Abstract
Inhibition of the deubiquitinating enzyme USP1 can induce synthetic lethality in tumors characterized by homologous recombination deficiency (HRD) and represents a novel therapeutic strategy for the treatment of BRCA1/2-mutant cancers, potentially including patients whose tumors have primary or acquired resistance to PARP inhibitors (PARPi). In this study, we present a comprehensive characterization of TNG348, an allosteric, selective, and reversible inhibitor of USP1. TNG348 induces dose-dependent accumulation of ubiquitinated protein substrates both in vitro and in vivo. CRISPR screens show that TNG348 exerts its antitumor effect by disrupting the translesion synthesis pathway of DNA damage tolerance through RAD18-dependent ubiquitinated PCNA. Although TNG348 and PARPi share the ability to selectively kill HRD tumor cells, CRISPR screens reveal that TNG348 and PARPi do so through discrete mechanisms. Particularly, knocking out PARP1 causes resistance to PARPi but sensitizes cells to TNG348 treatment. Consistent with these findings, combination of TNG348 with PARPi leads to synergistic antitumor effects in HRD tumors, resulting in tumor growth inhibition and regression in multiple mouse xenograft tumor models. Importantly, our data on human cancer models further show that the addition of TNG348 to PARPi treatment can overcome acquired PARPi resistance in vivo. Although the clinical development of TNG348 has been discontinued because of unexpected liver toxicity in patients (NCT06065059), the present data provide preclinical and mechanistic support for the continued exploration of USP1 as a drug target for the treatment of patients with BRCA1/2-mutant or HRD cancers.
Graphical Abstract
Introduction
Homologous recombination deficiency (HRD) is a DNA damage repair alteration frequently observed in ovarian and breast cancers and constitutes up to 50% and 20% of cases, respectively. Germline or somatic mutations in BRCA1 or BRCA2 genes are the most prevalent and pathologically significant alterations contributing to HRD and represent up to 15% of breast and ovarian cancer cases (1). HRD, through BRCA1/2 mutation or other means, is characterized by the inability of cells to repair DNA double-strand breaks (DSB) through homologous recombination (HR), increasing their reliance on alternate pathways of DNA repair. PARP inhibitors (PARPi) can exploit this deficiency to induce synthetic lethality in HRD tumors by causing DNA damage that is normally repaired by HR. PARPi blocks the base excision repair (BER) pathway, leading to the accumulation of DNA single-stranded breaks (SSB) that can be converted into DNA DSB during DNA replication. Importantly, normal nontumor cells retain the ability to repair DSB by HR and are largely spared by PARPi. Since their conception, additional mechanisms were identified contributing to the antitumor effect of PARPi in HRD cancers, including the trapping of PARP onto DNA (2, 3). The approval of PARPi in HRD and BRCA1/2-mutant cancers highlights the utility of synthetic lethal approaches in the targeted treatment of cancers. Despite the initial benefit of PARPi over historical standard-of-care therapies, their efficacy is limited by intrinsic and acquired drug resistance, highlighting the need for novel therapeutic strategies that can augment PARPi-based therapies (4, 5).
In addition to PARPi sensitivity, BRCA1/2-mutant cells have been shown to be dependent on translesion synthesis (TLS), a DNA damage tolerance pathway that allows DNA polymerases to replicate damaged DNA (6, 7). Although several factors are involved in TLS, the deubiquitinase (DUB) enzyme USP1 has been identified as having an essential function in HRD tumors (8–12). USP1 has several substrates, but its most consequential substrate in HRD tumors is ubiquitinated proliferating cell nuclear antigen (ub-PCNA). PCNA is ubiquitinated by RAD18–RAD6 when DNA replication forks encounter lesions that conventional DNA polymerases cannot bypass (13, 14). ub-PCNA promotes the switch to TLS polymerases, allowing DNA replication of damaged template DNA. Once this process is complete, USP1 is required for deubiquitination of PCNA and reversion to canonical DNA replication. Loss of USP1 function in BRCA1/2-mutant cells through genetic knockout (KO) or pharmacologic inhibition causes accumulation of mono– and poly–ub-PCNA and PCNA degradation. Ultimately, this results in elevated DNA damage, loss of replication fork protection, inhibition of DNA replication, and finally, cell death in HRD and BRCA1/2-mutant cells (10, 11). Owing to the different mechanisms of action, USP1 inhibitors (USP1i) and PARPi were shown to synergize both in vitro and in vivo in HRD models to drive strong antitumor responses (11), highlighting the potential of single agent and combined therapy with these two classes of inhibitors.
We initiated a small-molecule drug discovery program that led to the discovery of TNG348, a novel and potent inhibitor of USP1. Our data demonstrate that TNG348 is a highly selective and allosteric inhibitor of USP1, and the cryo-EM structure of USP1 in complex with TNG348 provided structural insights into the mode of inhibition. TNG348 synergizes with PARPi, and the combination causes strong antitumor effects in vivo, leading to tumor regression in several mouse xenograft tumor models. Importantly, our data demonstrate that TNG348 in combination with PARPi can overcome acquired PARPi resistance in vivo. Overall, our study supports the use of USP1is as a novel therapeutic strategy for the treatment of HRD cancers in combination with PARPi.
Materials and Methods
Compounds
Olaparib (Lot 109840) was purchased from MedChemExpress (HY-10162). MG132 (15) was purchased from VWR (80053-194). DMSO (D2650), methyl methanesulfonate (MMS) (129925), and cisplatin (232120) were purchased from Sigma-Aldrich. SN38 (16) was purchased from MedChemExpress (HY-13704), and RP3500 (17) was purchased from Selleck Chemicals (E1108). Saruparib [AZD5305 (18)] was purchased from MedChemExpress (HY-132167). ART558 (19) was purchased from MedChemExpress (HY-141520). I-138 (20) was synthesized as previously reported (11). TNG348 was synthesized as described in Supplementary Methods.
Cell culture and cell line engineering
Cell lines were maintained in incubators at 37°C with 5% CO2 atmosphere up to 8 weeks after thawing. MDA-MB-436, UWB1.289, and HCC1395 cells were obtained from ATCC in 2017 (HTB-130, CRL-2945, and CRL-2324), and COV362 cells were obtained from the European Collection of Animal Cell Cultures in 2017 (7071910). Information for other cell lines and cell-specific culture conditions can be found in Supplementary Table S1. All cell lines were authenticated by short tandem repeat analysis upon reception and were tested for Mycoplasma right after thawing and every 2 weeks throughout cell line maintenance using the Lonza MycoAlert Mycoplasma Detection Kit (Lonza Bioscience, LT07-118) and corresponding MycoAlert Assay Control Set (Lonza Bioscience, LT07-518). Lentivirus-mediated delivery was used to generate cell lines with stable Cas9 expression and subsequently used to deliver single-guide RNA (sgRNA) vectors. Lentivirus was produced by transfecting the sgRNAs or cDNA vector plasmids into Lenti-X cells. Cells were seeded in six-well plates or 10-cm dishes and allowed to adhere overnight. Transfection components included the plasmid, lentiviral packaging mix (Cellecta, CPCP-K2A), Lipofectamine 3000 (Thermo Fisher Scientific, L30000015), and Opti-MEM (Gibco, 31985-062). After 24 hours, the media was replaced with DMEM + 30% FBS. Media was collected 48 hours after transfection, filtered through a 0.45-μm membrane using a syringe, and stored at −80°C until usage. For infection, target cell lines were seeded and allowed to adhere overnight. Polybrene (Sigma-Aldrich, TR-1003-G) at 8 μg/mL was added along with virus. Media was replaced 24 hours after infection, and selection began 48 hours after infection with appropriate antibiotic [either blasticidin (Gibco, A1113903), puromycin (Gibco, A1113803), or geneticin (Gibco, 10-131-035)].
Protein expression and purification
Human USP1 and UAF1 proteins were expressed according to previously published procedures with minor modifications (21, 22). Full-length USP1 (G670A/G671A) containing a TEV-cleavable N-terminal His6-tag and full-length UAF1 containing a C-terminal FLAG tag were co-expressed in Sf21 cells. Following cell lysis using a high-pressure homogenizer and clarification of cell lysate by centrifugation (18,000 rpm, 60 minutes, 4°C), the USP1–UAF1 complex was purified by sequential affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) followed by anti-FLAG resin purification. The His6-tag was removed by digestion with TEV protease prior to anti-FLAG column affinity purification. Buffer for the Ni-NTA step consisted of 50 mmol/L Tris-HCl (pH 8.0), 500 mmol/L NaCl, 5% (v/v) glycerol, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), and 10 mmol/L imidazole. Protein was eluted with 500 mmol/L imidazole in this same buffer. For anti-FLAG chromatography, the buffer consisted of 50 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 5% (v/v) glycerol, and 1 mmol/L dithiothreitol (DTT), and the protein was eluted by using this same buffer containing 200 μg/mL FLAG peptide. Following the affinity purification steps, the protein complex was further purified by using anion exchange chromatography (Mono Q) and size-exclusion chromatography (Superdex 200). Purified protein at 2.1 mg/mL in the final column buffer [50 mmol/L 4-(2-hydroxyethyl)piperazine-1-ethane-sulfonic acid (HEPES), pH 7.5, 150 mmol/L NaCl, and 5% glycerol] was frozen as aliquots for future use. Ubiquitin vinyl sulfone (Ub-VS) was purchased from Boston Biochem (catalog #U-202).
