou, China), and the studies were conducted in accordance with the approved guidelines and regulations. Furthermore, this study is reported in accordance with ARRIVE guidelines.
Immunohistochemistry. After dewaxing and dehydration, the sections were blocked with 3% hydrogen peroxide (C804187; Macklin) to block endogenous peroxidase activity. Subsequently, sections were incubated with primary antibodies against caspase 3 (PA5-87536, 1:100, ThermoFisher), Bcl2 (120158, 1:50, Zen BioScience) and BAX (AF0120, 1:50, Affinity Biosciences) overnight at 4˚C, and biotinylated secondary antibodies (K21034M-Bio-100ul, 1:200, Solarbio) at 37˚C for 20 min, and incubated with 3,3′-diaminobenzidine (P0202; Beyotime Institute of Biotechnology). The images were captured by microscopy (Leica Microsystems GmbH). Three fields were randomly selected, and the average percentage of brown or dark yellow particles in the cytoplasm was determined.
Data processing of bioinformatics analysis. The OS-related chip data (GSE28423) were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo). The GSE28423 was uploaded by Department of Tumor Biology, Institute for cancer research, The Norwegian Radium Hospital, Oslo University Hospital. The differentially expressed genes in OS were analyzed with P < 0.05 and log fold change > 1.5 as the threshold using the limma package (Version 3.60.0, https://bioconductor.org/packages/release/bioc/html/limma.html ) in R software (R version 3.3.0, http://www.r-project.org/). The target miRNA of DHRS4-AS1 was predicted using StarBase (Version 2.0, https://starbase.sysu.edu.cn/starbase2/index.php) and compared with the upregulated miRNAs in the GSE28423 dataset. The dysregulated miRNAs were used for future analysis. Subsequently, the target mRNAs of dysregulated miRNAs were predicted using miRDB (Version 6.0, http://mirdb.org/), TargetScan (Release 8.0, http://www.targetscan.org/) and mirTarBase (Release 9.0 beta, https://mirtarbase.cuhk.edu.cn/). Finally, the common predicted target mRNAs were identified. Combined with the aforementioned key modules and lncRNA network, a DHRS4-AS1-dependent lncRNA subnetwork was constructed. The mRNAs were predicted using miRDB, TargetScan and mirTarBase, and all genes are shown in the turquoise module. Finally, the miRNA-mRNA-lncRNA regulatory network was visualized using Cytoscape software (Version 3.7.2, https://cytoscape.org/).
Receiver operating characteristic (ROC) curve analysis. By calculating the sensitivity and specificity, the diagnostic value of target mRNAs in patients with OS was evaluated using ROC curve analysis. The satisfactory discrimination performance was evaluated based on the area under the curve (AUC) of the ROC curve, which was > 0.7.
Survival, univariate and multivariate analyses. The overall survival of the patients with OS was predicted by Kaplan-Meier survival analysis using the survival R package (Version 2.40–5, https://github.com/therneau/survival). The differences in overall survival time between the two groups were compared using the stratified log-rank test. The Cox proportional hazards regression model was used to explore the univariate and multivariate hazard ratios and screen the OS prognostic factors. The independent clinicopathological factors and the potential prognostic biomarkers were subsequently detected by univariate and multivariate Cox proportional hazards regression model analysis. The patients were divided into high- and low-risk groups according to the risk score in the hazards model and the risk prediction ability of the target genes in the two groups was detected using a time-dependent ROC curve.
Statistical analysis. All data were analyzed using SPSS 22.0 software (IBM Corp.). The qualitative data are presented as counts (%) and the quantitative data are presented as the mean ± standard deviation. Multiple groups of data were analyzed using one-way analysis of variance. Tukey’s test was used to further explore the between-group differences shown by the ANOVA results. P < 0.05 was considered to indicate a statistically significant difference.
Results
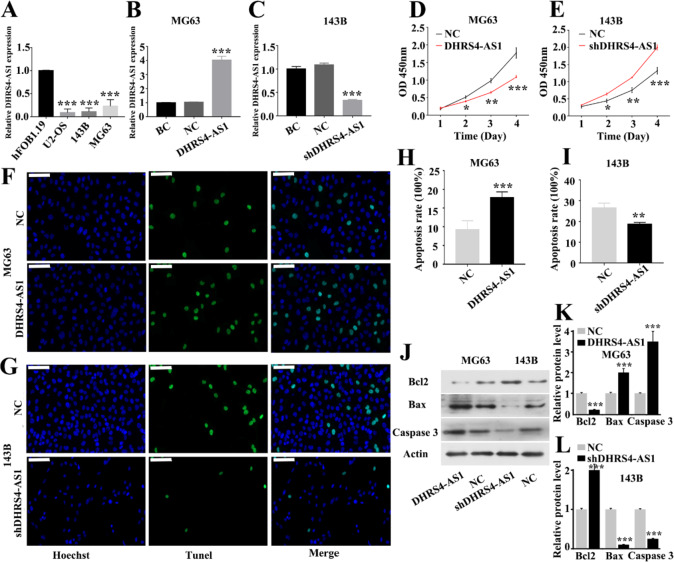
DHRS4-AS1 inhibits OS cell proliferation and promotes apoptosis in vitro. To confirm DHRS4-AS1 expression in OS, the difference in the expression levels of DHRS4-AS1 between OS cell lines and the hFOB1.19 human osteoblast cell line was first detected. DHRS4-AS1 expression was significantly downregulated in the MG63, 143B and U2-OS OS cell lines compared with the hFOB1.19 osteoblast cell line (Fig. 1A). Preliminary experimental screening revealed that MG63 and 143B cells were effective and stable in constructing DHRS4-AS1 overexpression (Fig. 1B) and DHRS4-AS1 silencing models (Fig. 1C). Therefore, the effects of DHRS4-AS1 on cell proliferation and apoptosis were subsequently validated using MG63 and 143B cells. CCK-8 assays revealed a decreased proliferation rate in the MG63 cells transduced with DHRS4-AS1 overexpression vectors compared with that of the NC group (Fig. 1D). Furthermore, TUNEL results demonstrated that upregulation of DHRS4-AS1 increased the apoptosis of MG63 cells (Fig. 1F and H). By contrast, downregulation of DHRS4-AS1 significantly inhibited the apoptosis of 143B cells and promoted cell proliferation (Fig. 1E, G and I). Subsequently, the protein expression levels of Bcl2, Bax and caspase 3 were detected in the cells of each group. The results suggested that overexpression of DHRS4-AS1 could upregulate Bax and caspase 3 expression and inhibit Bcl2 expression in MG63 cells. In 143B cells, low DHRS4-AS1 expression had the opposite regulatory effect (Fig. 1J-L).
Fig. 1.
Regulation of DHRS4-AS1 on OS cell proliferation and apoptosis in vitro. (A) Differential expression of DHRS4-AS1 in human osteoblast cell line and three OS cell lines, *Represent comparison with the hFOB1.19 group. (B) Overexpression expression of DHRS4-AS1 in MG63, *Represent comparison with the BC group. (C) DHRS4-AS1 silencing models in 143B cells, *Represent comparison with the BC group. (D) CCK-8 results showing proliferation level over 4 consecutive days for MG63 cells in each group, *Represent comparison with the NC group. (E) Proliferation level for 143B cells in each group, *Represent comparison with the NC group. (F) Tunel results showing apoptosis level for MG63 cells in each group. (G) Tunel results showing apoptosis level for 143B cells in each group. (H) Quantitative analysis of apoptosis rate of MG63 cells in different groups, *Represent comparison with the NC group. (I) Quantitative analysis of apoptosis rate of 143B cells in different groups, *Represent comparison with the NC group. (J) Apoptosis-related protein levels of MG63 and 143B cells in each group were determined using western blot. (K) Quantitative analysis of apoptosis-related protein in MG63 cells, *Represent comparison with the NC group. (L) Quantitative analysis of apoptosis-related protein in 143B cells, *Represent comparison with the NC group. *p<0.05, **p<0.01, ***p<0.001, the scale is 100 μm. BC and NC represent Blank Control and Negative Control respectively.