Cryo-EM structure determination
The purified USP1–UAF1 complex was covalently modified with ubiquitin by reacting the sample with Ub-VS overnight at 4°C using a 1:4 molar ratio of complex:Ub-VS. The sample was then purified using size-exclusion chromatography as described above. Following the addition of a 10-fold molar excess of TNG348 and incubation for 60 minutes 4°C, grids were prepared from 0.20 mg/mL protein samples. A sample was applied to a glow-discharged 1.2/1.3-μm Quantifoil gold grid, blotted for 3 seconds, and vitrified in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific).
Image data were collected using a 300 kV Titan Krios G4 electron microscope (Thermo Fisher Scientific) equipped with a Gatan K3 camera operating in a super resolution mode. All movies were automatically recorded using Electron Probe User at a magnification of 105 K with a physical pixel of 0.423 Å. A total dose of 51.3 e/Å2 was fractionated into 40 frames. A total of 6,490 movies were recorded with a defocus range from −1.0 to −2.0 μm, and the slit width of the Selectris X energy filter was set to be 20 eV. All movies were imported into CryoSPARC (23), and preliminary processing using patch motion with CryoSPARC’s own implementation and patch contrast transfer function correction was done. Following this step, 6,175 micrographs were selected for subsequent template-based picking and particle extraction. A total of 3,700,674 particles were extracted with 2 × 2 binning (150-pixel box size, 0.846 Å per pixel) and were then used for 2D classification followed by 3D classification. Altogether 151,407 particles from the best class were selected and re-extracted with unbinning (300-pixel box size, 0.423 Å per pixel) and then refined in 3D to yield a map with a 3.3 Å global resolution, which was used for model building. Initial models were generated by rigid body placement of UAF1, USP1, and ubiquitin proteins from previously published structures (PDB 5CVO and 7AY2) into the map. Subsequent refinement and model building were performed with Coot, CCP-EM, and PHENIX (24–26). Figures were generated using PyMOL (Schrodinger).
CRISPR-Cas9 screening
Cells expressing Cas9 were transduced with lentivirus carrying a DNA damage response–focused guide RNA (gRNA) library with unique molecular identifier barcodes (27). The DNA damage–focused library included gRNAs targeting 55 essential genes and 1,145 DNA damage response genes (28, 29). Each gene was targeted by six individual gRNAs from the Vienna BioCenter (30). The library additionally included 600 nontargeting and 72 intron-cutting gRNAs. The whole-genome library was assembled as previously published (11). The multiplicity of infection was set to 0.3 to optimize for single gRNA expression per cell. Cells were selected with puromycin for 4 days after transduction, and selection was confirmed by expression of red fluorescent protein marker present on the lentiviral vector via the NovoCyte Quanteon (Agilent) instrument equipped with NovoExpress v 1.6.1 software. Cells were then allowed to grow for 2 to 3 days without selection before being passaged and allowed to adhere overnight. Cells were then treated with indicated drug (day 0). Cells were subcultured every 2 to 3 days, ensuring library coverage of 1,000×. After the DMSO-treated arm had reached a cumulative doubling count of at least 10, the cells from each arm were harvested via trypsinization. Cell pellets were washed with PBS and frozen at −80°C. gDNA was later extracted using the DNeasy Blood & Tissue Kit (Qiagen, 69506) or with the ReliaPrep Large Volume HT gDNA Isolation System for COV362 and HCC1395 (Promega, A2751), following the manufacturer’s protocol. DNA sequencing and computational analysis were performed as previously described (11).
Cell viability assays
For colony formation assay, cells were seeded into 12-well plates and allowed to adhere overnight. After initial dosing, media was replaced every 2 to 3 days for 10 to 21 days, with DMSO as the normalizing agent. PBS was used to wash the cells, and 1% crystal violet solution (Sigma-Aldrich, V5265) was then used to stain the cells. After incubation on a plate rocker for 5 minutes, the crystal violet was removed, and cells were washed by submerging the plate in a container of water. The plates were set out to dry overnight and then imaged using the LI-COR Odyssey CLx and accompanying Image Studio software (LI-COR). Growth was assessed by densitometry, normalized to DMSO (100%) and 10 μmol/L MG132 (0%; ref. 15), and curve-fitted with a nonlinear regression.
For CellTiter-Glo viability assay, cells were seeded into 384-well plates (Corning) the day prior to treatment with compounds. Plates were dosed using a Tecan Digital Dispenser D300e (Hewlett Packard), with DMSO as the normalizing agent. After 7 days, plates were allowed to equilibrate to ambient room temperature (20°C–22°C). After 30 minutes of equilibration, CellTiter-Glo 2.0 reagent (Promega, G924C) was added to each well, with a subsequent incubation for 20 minutes at room temperature. Luminescence signals were measured using the EnVision Multimode Plate Reader (PerkinElmer) equipped using EnVision Manager v1.13.3009.1409 software (PerkinElmer). GraphPad Prism 10.1.1 was used to plot results with a three-parameter nonlinear regression curve-fitting model after normalizing signal to DMSO-treated (100%) and MG132-treated (0%) wells. Synergy was calculated using Genedata software equipped with the synergy package for the cell line panel and with SynergyFinder Plus for other experiments (31).
Chromatin fractionation
UWB1.289 cells were treated with either DMSO, TNG348, olaparib, or a combination of TNG348 + olaparib for 24 hours, with a replicate set receiving the addition of MMS to increase overall signal. After harvesting cells via trypsinization, the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific, 78840) was used to segment and collect cytoplasmic, nuclear soluble, and chromatin fractions using the manufacturer’s corresponding protocol.
Western blotting
Cells were washed with PBS (Gibco 14190-136) and lysed using RIPA buffer (Thermo Fisher Scientific, 23213) with protease inhibitor (Thermo Fisher Scientific, 1861278), EDTA (Thermo Fisher Scientific, 1861275), and universal nuclease (Thermo Fisher Scientific, 88702). Lysates were centrifuged at 21,000 g for 10 minutes at 4°C, and the bicinchoninic acid protein assay (Thermo Fisher Scientific, 23213) was used to quantify and normalize protein concentrations. Samples were heat-denatured in NuPAGE sample buffer and reducing agent (Invitrogen NP0007 and NP0009), loaded and run on SDS-PAGE using 4% to 12% gels (Invitrogen WG1402BOX), and transferred to nitrocellulose membranes (Bio-Rad 1704159). Membranes were blocked in 5% milk powder and incubated in primary and secondary antibodies diluted in 2% milk powder, washing in TBS-T (Tris buffer saline + 0.05% Tween-20) after each incubation. Membranes were visualized on the ChemiDoc using enhanced chemiluminescence substrate (Bio-Rad 1705060 and 1705062). For detailed information on antibodies used, see Supplementary Table S2. Protein quantifications were performed by densitometry using ImageJ software (v1.231; ref. 32).
In vitro selectivity profiling
TNG348 was screened at a single-point concentration of 10 μmol/L in a biochemical assay panel of 47 deubiquitinating enzymes (Ubiquigent DUBprofiler). TNG348 was screened at a single-point concentration of 10 μmol/L in the Eurofins KINOMEscan scanMAX kinase panel. This screening platform uses an active site–directed competition binding assay to quantitatively measure interactions between TNG348 and 468 human kinases and disease relevant mutant variants. TNG348 was profiled against 78 safety targets in the SAFETYscan E/IC50 ELECT service provided by Eurofins DiscoverX. The assays were performed utilizing the PathHunter enzyme fragment complementation technology, FLIPR-based cellular screening assays, KINOMEscan kinase binding assays, ion channel assays, and a variety of enzymatic assays. In these assays, TNG348 was tested in 10-point dilution series from a starting concentration of 10 μmol/L. Assay results were calculated as % activity relative to individual controls for each target in the SAFETYscan E/IC50 ELECT panel. Results showing an agonism or antagonism higher than 50% are considered to represent significant effects of the test compounds.
Biochemical characterization and TNG348 IC50 measurement for recombinant wild-type and V156K mutant USP1 enzymes
The activity of deubiquitinating enzyme USP1/UAF1 was measured by the increase in fluorescence due to enzyme-catalyzed cleavage of the fluorogenic substrate Ubiquitin-Rhodamine110 (Ub-Rhod-110) from Ubiquigent (catalog number 60-0117-BUL). Although the Ub-Rhod-110 is essentially nonfluorescent, the cleavage results in a dequenched Rhodamine 110-Glycine, which exhibits intense fluorescence at 540 nm when excited at 480 nm.
Steady-state kinetic parameters measurement for wild-type USP1 and V156K USP1 mutant
Initial rates of cleavage of Ub-Rhod-110 substrate catalyzed by 0.3 nmol/L of recombinant wild-type (WT) or V156K mutant USP1 enzyme were measured in 20 μL of reaction mixture in a 384-well black plate (Greiner Bio-One, 781076). The assay buffer contains 50 mmol/L HEPES, pH 7.0, 0.5 mmol/L EDTA, 100 mmol/L NaCl, 10 mmol/L Tris(2-carboxyethyl)phosphine HCl, 0.01% BSA, and 0.005% Tween-20. Ub-Rhod-110 substrate was titrated from 12.5 to 0.012 μmol/L with a twofold dilution. The reactions were recorded on a SpectraMax Paradigm microplate reader in kinetic mode for 90 minutes (Ex: 480 nm/Em: 540 nm). Steady-state kinetic parameters Vmax and KM were obtained from the data analysis using the Michaelis–Menten equation in GraphPad Prism software.