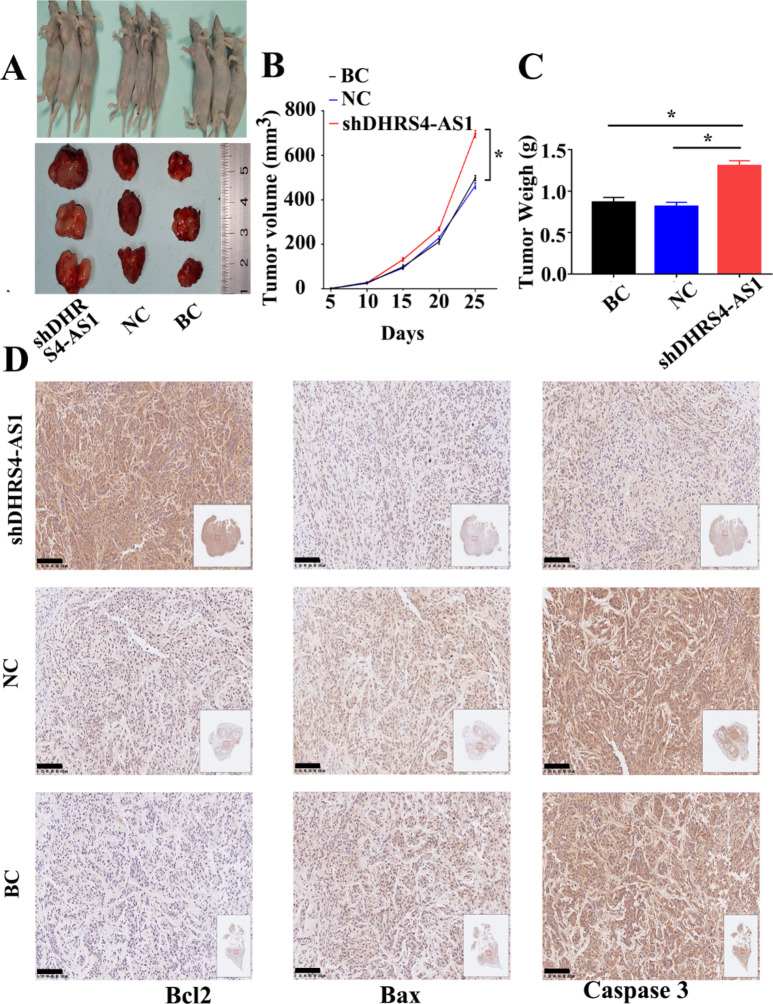
DHRS4-AS1 inhibits OS cell proliferation and promotes apoptosis in vivo. To confirm these findings in vivo, 143B cells were implanted into nude mice, and the effects of DHRS4-AS1 were assessed after 25 days of treatment (Fig. 2A). The tumor size (Fig. 2B) and weight (Fig. 2C) were measured. Compared with those of the control group, low DHRS4-AS1 expression increased the size and weight of tumors. The preliminary qualitative analysis by immunohistochemical staining indicated that the expression levels of Bcl2 in the shDHRS4-AS1 group were higher than those in the control group, while the expression levels of Bax and caspase 3 in the shDHRS4-AS1 group were lower than those in the control group (Fig. 2D).
Fig. 2.
Regulation of DHRS4-AS1 on OS cell proliferation and apoptosis in vivo. (A) Representative photo of nude mouse tumors. (B) The tumor size were measured after different treatments. (C) The weight were measured. (D) Immunohistochemical staining of apoptosis related proteins in each group. *p<0.05, the scale is 100 μm. BC and NC represent Blank Control and Negative Control respectively.
Comprehensive analysis identified DHRS4-AS1 as a ceRNA for miR-362-5p/NPEPPS in OS. To further study the specific mechanism by which DHRS4-AS1 regulates the progression of OS cells, a bioinformatics method was used to predict the potential downstream target genes of DHRS4-AS1. First, differentially expressed miRNAs were obtained by analyzing the OS GSE28423 dataset, the target miRNA of DHRS4-AS1 was predicted using the StarBase database, and comparison with the differentially expressed miRNAs showed that miR-330-3p and miR-362-5p might be the potential targets of DHRS4-AS1 (Fig. 3A). Subsequently, the target mRNAs of miR-330-3p and miR-362-5p were predicted by miRDB, TargetScan and miRTarBase. The prediction results of the three databases were overlappeded to obtain 30 target genes (Fig. 3B). Combined with the aforementioned key lncRNA network, a DHRS4-AS1-dependent lncRNA subnetwork was constructed (Fig. 3C). To further screen the target genes of DHRS4-AS1, a signature model and risk score were constructed based on single-factor regression analysis, clinical characteristics and the genes in the aforementioned modules. Among them, only the P-values of capping actin protein of muscle Z-line subunit α1 (CAPZA1), F-box and leucine rich repeat protein 17 (FBXL17) and NPEPPS were < 0.05 in univariate Cox regression analysis (Fig. 3D). The P-value of programmed cell death 4 (PDCD4) was < 0.05 in the multivariate Cox regression analysis (Fig. 3E). Based on the risk score, the Kaplan-Meier method was used to testthe association between high risk and low survival. The AUC value of the time-dependent ROC prediction model was 0.73 (> 0.7), and the prediction model was relatively good (Fig. 3F).
Fig. 3.
Prediction and Functional enrichment analysis of DHRS4-AS1 target genes. (A) Venn diagram representing the number of up-regulated miRNAs in GSE28423 and potential target genes of DHRS4-AS1 predicted by starbase. (B) Venn diagram representing the number of target mRNAs of miR-330-3p and miR-362-5p predicted by miRDB, TarScan and mirTarBase. C DHRS4-AS1-dependent lncRNA subnetwork. (D-E) univariate Cox regression and multivariate Cox regression of potential target genes. (F) Verify the accuracy of the model based on risk score (AUC > 0.7).
Based on the aforementioned results, the key target genes were identified by analyzing the association between lncRNAs and target genes. The CAPZA1, FBXL17 and NPEPPS genes were expressed at low levels in OS tissues and had a high risk of death, and PDCD4 was expressed at high levels and had a high risk of death. There was no difference in CAPZA1 expression. Subsequently, the association between survival and target gene expression was analyzed. The results demonstrated that FBXL17 (Fig. 4B) and NPEPPS (Fig. 4C) were expressed at low levels in OS tissues and had a high risk. CAPZA1 (Fig. 4A) and PDCD4 (Fig. 4D) showed no difference in prognostic effects. Finally, the GSE126209 dataset was used to verify the change in expression of the aforementioned predicted target genes. Among them, only NPEPPS was differentially expressed at low levels, so FBXL17 was screened out. The AUC value of NPEPPS in ROC detection was > 0.8, indicating NPEPPS has high sensitivity and specificity for predicting mortality in osteosarcoma (Fig. 4E and F).
Fig. 4.
Construction of DHRS4-AS1 dependent ceRNA network and screening of target genes of DHRS4-AS1. (A-D) Identification of interaction correlation and survival analysis of target genes. (E-F) Dataset validation analysis of NPEPPS and FBXL17 based on GSE126209 dataset.
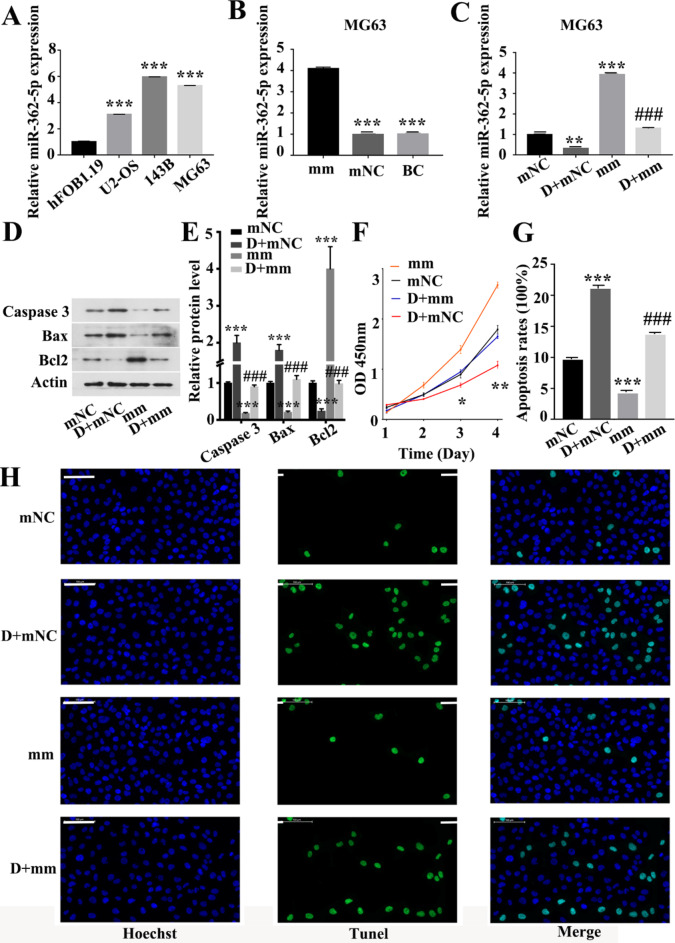
DHRS4-AS1 directly interacts with miR-362-5p and represses its expression. After obtaining the potential target gene of DHRS4-AS1, the expression levels of miR-362-5p in OS cell lines and the hFOB1.19 human osteoblast cell line were first verified by qPCR. The results demonstrated that the expression levels of miR-362-5p in OS cells were significantly upregulated (Fig. 5A). After overexpressing miR-362-5p in OS MG63 cells (Fig. 5B), DHRS4-AS1 and miR-362-5p mimic were co-transfected into MG63 cells and the regulation of miR-362-5p by DHRS4-AS1 was examined. miR-362-5p expression could be negatively regulated by DHRS4-AS1 (Fig. 5C). To determine whether DHRS4-AS1 mediates OS cell proliferation and apoptosis via miR-362-5p, the expression levels of cell apoptosis-related proteins were examined by western blotting. The results showed that overexpression of DHRS4-AS1 upregulated Bax and caspase 3 expression and downregulated Bcl2 expression. Furthermore, overexpression of miR-362-5p resulted in the opposite trend. Overexpression of miR-362-5p attenuated the protein expression changes mediated by upregulation of DHRS4-AS1 in MG63 cells (Fig. 5D and E). The present study determined the effect of miR-362-5p on OS cell proliferation and apoptosis. The results demonstrated that overexpression of miR-362-5p could promote the proliferation of OS cells and inhibit the apoptosis of OS cells. Furthermore, overexpression of DHRS4-AS1 inhibited the decrease in proliferation and increase in apoptosis caused by upregulation of miR-362-5p (Fig. 5F-H). These results indicated that DHRS4-AS1 and miR-362-5p had opposite regulatory effects on the proliferation and apoptosis of OS cells, and DHRS4-AS1 could inhibit the regulatory effect of miR-362-5p on the proliferation and apoptosis of OS cells.