Michaelis–Menten equation:
Here, Y is the initial velocity and X is the concentration of Ub-Rhod-110 substrate.
TNG348 IC50 measurement against WT and V156K USP1 mutant
To measure the inhibition of TNG348 against WT and V156K USP1 mutant, the reactions were performed in a total volume of 20 μL with final 100 nmol/L Ub-Rhod-110 substrate. TNG348 ranged from 10 to 0.0005 μmol/L with a 10-point threefold dilution. In brief, final 0.15 nmol/L WT enzyme or 0.3 nmol/L V156K mutant was preincubated with TNG348 at room temperature for 30 minutes. Substrate was added subsequently to initiate the reaction. Following incubation for 40 minutes, final 20 μmol/L of ML323 was added to stop the reactions. The reactions were recorded on a SpectraMax Paradigm microplate reader in endpoint mode (Ex: 480 nm/Em: 540 nm). Dose–response curves are fitted by XLfit as %inhibition versus log (compound concentration) using a four-parameter logistic model 205 y = A + [(B − A)/(1 + ((C/x)∧D))] with fixed 0% and 100% inhibition limits to calculate the IC50. A: bottom = 0%; B: top = 100%; C: relative IC50; D: hill slope.
In vivo experiments
For cell line–derived xenograft studies, MDA-MB-436 cells were grown with L15 media with 10% FBS + 10 μg/mL insulin + 16 μg/mL glutathione and were then implanted subcutaneously into the mice [MDA-MB-436, 1 × 106 cells with 50% Matrigel (Corning) to NOD-SCID female mice (Beijing AKYB)]. BR-05-0028 and olaparib-resistant-BR-05-0028 (olaparib-R-BR-05-0028) patient-derived xenograft (PDX) studies, harboring BRCA1 Q544* mutation [triple-negative breast cancer (TNBC)] were performed at WuXi. BR-05-0028 was originally established from a surgically resected clinical sample by WuXi and implanted in nude mice to establish a tumor model. Olaparib-R-BR-05-0028 was generated by continuous treatment of olaparib at 100 mg/kg every day by oral gavage until resistance occurred. Tumors grew under olaparib treatment were passaged/dosed for several rounds in mice until a stable resistance phenotype established. PA1338 PDX studies harboring BRCA1 Y1845fs mutation (pancreatic cancer) were performed at Crown Bioscience. ST4139 and ST1213 PDX studies, harboring E3167fs and RAD51C R193*, respectively, were performed at XenoSTART. CTG-0012 PDX studies harboring BRCA1 Y682* mutation (TNBC) was performed at Champions Oncology. HBCx-11, a HRD+ model defined by an HRD score (GIS) > 42, and HBCx-14 PDX model, harboring BRCA1 K654fs mutation, were both TNBC models performed at Xentech. Tumor fragments were implanted subcutaneously into female BALB/c nude mice (Beijing Vital River) for BR-05-0028 and olaparib-R-BR-05-0028 PDX studies; BALB/c nude mice (GemPharmatech) for PA1338 PDX studies; Athymic Nude Crl:NU(NCr)-Foxn1nu mice (Charles River Laboratories) for ST4139 and ST1213 PDX studies; and Athymic Nude-Foxn1nu mice (Envigo) mice for CTG-0012, HBCx-11, and HBCx-14 PDX studies. Animal studies were conducted with approval from the Institutional Animal Care and Use Committee or applicable Animal Ethics Committee of WuXi AppTec, Champions Oncology, XenoSTART, Crowns Bioscience, and Xentech, following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care. Mice were maintained in a controlled, specific pathogen–free environment at 20°C to 26°C, 30% to 70% humidity, and 12 hours light-to-dark cycle. Tumor volume was measured twice weekly, both length and width were measured by a caliper, and tumor volume was calculated using the formula volume = ½ (length × width2). Animal body weight and mice condition were monitored throughout the study. Mice were randomized into treatment groups when the mean tumor volume reached approximately 150 to 300 mm3. Animals were treated from the day of the randomization. TNG348, olaparib, niraparib, and saruparib were administered by oral gavage once daily or twice a day at an 8:16-hour interval. TNG348 was prepared fresh daily and formulated in 5% dimethylacetamide (DMA) + 30% polyethylene glycol 400 + 20% Solutol HS-15 + 45% water with pH adjusted to 3.0 to 4.0 with 1 mol/L HCl. Niraparib was formulated in 0.5% w/v methylcellulose. Olaparib was formulated in 10% DMSO + 90% [10% (2-Hydroxypropyl)-β-cyclodextrin (HP-β-CD) in PBS]. Saruparib was formulated in water/HCl pH 3.5 to 4. In experiments testing pharmacodynamic and exposure relationships, tumors were collected from animals at indicated timepoints after last dose and flash-frozen. Proteins were extracted using RIPA buffer, homogenized with TissueLyser II (QIAGEN), and subjected to Western blotting.
Pharmacokinetics
Blood samples were collected via the jugular vein or the vena cava into tubes containing anticoagulant potassium (K2) EDTA, at 0 (predose) and 2, 4, and 8 hours after last dose. Blood samples were processed to plasma and analyzed using the LC-MS/MS method. Pharmacokinetic parameters were estimated using a noncompartmental analysis consistent with the oral gavage route of administration. Pharmacokinetic parameters were generated from composite concentrations in mouse plasma using nominal sampling times relative to the start of the last dose administration.
Data availability
The cryo-EM data generated in this study are publicly available in RCSB protein data bank at PDB ID: 9EBS. CRISPR screening data generated in this study are publicly available in Gene Expression Omnibus at GSE284239. All other data are available upon request.
Results
TNG348 is a reversible allosteric USP1i suitable for oral administration
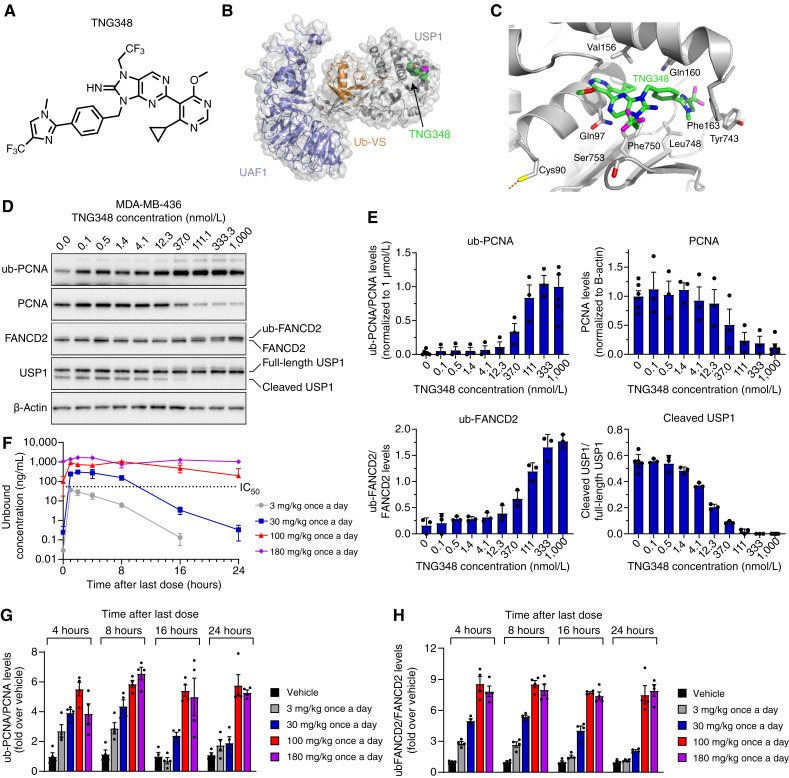
TNG348 is a substituted 8-iminopurine derivative (Fig. 1A) that inhibits the DUB activity of USP1, in complex with its stochiometric partner UAF1, with an IC50 value of 98.5 nmol/L when assayed using Ub-Rhod-110 as a substrate (Supplementary Fig. S1A and S1B). To gain insights on the compound’s mode of action, we determined the cryo-EM structure of TNG348 bound to the USP1 + UAF1 + Ub-VS complex at an overall average resolution of 3.3 Å (Fig. 1B and C; Supplementary Table S3). Similar to the previously published structure of the USP1i ML323 (33), we found that TNG348 interacts noncovalently with the enzyme by binding to a cryptic allosteric pocket in USP1. This pocket is formed by displacement of a portion of the N-terminal polypeptide of USP1 upon compound binding (encompassing residues Leu77-Asn88) and disruption of a loop region containing residues Leu165-Gln177 and Asp751-Val755. The net effect of this altered conformation is to disrupt the precise positioning of the USP1 protease catalytic triad (Cys90, His593, and Asp751), thereby preventing enzyme activity. In agreement with the noncovalent allosteric mode of binding, compound and substrate double titration experiments revealed that TNG348 is a noncompetitive inhibitor of USP1/UAF1 (Supplementary Fig. S1C), and jump dilution experiments indicate that TNG348 is a reversible inhibitor (Supplementary Fig. S1D).
Figure 1.