Fig. 5.
DHRS4-AS1 directly interacts with miR-362-5p and represses its expression. (A) The expression level of miR-362-5p in osteosarcoma cell lines and the human osteoblast cell line, *Represent comparison with the Hfob1.19 group. (B) The expression level of miR-362-5p in MG63 cells was increased after transfection with miR-362-5p mimic, *Represent comparison with the mm group. (C) After co-transfection with DHRS4-AS1 and miR-362-5p mimic, DHRS4-AS1 reversed the high expression of miR-362-5p in MG63 cells, *Represent comparison with the mNC group, # represents comparison with the mm group. (D-E) Expression levels of apoptosis-related proteins in MG63 cells, *Represent comparison with the mNC group, # represents comparison with the mm group. (F) cck8 assay was performed to detect the proliferation and apoptosis levels of MG63 cells after co-transfection with DHRS4-AS1 and miR-362-5p mimic, *Represent comparison with the mNC group, # represents comparison with the mm group. (G) Quantification of apoptosis rate in MG63 cells after co-transfection with DHRS4-AS1 and miR-362-5p mimic, *Represent comparison with the mNC group, # represents comparison with the mm group. (H) Tunel detection of apoptosis in MG63 cells after co-transfection with DHRS4-AS1 and miR-362-5p mimic. *p<0.05, **p<0.01, ***p<0.001, ###p<0.001, the scale is 100 μm. mm represent miR-362-5p mimic group. mNC represent the mimic Negative Control group. D + mm represents the DHRS4-AS1 + miR-362-5p mimic group, indicating that both DHRS4-AS1 and miR-362-5p are overexpressed. D + mNC represents the DHRS4-AS1 + mimic Negative Control group, indicating that DHRS4-AS is overexpressed but not expressing miR-362-5p. BC represents Blank Control group.
miR-362-5p mediates the proliferation and apoptosis of OS cells through NPEPPS. To determine whether miR-362-5p mediates OS cell proliferation and apoptosis via NPEPPS, low NPEPPS expression in OS cells was first confirmed by western blotting (Fig. 6A and B). NPEPPS was overexpressed in MG63 OS cells using pCMV NPEPPS (Fig. 6C and D). After miR-362-5p and NPEPPS were overexpressed in MG63 cells, the expression levels of cell apoptosis-related proteins were examined by western blotting. The results demonstrated that overexpression of miR-362-5p downregulated the expression levels of NPEPPS, Bax and caspase 3, and upregulated Bcl2 expression in MG63 cells, and this trend was reversed by overexpression of NPEPPS (Fig. 6E and F). Subsequently, the effect of NPEPPS on OS cell proliferation and apoptosis was determined. The results demonstrated that overexpression of NPEPPS could promote apoptosis and inhibit proliferation of MG63 cells. Furthermore, overexpression of NPEPPS attenuated the changes in apoptosis and proliferation mediated by overexpression of miR-362-5p (Fig. 6G-I).
Fig. 6.