TNG348 is an allosteric USP1i. A, Chemical structure of TNG348. B, Cryo-EM structure of TNG348 (green) bound to the USP1 + UAF1 + Ub-VS complex (gray, purple, and orange, respectively). C, TNG348 binding pocket in USP1; selected residues in close contact with the inhibitor are labeled. The catalytic cysteine 90 is depicted at the lower left; the dashed orange line indicates its covalent attachment to the Ub-VS. D and E, TNG348 causes dose-dependent accumulation of USP1 substrates. D, MDA-MB-436 cells were treated for 24 hours with indicated doses of TNG348, and samples were collected for Western blot analysis. E, Combined densitometric quantifications of Western blot signals of three independent experiments shown in D. F–H, Plasma exposure and induction of USP1 substrates upon TNG348 dosing correlate in vivo. Mice-bearing MDA-MB-436 tumors were exposed to TNG348 at indicated doses for 5 days. F, Plasma samples from three to four mice were taken at indicated timepoints after last dose; TNG348 levels were quantified by mass spectrometry. TNG348 concentrations were corrected with plasma protein binding from NOD-SCID mice. The dashed line represents the average viability IC50 of 68.3 nmol/L. G and H, Tumors were harvested from three to four mice for each condition and processed for Western blotting against ub-PCNA and PCNA (G) and ub-FANCD2 (H). Protein levels were measured by densitometry of three to four different mice, and error bars represent the SEM.
USP1 is a cysteine protease that removes ubiquitin from multiple cellular substrates, including ub-PCNA and ub-FANCD2, and separately cleaves itself at a ubiquitin-like motif (10, 11, 34). Consistent with its biochemical activity, TNG348 treatment of MDA-MB-436 cancer cells, which is a BRCA1-mutant and TNBC cell line, resulted in a dose-dependent accumulation of ub-PCNA and ub-FANCD2, as well as reduction of the USP1 autocleaved fragment (Fig. 1D and E). The cellular pharmacodynamic potency of TNG348 was further assessed by AlphaLISA-based measurement of ub-PCNA induction in MDA-MB-436 cells and yielded an average IC50 of 98.6 nmol/L (Supplementary Fig. S1B and S1E). To evaluate the cellular anticancer activity of TNG348 and its synthetic lethality with BRCA1/2 mutations, 10-day viability assays were performed in MDA-MB-436 (BRCA1 mutant) and HCC1954 (BRCA1 WT) cell lines. Our data suggest that TNG348 induces a dose-dependent loss of viability in the BRCA1-mutant MDA-MB-436 cell line with an average IC50 of 68.3 nmol/L while having no impact on the BRCA1 WT HCC1954 cell line (Supplementary Fig. S1B and S1F).
Oral administration of TNG348 led to dose-dependent exposures in MDA-MB-436 tumor-bearing mice in both once daily and twice daily treatment regimens (Fig. 1F; Supplementary Fig. S1G). Doses of 30 mg/kg every day or 20 mg/kg twice a day resulted in free exposures above the TNG348 MDA-MB-436 in vitro IC50 for more than 8 hours, and higher doses (100 mg/kg every day or 80 mg/kg twice a day ) allowed exposures above IC50 for 24 hours after last dose. Ub-PCNA, ub-FANCD2, and cleaved USP1 levels in tumors correlated with exposure over time, with the highest doses maintaining maximal levels for 24 hours (Fig. 1G and H; Supplementary Fig. S1H–S1J). Overall, these data demonstrate that TNG348 acts as a potent inhibitor of USP1 both in vitro and in vivo.
TNG348 is a selective inhibitor of USP1
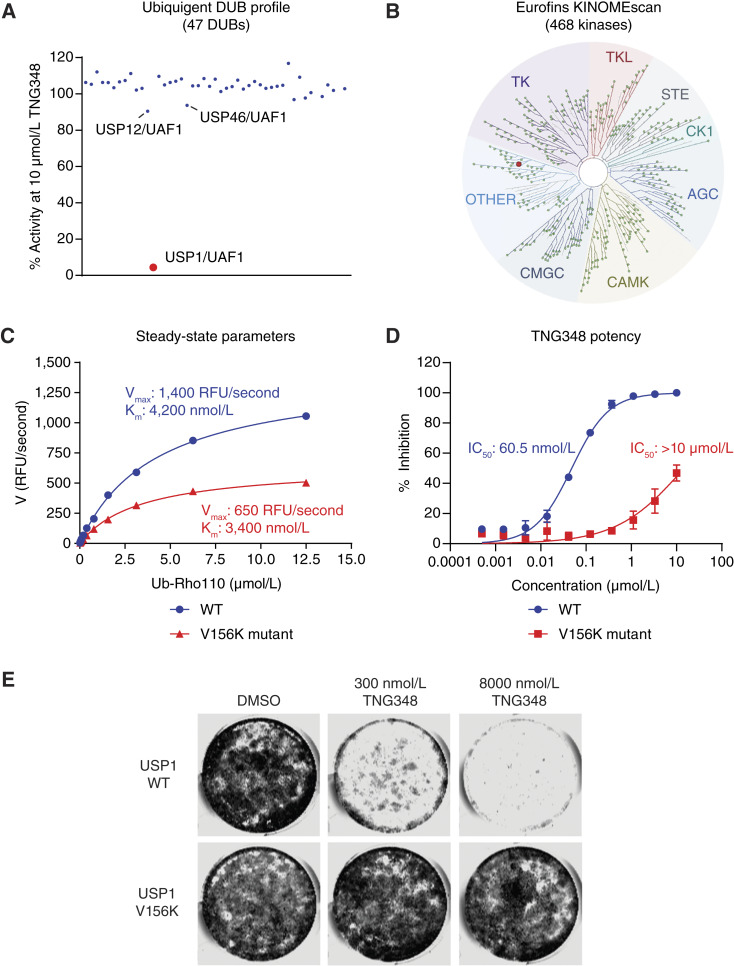
We next sought to characterize the selectivity of TNG348 by testing its activity in a panel of 47 DUBs (Ubiquigent), including USP12 and USP46, the two members of the USP subfamily that also interact with UAF1/WDR48 (35). Strikingly, USP1 is the only DUB that was significantly inhibited by TNG348 (Fig. 2A). Next, we assayed the off-target effects of TNG348 on kinases in the KINOMEscan scanMAX panel (Eurofins). TNG348 was found to be selective against 467 of the 468 kinases tested, only showing a significant binding interaction with ULK3 kinase (89% inhibition at 10 μmol/L; Fig. 2B; Supplementary Fig. S2A). When tested against 78 safety targets in the SAFETYscan E/IC50 ELECT (Eurofins DiscoverX), TNG348 was found to be inactive as an agonist or antagonist in 77 of 78 assays in the panel. It only displayed modest antagonism against the 5-HT2B receptor with an IC50 of 1.8 μmol/L and a maximum response of 91.5% at 10 μmol/L (Supplementary Fig. S2B). These data indicate that TNG348 is highly selective for USP1 in vitro.
Figure 2.
TNG348 is a selective USP1i. A, TNG348 only inhibits USP1 in vitro in a panel of 47 protein DUBs. Activity of TNG348 was tested at 10 μmol/L against the Ubiquigent DUBprofiler panel. USP12, USP46, and USP1 (in red), which all require UAF1 for activity, are labeled. B, TNG348 was assayed in vitro against the Eurofins KINOMEscan panel of 468 kinases at a single-point 10 μmol/L concentration. Only ULK3 (in red) was found to be inhibited at 89% of control binding. C, The activity of recombinant USP1 WT or V156K mutant protein was assayed in vitro. The V156K mutant shows ∼50% reduction in activity but maintains similar Km for Ub-Rhod-110 substrate to WT enzyme. RFU, Relative fluoresence units. D, The USP1 V156K mutant protein is not inhibited by TNG348 in in vitro ub-Rhod-110 assay (TNG348 lost >200-fold potency). E, The USP1 V156K mutant completely abolishes the antitumor growth effect of TNG348. Endogenous USP1 was knocked out with two USP1 gRNAs in Cas9-expressing MDA-MB-436 cells, and CRISPR-resistant WT or V156K mutant cDNA was concurrently expressed. The viability at the indicated TNG348 dose was monitored in 14-day clonogenic assays.
The cellular activity of TNG348 depends on USP1 binding
To further validate the on-target activity of TNG348, we generated USP1 mutants that were predicted to prevent TNG348 inhibitor binding. The cryo-EM structure revealed several residues on USP1 that coordinate the interaction with TNG348, namely, V156, Q97, Y743, and Q160. The residue V156 seems to form a hydrophobic interaction with the pyrimidine side chain on TNG348, and mutating this residue to a lysine is expected to hamper the interaction by changing the binding pocket polarity at this site as well as creating steric hindrance due to the longer side chain. Notably, the purified USP1 V156K mutant protein behaved like the WT enzyme with a similar KM for Ub-Rhod-110 substrate and only slightly reduced catalytic activity (Vmax, Fig. 2C). However, consistent with the predicted impact of the V156K mutation, TNG348 was >200-fold less potent against this mutant form of USP1 (Fig. 2D), confirming the binding mode revealed by the Cryo-EM structure. We next generated a CRISPR-resistant USP1 cDNA carrying the V156K mutation and co-expressed this construct together with sgRNAs to simultaneously knockout the endogenous USP1 gene in the BRCA1-mutant MDA-MB-436 cell line (Supplementary Fig. S2C). In these USP1 KO cells, the expression of the WT cDNA led to sensitivity to TNG348, indicating that although the cDNA-mediated USP1 expression levels were higher than the endogenous levels, TNG348 inhibitor treatment still caused growth defects in these cells. Interestingly, cells carrying the USP1 V156K cDNA lost sensitivity to TNG348, completely abrogating the viability effect of TNG348 even at high concentrations (Fig. 2E). This was associated with lack of ub-PCNA accumulation and presence of the USP1 cleavage fragment in the V156K mutant upon USP1 inhibition (Supplementary Fig. S2D). We also expressed the Q97E, Y743A, and Q160E mutations, but these cDNA constructs failed to improve the viability of USP1 KO cells, indicating these residues are critical for protein function (Supplementary Fig. S2E). Overall, our data support the conclusion that antitumor activity of TNG348 is driven by the selective, on-target activity of USP1 inhibition.