MiR-362-5p mediates the proliferation and apoptosis of OS cells through NPEPPS. (A-B) The expression level of NPEPPS in osteosarcoma cell lines and the human osteoblast cell line, *Represent comparison with the Hfob1.19 group. (C-D) NPEPPS was overexpressed in MG63 OS cells, *Represent comparison with the pN group. (E-F) the expression level of NPEPPS and poptosis related protein in MG63 cells after cotransfected miR-362-5p mimic and NPEPPS, *Represent comparison with the NC group. (G) The Tunel assay was used to successfully detect apoptosis in MG63 cells after co-transfection with miR-362-5p mimetics and NPEPPS. (H) The CCK-8 assay was employed to determine the proliferation and apoptosis levels of MG63 cells co-transfected with miR-362-5p mimetics and NPEPPS, *Represent comparison with the NC group. (I) The apoptosis rate in MG63 cells co-transfected with miR-362-5p mimic and NPEPPS was quantified, *Represent comparison with the NC group, # represents comparison with the pN group. *p<0.05, **p<0.01, ***p<0.001, #p<0.01, ##p<0.001, ###p<0.001, the scale is 100 μm. NC represents Negative Control group. BC represents Blank Control group. mm represent miR-362-5p mimic group. pN represents the pCMV NPEPPS group, which overexpresses NPEPPS using the pCMV NPEPPS vector. mm + pN represents the miR-362-5p mimetic + pCMV NPEPP group, indicating the overexpression of both miR-362-5p and NPEPPS using miR-362-5p mimetic transfection and pCMV NPEP vector.
Discussion
There is still limited research on DHRS4-AS1 and, to the best of our knowledge, no studies have been conducted on its expression in OS, which attracted our interest. Therefore, in the present study, qPCR was first used to detect the expression levels of DHRS4-AS1 in OS cell lines. The results suggested that, compared with in human osteoblasts, DHRS4-AS1 was significantly downregulated in three OS cell lines. The present study confirmed the downregulation of DHRS4-AS1 expression in OS cells, which is consistent with existing literature reporting that DHRS4-AS1 expression is downregulated in tumor tissues12,15. Zhou et al.17demonstrated that lncRNA DHRS4-AS1 inhibited liver cancer cell proliferation and promoted apoptosis via the miR-522-3p/SOCS5 signaling axis. Another study demonstrated that lncRNA DHRS4-AS1 inhibited the proliferation, migration and invasion of endometrial cells in endometriosis by regulating microRNA-139-5p expression18. A study by Wang et al.13 revealed that DHRS4-AS1 served a role in inhibiting the proliferation and invasion of clear cell renal cell carcinoma, arresting the cell cycle process, and promoting cell apoptosis. The present experiments revealed similar results, with the overexpression of DHRS4-AS1 inhibiting cell proliferation and increasing the level of cell apoptosis.
At present, most cancer treatment methods aim to eliminate malignant cells by inducing cell apoptosis. Among them, Bcl-2 is one of the key resistance targets. Bcl2 is an anti-apoptotic protein that inhibits cell apoptosis. Zhang et al.19demonstrated that miR-146a suppressed the proliferation of MC3T3-E1 mouse osteoblasts and induced apoptosis by reducing Bcl2 expression. Other research has shown that miRNA-449a promoted OS cell apoptosis by downregulating Bcl2 expression20. Bax and caspase 3 proteins have pro-apoptotic effects. Zou et al.21 revealed that deoxyhydroxyroasted saponin induced apoptosis in OS cells by activating the apoptosis-related proteins Bax and caspase 3. The present results demonstrated that overexpression of DHRS4-AS1 could inhibit Bcl2 and promote the expression of Bax and caspase 3, indicating that DHRS4-AS1 could promote OS cell apoptosis.
Based on this, we hypothesized that DHRS4-AS1 expression is downregulated in OS, and DHRS4-AS1 serves as a tumor suppressor gene that regulates the biological functions of OS cells. However, the regulatory mechanism behind this remains unclear. Therefore, in order to further investigate the regulatory mechanism of DHRS4-AS1 in the development of OS, bioinformatics methods were employed to analyze differential genes in gene chips and perform enrichment analysis. Bioinformatics screening was used to predict potential target genes of DHRS4-AS1, which were then validated and studied. Ultimately, only NPEPPS was found to be downregulated and differentially expressed in OS. NPEPPS is a cytosolic aminopeptidase that is highly expressed in the central nervous system. Several studies have reported that NPEPPS is a neurodegenerative regulatory protein induced by τ protein. NPEPPS exerts a neuroprotective effect by directly hydrolyzing τ protein22–24, and the mechanism may involve NPEPPS regulating τ protein by post-translational modification25. This effect is related to the autophagy system and some neurotoxic targets26. At present, research on NPEPPS is limited and, to the best of our knowledge, there is no research report on NPEPPS in human tumors. The results of the present study demonstrated significant downregulation of NPEPPS expression in OS. Overexpression of NPEPPS significantly inhibited the proliferation of OS cells and increased the level of cell apoptosis, and thus, NPEPPS is a potential inhibitor of OS progression. Therefore, this experiment has added to the understanding of the role of NPEPPS in OS.
To the best of our knowledge, the present study was the first to report the regulatory role of the DHRS4-AS1/miR-362-5p/NPEPPS axis in OS. However, there is still limited research on the mechanism of miR-362-5p in OS. Therefore, the present study investigated the regulatory effect of miR-362-5p on OS cell proliferation and apoptosis. The present results indicated that miR-362-5p could reverse the role of NPEPPS in promoting Bax and caspase 3 and inhibiting Bcl-2, thereby promoting OS cell proliferation and inhibiting apoptosis. miR-362-5p has been reported to promote tumor growth in various cancer types27–30, which is consistent with the present findings.
Although a lot of work has been done, there was still room for further research. First, we have not collected enough clinical specimens for the experiment in the past two years, and currently there are less than 10 cases. because of the insufficient clinical sample size, the present study did not include investigations of clinical cases. Second, the research on the cellular function and molecular mechanism of DHRS4-AS1 is not comprehensive, and further research is needed to verify the role of DHRS4-AS1 in the occurrence and development of OS. Furthermore, cell lines are unable to fully replicate the intricacies of the in vivo environment. Further in vivo model studies are required to validate the findings presented here.
Conclusions
DHRS4-AS1 served as a tumor suppressor gene that regulated the proliferation and apoptosis of OS cells. DHRS4-AS1 exerted its tumor suppressor effect by regulating the DHRS4-AS1/hsa-miR-362-5p/NPEPPS axis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
ZZT, ZHL and GFW performed experiments and completed the writing of the first draft of the article. LXZ proposed ideas and analyzed the data and perfected the article. JJL and JYT analyzed and interpreted data. All authors read and approved the final manuscript. ZZT, ZHL and GFW contribute equally to this work.
Funding
This research was funded by the president funding of Zhujiang Hospital (yzjj2021qn04).
Data availability
The datasets generated and/or analysed during the current study are available in the science data bank repository with the primary accession code CC BY-NC-SA 4.0. https://www.scidb.cn/detail? dataSetId=46440f74023a470e919a8068cf7fabcf.
Declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Ethics approval
The study was approved by the Ethics Committee of the Zhujiang Hospital of Southern Medical University (LAEC-2022-043).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhouzhou Tang, Zhihao Li and Guofeng Wu contributed equally.
References
- 1.Sadykova, L. R. et al. Epidemiology and risk factors of Osteosarcoma. Cancer Invest.38, 259–269. 10.1080/07357907.2020.1768401 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Chen, C. et al. Immunotherapy for osteosarcoma: fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett.500, 1–10. 10.1016/j.canlet.2020.12.024 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Lin, W. et al. Analysis of miR-148b expression differences in stage-I and II parosteal osteosarcoma. Oncol. Lett.16, 998–1002. 10.3892/ol.2018.8709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park, J. A. & Cheung, N. V. Promise and challenges of T Cell Immunotherapy for Osteosarcoma. Int. J. Mol. Sci.2410.3390/ijms241512520 (2023). [DOI] [PMC free article] [PubMed]
- 5.Heymann, M. F., Brown, H. K. & Heymann, D. Drugs in early clinical development for the treatment of osteosarcoma. Expert Opin. Investig. Drugs. 25, 1265–1280. 10.1080/13543784.2016.1237503 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Chen, M. et al. Decreased expression of lncRNA VPS9D1-AS1 in gastric cancer and its clinical significance. Cancer Biomark. A. 21, 23–28. 10.3233/CBM-170172 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Bracken, C. P., Scott, H. S. & Goodall, G. J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet.17, 719–732. 10.1038/nrg.2016.134 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Li, Z. et al. Long non-coding RNA ATB promotes malignancy of esophageal squamous cell carcinoma by regulating miR-200b/Kindlin-2 axis. Cell Death Dis.8, e2888. 10.1038/cddis.2017.245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, Y. et al. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci. Rep.6, 20961. 10.1038/srep20961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsunaga, T. et al. Characterization of human DHRS4: an inducible short-chain dehydrogenase/reductase enzyme with 3beta-hydroxysteroid dehydrogenase activity. Arch. Biochem. Biophys.477, 339–347. 10.1016/j.abb.2008.06.002 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Huang, D. Y. & Ichikawa, Y. Purification and characterization of a novel cytosolic NADP(H)-dependent retinol oxidoreductase from rabbit liver. Biochim. Biophys. Acta. 1338, 47–59. 