TNG348 activity is most active in BRCA mutant and HRD+ breast and ovarian cancer cells and acts through a distinct mechanism from PARPi
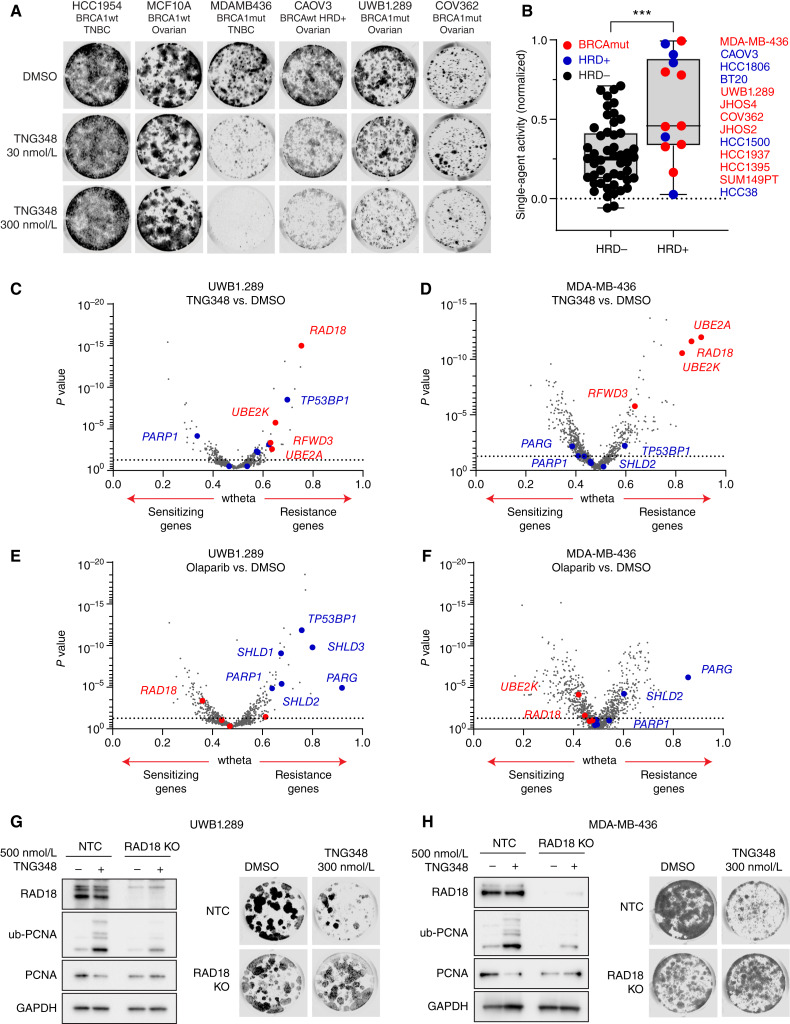
USP1 has previously been reported as a synthetic lethal interactor of BRCA1 (10, 11). We wanted to assess whether this phenomenon could be recapitulated by USP1 inhibition via TNG348, and whether TNG348 affected BRCA1/2 WT and HRD models. To this end, we tested TNG348 single-agent activity by clonogenic assays in a panel, comprising 62 breast and ovarian cancer cell lines (examples on Fig. 3A). We then calculated and normalized dose–response AUC for each cell line and stratified them based on their BRCA1/2 mutation or HRD+ status. The HRD+ status was determined with scarHRD (36), and a threshold score of 65 was used to call HRD+ status. Except for SUM149PT, all BRCA-mutant cell lines had a score above 65. We found that the six most sensitive cell lines to TNG348 were BRCA1/2-mutant and HRD+ cell lines (Fig. 3A and B; Supplementary Table S4). Interestingly, the seven other BRCAmut/HRD+ cell lines showed lower sensitivity, suggesting that the synthetic lethality of USP1 with HRD is not fully penetrant.
Figure 3.
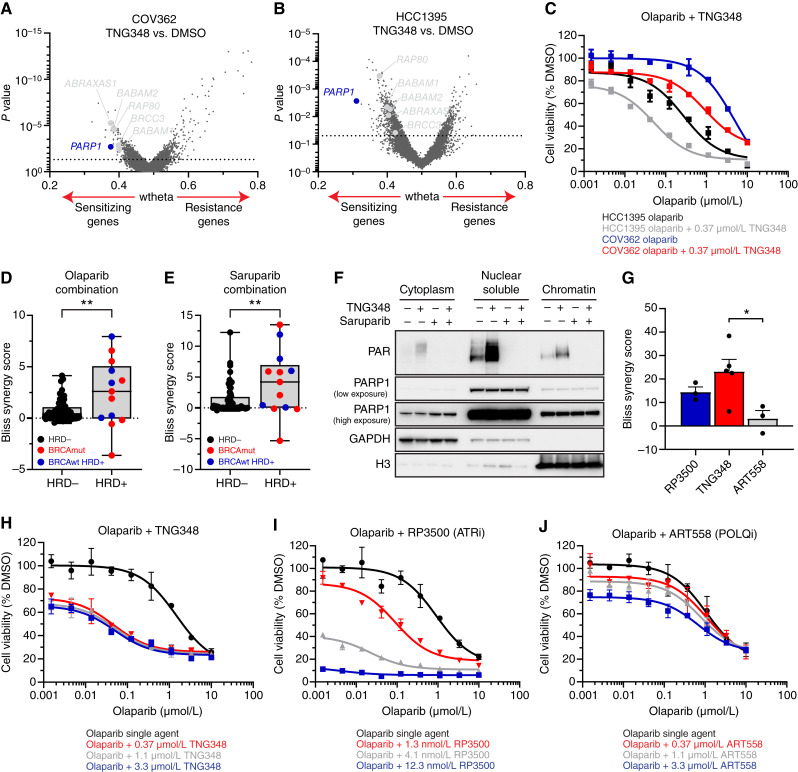
TNG348 activity is enriched in HRD cell lines and acts through a distinct mechanism from PARPi. A and B, TNG348 activity is elevated in BRCA-mutant or HRD cells. TNG348 activity was assayed in a panel of 62 breast and ovarian cell lines by 10–21-day clonogenic assays using 10-point threefold serial dilutions of TNG348 with an 8 μmol/L top dose. Examples are shown in A. B, The average AUC from biological duplicates of technical duplicates was calculated from nonlinear regression of the dose–response and normalized so that the highest possible response is assigned a value of 1 and no response, 0. Cell lines were grouped according to the HRD status; HR proficient (HRD negative, HRD−) cells without BRCA1/2 mutations in black, and cell lines that carry a BRCA1/2 mutation or that are BRCAwt but HRD positive (HRD+, ScarHRD > 65) in red and blue, respectively. Cells line names are shown on the right from most sensitive (top) to least sensitive (bottom) to TNG348. Statistical significance was calculated with a two-tailed unpaired Student t test. ***, P < 0.001. C and D, Volcano plot showing CRISPR screening results comparing DMSO and 200 (C) or 100 nmol/L (D) TNG348-treated Cas9-expressing UWB1.289 and MDA-MB-436 cells. Dashed line: P value = 0.05. E and F, Volcano plots showing CRISPR screening comparing DMSO and 1 μmol/L (E) or 10 nmol/L (F) olaparib-treated Cas9-expressing UWB1.289 and MDA-MB-436 cells. A focused gRNA library targeting DNA damage response genes was used for C–F. Labeled in red are genes known to cause resistance to USP1i when lost or involved in the ub-PCNA pathway. In blue are genes known to confer resistance to PARPi when knocked out. G and H, RAD18 suppresses the TNG348 sensitivity of UWB1.289 (G) and MDA-MB-436 (H) cell lines. Endogenous RAD18 was knocked out in Cas9-expressing cells, and the KO efficiency was validated, along with the impact on ub-PCNA levels, by Western blotting after 24 hours DMSO or TNG348 treatment (left). The impact on viability was monitored by clonogenic assays (right). NTC, Non-target control.
We assessed whether cells respond similarly to TNG348 and other published USP1is and how it compared with PARPi. To this end, we performed unbiased DNA repair–focused CRISPR screens to identify which genes, when lost, could confer resistance or sensitivity to TNG348 or olaparib in the USP1i-sensitive UWB1.289 and MDA-MB-436 cell lines (Fig. 3C–F). Consistent with previous reports, we found that loss of several PCNA ubiquitination–promoting genes, including RAD18, UBE2K, UBE2A, and RFWD3, caused resistance to TNG348 in both cell lines (Fig. 3C and D; Supplementary Tables S5 and S6; refs. 11, 12). We further validated that the loss of RAD18 reduced ubiquitination of PCNA and abrogated the sensitivity to TNG348 in both cell lines (Fig. 3G and H). These data confirm that the antitumor mechanism of TNG348 is dependent on RAD18-initiated PCNA ubiquitination.
In contrast and consistent with published data, knockout of shieldin subunits, TP53BP, PARP1, and PARG, conferred resistance to olaparib and loss of RAD18 or any of the other genes involved in the ub-PCNA pathway did not affect olaparib sensitivity (Fig. 3E and F; Supplementary Tables S7 and S8; refs. 3, 37–41). Conversely, loss of shieldin subunits, PARP1 or PARG, did not provide growth advantages in the presence of TNG348 in both cell lines. In fact, loss of PARP1, as well as other factors involved in BER, sensitized UWB1.289 cells to TNG348 (Fig. 3C and D; Supplementary Tables S5 and S6). We note that TP53BP1 KO conferred resistance to TNG348 in UWB1.289 and to some extent in MDA-MB-436 cells, indicating that there can be overlap of resistance mechanisms between TNG348 and PARPi. Interestingly, genes coding for proteins associated with the BRCA1-A complex were identified as sensitizers to TNG348, but not olaparib, suggesting that the dependency on BRCA1 functionality further differs between USP1i and PARPi (Supplementary Fig. S3A–S3D). Overall, these data indicate that TNG348 acts through a mechanism that is globally distinct from that of PARPi.
TNG348 synergizes with PARPi and other DNA-damaging agents in HRD+ and BRCA-mutant cell lines
Given that several HRD cell lines were largely unaffected by TNG348, we asked what conferred resistance to these cell lines by performing whole-genome CRISPR screens in two TNG348-resistant, BRCA1-mutant cell lines, COV362 and HCC1395, and looked for genes that sensitize cells to TNG348. Although several hits were identified using this approach, most were also identified in the UWB1.289 and MDA-MB-436 screens, including genes encoding for subunits of the BRCA1-A complex (Fig. 4A and B; Supplementary Tables S9 and S10). Interestingly, however, PARP1 loss was found as a common sensitizer to TNG348 in both cell lines, suggesting that PARP function may suppress TNG348 sensitivity. Consistent with this finding, TNG348 potentiated the effect of olaparib and reduced its IC50 by fivefold in COV362 and HCC1395 cells, despite the lack of response to TNG348 single agent (Fig. 4C). To further explore the synergy between TNG348 and PARPi, we tested the combination of TNG348 with PARP1/2 and PARP1-selective inhibitors in a panel of cell lines. BRCA1 mutant and HRD+ cell lines showed generally stronger synergy compared with BRCAwt cells, as measured by the Bliss synergy score, when TNG348 was combined with either olaparib or saruparib (AZD5305, Fig. 4D and E; Supplementary Fig. S4A–S4E; Supplementary Tables S11 and S12; ref. 18). We note that additional cell lines insensitive to either PARPi or TNG348 in this assay showed strong synergy (e.g., JHOS2 and BT20). Cells that were especially sensitive to either single-agent treatment showed a low synergy score (e.g., MDAMB436 and JHOS4), owing to the challenge of observing synergy when single-agent response cannot technically be improved (Supplementary Fig. S4A–S4D). Overall, we found that most HRD+ cell lines (10/13 lines) were sensitive to either single agent or their combination.
Figure 4.
TNG348 synergizes with PARPi in BRCA1/2-mutant and HRD cell lines. A and B, Volcano plots of whole-genome CRISPR screens in BRCA1mut COV362 (A) and HCC1395 (B) cell lines comparing gene mutations that sensitize (left) or cause resistance (right) to TNG348. PARP1 is labeled in blue, and members of the BRCA1-A complex are labeled in gray. Dashed line: P value = 0.05. C, TNG348 synergizes with olaparib in COV362 and HCC1395 cell lines. Viability was measured by CellTiter-Glo after 7 days of growth. D and E, Combination of TNG348 with olaparib (D) or saruparib (E) was tested in a panel of 69 breast and ovarian cell lines. The average Bliss synergy score for each cell line was calculated across an entire 9 × 9 matrix, and data are presented as the average of two biological replicates. Cell lines were categorized according to the HRD status; HR-proficient (HRD−) cells without BRCA1/2 mutations (black), and cell lines that carry a BRCA1/2 mutation or that are BRCAwt but HRD+ (ScarHRD > 65) in red and blue, respectively. Statistical significance was calculated with a two-tailed unpaired Student t test. **, P value < 0.01. F, TNG348 induces PARylation. UWB1.289 cells were treated for 24 hours with indicated drugs, and samples were collected for subcellular fractionation experiments followed by Western blotting with indicated antibodies. G–J, Olaparib synergy comparison among ART558 (POLQi), RP3500 (ATRi), and TNG348 in the UWB1.289 cell line. G, Average Bliss synergy score was calculated over a 4 × 4 portion of the dose–response matrices (0.04–1.1 μmol/L for olaparib, TNG348, and ART558 and 0.45–12.3 nmol/L for RP3500). H–J, Viability assay examples of olaparib combination with TNG348 (H), RP3500 (I), and ART558 (J) plotted in G. Error bars: SEM.
The screen and cell line panel data suggest that PARP function may suppress TNG348 sensitivity in HRD cells. USP1 inhibition was reported to induce the formation of DNA SSB, potentially leading to the recruitment of PARP1 for repair (11, 12, 42). The antitumor activity of PARPi results from both catalytic inhibition of PARylation and trapping of PARP enzymes onto chromatin at sites of DNA damage, suggesting that either or both of these effects underlie the synergy between TNG348 and PARPi. To assess these possibilities, we monitored PARP1 and PAR levels in different cellular compartments following treatment with TNG348 or the strong PARP1 trapper saruparib in the BRCA-mutant UWB1.289 cell line (18). We did not observe increased PARP1 trapping on chromatin after co-treatment with TNG348 and saruparib, even upon the addition of MMS (Fig. 4F; Supplementary Fig. S4F). However, PAR signal was significantly induced after TNG348 treatment in a PARP-dependent manner. As PARPi are known to induce DNA damage, we wondered if TNG348 could synergize with other DNA damaging agents known to be more efficacious in BRCA1/2-mutant or HRD+ tumors, such as platinum-based cisplatin or the topoisomerase I poison SN38 (16, 43, 44). Although TNG348 enhanced the antitumor growth effect of both cisplatin and SN38 in the BRCA1-mutant UWB1.289 cell line (2–3-fold IC50 reduction), TNG348 induced a much deeper potentiation of olaparib activity, with as much as 50-fold reduction of its IC50 (Supplementary Fig. S4G–S4I). These results suggest that the response to TNG348 induces PARP-mediated repair that may underlie TNG348 resistance in a subset of cell lines and support the potential of TNG348–PARPi combinations.
Next, we compared the selectivity and efficacy of TNG348 with other targeted therapies working through alternative mechanisms of action independently shown to synergize with PARPi in BRCA-mutant tumor cell lines, namely, POLQ and ATR inhibitors. POLQ (Polθ) was identified as a synthetic lethal target in BRCA mutant tumors through the elimination of the θ-mediated end joining mechanism (19, 45), and ATR inhibitors (ATRi) were shown to overcome PARPi resistance (46). All three inhibitors showed synergy with olaparib in the BRCA1-mutant UWB1.289 cell line, with the TNG348 combination showing the highest levels of synergy, followed by the ATRi RP3500 (17) and the POLQi ART558 (Fig. 4G–J; ref. 19). RP3500 provided the deepest growth inhibition effect in combination with PARPi, owing to significantly stronger single-agent activity at higher doses. This is consistent with the essential nature of ATR, and we note that RP3500 treatment resulted in complete loss of viability and could synergize with olaparib at higher doses in the BRCA WT MCF10A cell line, whereas TNG348 or ART558 did not (Supplementary Fig. S4J–S4L). This suggests that TNG348 and ART558 confer enhanced selectivity for BRCA1/2mut and HRD+ in combination with PARPi compared with ATRi and that TNG348 confers improved anticancer activity.