10.1016/s0167-4838(96)00183-5 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Utnes, P., Lokke, C., Flaegstad, T. & Einvik, C. Clinically relevant Biomarker Discovery in High-Risk recurrent neuroblastoma. Cancer Inform.18, 1176935119832910. 10.1177/1176935119832910 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, C. et al. The downregulated long noncoding RNA DHRS4-AS1 is protumoral and associated with the prognosis of clear cell renal cell carcinoma. OncoTargets Therapy. 11, 5631–5646. 10.2147/OTT.S164984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luan, F. et al. An autophagy-related long non-coding RNA signature for glioma. FEBS open. Bio. 9, 653–667. 10.1002/2211-5463.12601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan, F. et al. LncRNA DHRS4-AS1 inhibits the stemness of NSCLC cells by sponging mir-224-3p and upregulating TP53 and TET1. Front. cell. Dev. Biology. 8, 585251. 10.3389/fcell.2020.585251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods25, 402–408. 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Zhou, Y. et al. LncRNA DHRS4-AS1 ameliorates hepatocellular carcinoma by suppressing proliferation and promoting apoptosis via miR-522-3p/SOCS5 axis. Bioengineered12, 10862–10877. 10.1080/21655979.2021.1994719 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui, X., Zhou, S. & Lin, Y. Long non-coding RNA DHRS4 antisense RNA 1 inhibits ectopic endometrial cell proliferation, migration, and invasion in endometriosis by regulating microRNA-139-5p expression. Bioengineered13, 9792–9804. 10.1080/21655979.2022.2060781 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, B. et al. MiR-146a inhibits proliferation and induces apoptosis in murine osteoblastic MC3T3-E1 by regulating Bcl2. Eur. Rev. Med. Pharmacol. Sci.21, 3754–3762 (2017). [PubMed] [Google Scholar]
- 20.Chen, J. et al. miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biology: J. Int. Soc. Oncodevelopmental Biology Med.36, 8221–8229. 10.1007/s13277-015-3568-y (2015). [DOI] [PubMed] [Google Scholar]
- 21.Zou, J. et al. Deoxyelephantopin induces reactive oxygen species-mediated apoptosis and autophagy in human osteosarcoma cells. Cell. Physiol. Biochemistry: Int. J. Experimental Cell. Physiol. Biochem. Pharmacol.42, 1812–1821. 10.1159/000479537 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Karsten, S. L. et al. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron51, 549–560. 10.1016/j.neuron.2006.07.019 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Yanagi, K. et al. Involvement of puromycin-sensitive aminopeptidase in proteolysis of tau protein in cultured cells, and attenuated proteolysis of frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) mutant tau. Psychogeriatrics: Official J. Japanese Psychogeriatr. Soc.9, 157–166. 10.1111/j.1479-8301.2010.00307.x (2009). [DOI] [PubMed] [Google Scholar]
- 24.Ren, G. et al. Cu, Zn-superoxide dismutase 1 (SOD1) is a novel target of puromycin-sensitive aminopeptidase (PSA/NPEPPS): PSA/NPEPPS is a possible modifier of amyotrophic lateral sclerosis. Mol. Neurodegeneration. 6, 29. 10.1186/1750-1326-6-29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta, S. et al. Degradation of tau protein by puromycin-sensitive aminopeptidase in vitro. Biochemistry45, 15111–15119. 10.1021/bi061830d (2006). [DOI] [PubMed] [Google Scholar]
- 26.Menzies, F. M. et al. Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum. Mol. Genet.19, 4573–4586. 10.1093/hmg/ddq385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, P. et al. MiR-362-5p promotes the malignancy of chronic myelocytic leukaemia via down-regulation of GADD45alpha. Mol. Cancer. 1410.1186/s12943-015-0465-3 (2015). [DOI] [PMC free article] [PubMed] [Retracted]
- 28.Wu, K. et al. Mir-362-5p inhibits proliferation and migration of neuroblastoma cells by targeting phosphatidylinositol 3-kinase-C2beta. FEBS Lett.589, 1911–1919. 10.1016/j.febslet.2015.05.056 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Wei, X. et al. MiR-362-5p, which is regulated by long non-coding RNA MBNL1-AS1, promotes the cell proliferation and tumor growth of bladder Cancer by targeting QKI. Front. Pharmacol.11, 164. 10.3389/fphar.2020.00164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, F. et al. Mir-362-5p promotes cell proliferation and cell cycle progression by targeting GAS7 in acute myeloid leukemia. Hum. Cell. 33, 405–415. 10.1007/s13577-019-00319-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the science data bank repository with the primary accession code CC BY-NC-SA 4.0. https://www.scidb.cn/detail? dataSetId=46440f74023a470e919a8068cf7fabcf.