TNG348 drives strong antitumor activity in combination with PARPi in HRD+ models and can overcome PARPi resistance
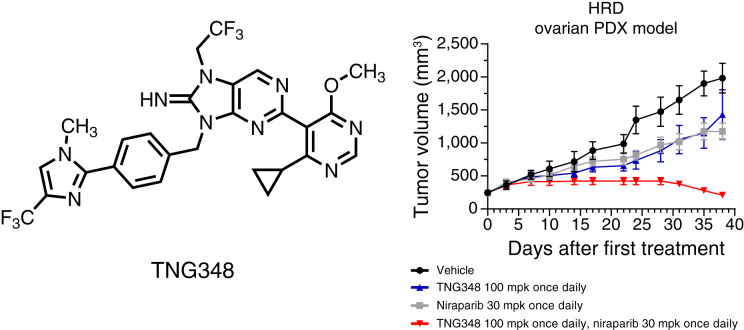
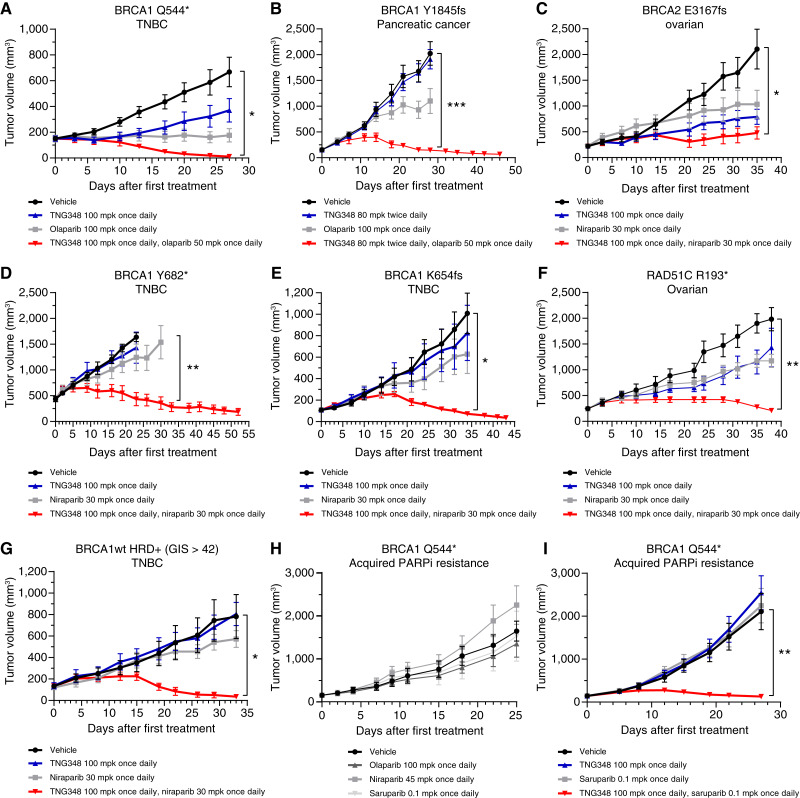
To assess whether the antitumor effect of TNG348 observed in vitro translated to in vivo models, the activity of TNG348, PARPi, or their combination was monitored in mice-bearing PDXs. Models from multiple patients with breast, ovarian, and pancreatic cancers carrying either a BRCA1, BRCA2, or RAD51C pathogenic mutation or that are BRCAwt and HRD+ were assayed (Fig. 5A–G; Supplementary Tables S13 and S14). Overall, single-agent TNG348 was well tolerated, and combination with either olaparib or niraparib did not lead to body weight loss in mice (Supplementary Fig. S5A and S5B). As single agents, TNG348 and PARPi had varying levels of tumor growth inhibition, from no effect to partial tumor growth inhibition. Strikingly, the combination of TNG348 at 100 mg per kilogram (mpk) every day or 80 mpk twice a day with olaparib or niraparib consistently led to improvements in tumor growth inhibition compared with single agents across all HRD+ models. Similar antitumor activity was observed when combining lower doses of TNG348 with PARPi, suggesting that the combination can provide synergistic benefit even if maximal suppression of USP1 activity is not maintained throughout the 24-hour between doses (Supplementary Fig. S5C–S5E, see Fig. 1F–H). Importantly, some of these xenograft models were generated from patients who had undergone several lines of chemotherapy, suggesting that combination benefits can be observed even in heavily pretreated cancers (Fig. 5C and D, see Supplementary Table S14 for details). Moreover, the growth of a BRCAwt ovarian model that does not carry any pathogenic mutations in HR genes and is not sensitive to niraparib was not significantly affected by the TNG348 and niraparib combination (Supplementary Fig. S5F). It should be noted that the dose of olaparib was reduced from 100 to 50 mpk every day to match the plasma exposure when combined with TNG348. No significant differences between 100 mpk every day single agent and combination at 50 mpk every day were observed, and niraparib exposure was not affected by TNG348 combination (Supplementary Fig. S5G). These data are consistent with the observed synergy between TNG348 and PARPi in vitro and highlight the potential of their combination in driving remarkable antitumor responses in HRD+ cancers.
Figure 5.
TNG348 drives strong in vivo antitumor activity in combination with PARPi and can overcome PARPi resistance. A–G, The growth of PDX models BR-05-0028 (A), PA1338 (B), ST4139 (C), CTG-0012 (D), HBCx-11 (E), ST1213 (F), and HBCx-14 (G) was monitored in mice during vehicle, TNG348, PARPi, or combination treatment. Average tumor volume from at least four mice from each arm is shown. Error bars represent SEM. See Supplementary Table S14 for additional information on models. H and I, TNG348 can resensitize tumors to PARPi. The model from A was subjected to prolonged treatment to olaparib until tumor regrowth was observed. A tumor regrowing from a single animal was further passaged under constant olaparib pressure to create an olaparib-resistant model. H This model is cross-resistant to several PARPi. I, Tumor growth following vehicle, TNG348, olaparib, or combination treatment in the PARPi-resistant model was monitored for 27 days. Error bars represent SEM of eight different animals for each arm. mpk, milligrams per kilogram. Statistical significance was calculated using two-way ANOVA followed by the Tukey multiple comparisons test. Only vehicle to combination comparison is shown, see Supplementary Table S13 for other comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Development of PARPi resistance caused by prolonged treatment with PARPi is often observed clinically and hampers long-term efficacy (2). Previous publications have suggested alternative mechanisms of action of USP1i that may allow USP1 inhibition to overcome PARPi resistance in vitro (10–12). Here, we used a BRCA1-mutant PDX model (Fig. 5A) that was rendered resistant to olaparib by maintaining olaparib pressure in vivo until tumor growth was recovered to generate a model of acquired PARPi resistance that is cross-resistant to several PARPi (Fig. 5H). The mechanism of resistance could not be identified but does not result from MDR1 overexpression, BRCA1 reversion, or loss of any of the genes typically associated with PARPi resistance (TP53BP1, SHLD1-3, PARG, etc.). The model was also cross-resistant to TNG348 single-agent treatment, but, strikingly, combination with either saruparib or olaparib resulted in strong and synergistic antitumor activity (Fig. 5I; Supplementary Fig. S5H). Taken together, these data suggest that combination of TNG348 with PARPis can drive deeper antitumor responses in BRCA-mutant or HRD+ tumors and overcome acquired PARPi resistance.
Discussion
In this study, we characterized TNG348 as a novel, allosteric, and selective USP1i that reversibly binds to a cryptic pocket on USP1. Although TNG348 was most active in HRD+ cells, we show that it provided markedly improved antitumor benefits and selectivity when combined with PARPis, leading to regression in several in vivo HRD+ PDX models. Our data further reveal that TNG348 can resensitize tumors with acquired PARPi resistance to PARPis.
Using unbiased CRISPR screening and single-gene validation, we show that the antitumor effect of TNG348 is driven by RAD18 and ub-PCNA, which is consistent with the functions of USP1 and the activity of other USP1is. Moreover, in agreement with previous studies showing that USP1 loss causes synthetic lethality in BRCA-mutant tumors, we found that the most sensitive cell lines to TNG348 are HRD+ (10–12). Surprisingly, a significant proportion of HRD+ cell lines seemed largely resistant to TNG348, indicating that HRD+ status does not fully predict USP1i sensitivity. We note that limitations exist with regards to the HRD scar classification used in this study as it reflects events in cells’ growth history rather than current functional defects. However, most TNG348-sensitive and -resistant BRCA1-mutant or HRD+ cell lines were characterized elsewhere as deficient for HR in functional assays, suggesting that the observed resistance results from incomplete penetrance rather than misclassification of cell lines (47–50). Moreover, some HR proficient cell lines were intermediately sensitive to TNG348, indicating that HRD status does not fully capture the drivers of TNG348 sensitivity and that additional DNA damage repair alteration biomarkers may be associated with USP1 dependency. This is consistent with previous findings that a subset of HR-proficient non–small cell lung cancer cell lines is sensitive to USP1 inhibition (11). It is possible that sensitivity to TNG348 results from composite features, with HRD being one component that is insufficient to drive sensitivity on its own. To assess what would be the other components, we performed whole-genome CRISPR screens in TNG348-resistant, BRCA1-mutant cell lines which revealed that despite the absence of BRCA1, residual BRCA1-A complex function may protect against USP1 inhibition. Interestingly, this complex has been shown to regulate HR and modulate histone ubiquitination surrounding DSBs (51, 52). However, how the BRCA1-A complex affects USP1 inhibition response warrants further research.
Several other genes were identified in CRISPR screens presented in this study, but the most consistent and actionable hit we uncovered in both TNG348-sensitive and -resistant cells is PARP1. This was further validated in a panel of cell lines in which PARPi and TNG348 were either effective as single agents or synergized in most HRD cell lines even if insensitive to either inhibitor. Although we show that TNG348 induces PARylation, implying that the DNA damage induced by USP1 inhibition requires PARP activity for repair, the mechanism through which USP1 inhibition by TNG348 synergizes with PARPi is likely multifactorial. Evidence suggests that HRD cells are dependent on both BER and TLS for survival (2, 6, 7), and combined PARP and TLS inhibition was shown to drive deeper responses in BRCA1-mutant cells (7). PARPi and USP1i were also shown to induce SSB formation, and their combination exacerbated this effect (11, 12, 42, 53). An additional factor contributing to the TNG348-PARPi synergy may be impediment of the response to single-ended DSB. Our data show that TNG348 improves the potency of topoisomerase I inhibitor SN38, a class of inhibitor that generates single-ended DSB. Single-ended DSB is also formed by PARPi when DNA replication forks encounter unrepaired SSB. Single-ended DSB induced by topoisomerase I inhibition causes DNA replication fork translocation to a reversed structure and exposes a free double-stranded DNA end, which, in turn, can be degraded upon USP1 inhibition, leading to fork collapse (10). This is consistent with the finding that RFWD3, which has been shown to promote fork reversal (54), confers resistance to TNG348 when lost in the BRCA1-mutant cell lines MDA-MB-436 and UWB1.289 (Fig. 3C and D and ref. 12). USP1 also regulates the Fanconi anemia repair pathway through deubiquitination of FANCD2 and FANCI (55, 56). This pathway is essential for the repair of interstrand cross-links caused by platinum compounds such as cisplatin and is also involved in the response to PARPi. The finding that TNG348 can improve the response to cisplatin suggests that it may also disrupt the Fanconi anemia pathway and that this may contribute to the synergy with PARPi. In support of this, RAD18, which fully abrogates USP1i sensitivity when lost, was found to regulate FANCD2 function and ubiquitination through a ub-PCNA–dependent mechanism (57, 58). The contribution of each individual synergy mechanism may differ between tumor types, but the pleotropic and synthetic lethal nature of the TNG348 plus PARPi combination may contribute to its versatility in treating HRD+ tumors. Indeed, resistance to either TNG348 or PARPi single agents, whether intrinsic or acquired, was overcome when combining both classes of inhibitors. In every in vivo model we tested, the combination resulted in stronger antitumor effects irrespective of treatment history, frequently leading to tumor regression. This suggests that TNG348 in combination with PARPi could be effective to treat patients with HRD+ tumors that do not or no longer respond to PARPi.
We note that three HRD+ cell lines were insensitive to TNG348 and PARPi combination. Although this may be a result from the aforementioned limitations to HRD scar, it is possible that genetic alterations conferring resistance to the combination exist. For example, TP53BP1 loss was identified as a common resistor in both MDA-MB-436 and UWB1.289 cell lines, suggesting that there may be instances of overlapping mechanisms of resistance to both classes of drugs. Conversely, the HBCx-11 model, which was shown to retain HR functionality through the ability to form RAD51 foci, despite harboring a pathogenic BRCA1-mutation and HRD+ score (Fig. 5E; Supplementary Table S14; ref. 59), was highly sensitive to the TNG348 and PARPi combination. Additionally, the combination synergized in several cell lines not characterized as HRD+. These data suggest that other DNA damage repair alterations, in addition to HRD, may govern TNG348 plus PARPi response. It will be essential to understand what underlies response to the combination to optimize the treatment conditions.
Unfortunately, the clinical development of TNG348 has been terminated because of the unanticipated observation of liver effects consistent with drug-induced liver injury during the phase I trial (NCT06065059). Although the cause of liver toxicity is unclear, neither in vivo studies nor the off-targets identified in this study, ULK3 and the 5-HT2B receptor antagonism, are likely to be responsible. ULK3 and the 5-HT2B receptor are weakly inhibited by TNG348, ULK3 KO mice are viable without body weight loss, and 5-HT2B antagonists have not been associated with liver damage. Importantly, liver toxicity is unlikely to be on-target because another USP1i has been well tolerated in clinical trials (42, 60). Though the clinical development of TNG348 has been discontinued, this study provides strong preclinical support for the use of USP1is in combination with PARPi in BRCA1/2 mutant or other HRD tumors in the clinic.
Supplementary Material
Supplementary methods include the chemical synthesis procedure of TNG348 and additional biochemical assay protocols not included in the main text.
Supplementary Figure S1 shows additional biochemical and in vivo exposure data to support Figure 1
Supplementary Figure S2 shows additional selectivity data from Eurofins KinomeScan and DiscoverX Safety Scan to support Figure 2. It additionally shows western blot controls for USP1 KO and cDNA expression, as well as the impact of the USP1 V156 K mutation on USP1 substrate and other mutations on the cell growth.
Supplementary Figure S3 shows CRISPR screening data suggesting that loss of the BRCA1-A complex sensitizes cells to TNG348, but not olaparib
Supplementary Figure S4 shows additional data supporting Figure 4. This includes an alternative representation for the combination cell line panels shown in Figure 4D-E, examples of combination between TNG348 and saruparib, olaparib, cisplatin or SN38, and the viability effects in MCF10A of TNG348, RP3500 and ART558 alone or in combination with olaparib.
Supplementary Figure S5 shows additional data supporting Figure 5 including mouse body weight following TNG348, PARPi or combination treatment, additional in vivo experiments with lower TNG348 doses, an in vivo experiment in a BRCA1/2wt PDX model, and plasma concentration comparisons between TNG348 or PARPi alone versus their combination.
Supplementary Table S1 shows the list of cell lines and culture conditions used in this study
Supplementary Table S2 shows the list of antibodies used in this study
Supplementary Table S3 shows information on Cryo-EM data collection, refinement and validation
Supplementary Table S4 lists the area under the curve values for each cell line in the TNG348 single agent cell line panel shown in Figure 3A-B.
Supplementary Table S5 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the UWB1.289 cell line from Figure 3C
Supplementary Table S6 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the MDA-MB-436 cell line from Figure 3D
Supplementary Table S7 lists the wtheta and p-values for each gene in the olaparib vs DMSO CRISPR screen in the UWB1.289 cell line from Figure 3E
Supplementary Table S8 lists the wtheta and p-values for each gene in the olaparib vs DMSO CRISPR screen in the MDA-MB-436 cell line from Figure 3F
Supplementary Table S9 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the COV362 cell line from Figure 4A
Supplementary Table S10 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the HCC1395 cell line from Figure 4B
Supplementary Table S11 lists the bliss synergy score values for each cell line in the TNG348 and olaparib combination cell line panel shown in Figure 4D.
Supplementary Table S12 lists the bliss synergy score values for each cell line in the TNG348 and saruparib combination cell line panel shown in Figure 4E.
Supplementary Table S13 shows the statistical analysis of the different treatment groups from the different in vivo PDX studies
Supplementary Table S14 shows additional information on the PDX used in this study, including model histology, clinical subtype, relevant mutation and treatment history
Acknowledgments
We thank the scientists from ChemPartner, Pharmaron, Xentech, XenoSTART, WuXi AppTec, Crown Bioscience, Champions Oncology, Biortus, and Ubiquigent for their contributions to this work. We thank Dr. Adam Crystal and Prof. Christopher Lord for critical reading of the manuscript.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors’ Disclosures
A. Simoneau reports other support from Tango Therapeutics outside the submitted work. C.B. Pratt reports other support from Tango Therapeutics outside the submitted work. S.S. Rajeswaran reports other support from Tango Therapeutics outside the submitted work. H.-J. Wu reports other support from Tango Therapeutics outside submitted work. C.G. Comer reports other support from Tango Therapeutics outside submitted work. S. Sudsakorn reports other support from Tango Therapeutics outside the submitted work. W. Zhang reports other support from Tango Therapeutics outside the submitted work. S. Liu reports other support from Tango Therapeutics outside submitted work. S.R. Meier reports other support from Tango Therapeutics outside the submitted work. A.H. Choi reports other support from Tango Therapeutics outside the submitted work. T. Khendu reports other support from Tango Therapeutics outside the submitted work. H. Stowe reports other support from Tango Therapeutics outside the submitted work. B. Shen reports other support from Tango Therapeutics outside the submitted work. D.A. Whittington reports other support from Tango Therapeutics and personal fees and other support from Sesame Therapeutics outside the submitted work. Y. Chen reports other support from Tango Therapeutics outside the submitted work. Y. Yu reports personal fees from Tango Therapeutics during the conduct of the study, as well as holding Tango Therapeutics stocks and options. W.D. Mallender reports other support from Tango Therapeutics outside the submitted work. T. Feng reports other support from Tango Therapeutics outside the submitted work. J.N. Andersen reports other support from Tango therapeutics during the conduct of the study. J.P. Maxwell reports other support from Tango Therapeutics outside submitted work. S. Throner reports other support from Tango Therapeutics outside the submitted work, as well as a patent for WO2022197892 pending.
Authors’ Contributions
A. Simoneau: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. C.B. Pratt: Data curation, investigation, methodology, writing–review and editing. H.-J. Wu: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft. S.S. Rajeswaran: Investigation, methodology, writing–review and editing. C.G. Comer: Validation, investigation, methodology. S. Sudsakorn: Data curation, formal analysis, validation, methodology. W. Zhang: Data curation, formal analysis, methodology, writing–review and editing. S. Liu: Formal analysis, methodology. S.R. Meier: Resources, data curation, formal analysis. A.H. Choi: Data curation, formal analysis. T. Khendu: Data curation, formal analysis. H. Stowe: Resources, validation. B. Shen: Resources, validation. D.A. Whittington: Conceptualization, resources, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Chen: Resources, supervision, writing–review and editing. Y. Yu: Resources, data curation, formal analysis. W.D. Mallender: Resources, data curation, supervision, writing–review and editing. T. Feng: Conceptualization, resources, supervision, investigation, methodology, project administration, writing–review and editing. J.N. Andersen: Conceptualization, resources, supervision, project administration, writing–review and editing. J.P. Maxwell: Conceptualization, resources, data curation, formal analysis, supervision, validation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Throner: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Nguyen L, Martens JWM, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun 2020;11:5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 2017;355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murai J, Huang SyN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 2012;72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 5. Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 6. Taglialatela A, Leuzzi G, Sannino V, Cuella-Martin R, Huang J-W, Wu-Baer F, et al. REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol Cell 2021;81:4008–25.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tirman S, Quinet A, Wood M, Meroni A, Cybulla E, Jackson J, et al. Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells. Mol Cell 2021;81:4026–40.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 2007;28:786–97. [DOI] [PubMed] [Google Scholar]
- 9. Huang TT, Nijman SMB, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006;8:341–7. [DOI] [PubMed] [Google Scholar]
- 10. Lim KS, Li H, Roberts EA, Gaudiano EF, Clairmont C, Sambel LA, et al. USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol Cell 2018;72:925–41.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simoneau A, Engel JL, Bandi M, Lazarides K, Liu S, Meier SR, et al. Ubiquitinated PCNA drives USP1 synthetic lethality in cancer. Mol Cancer Ther 2023;22:215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. da Costa AA, Somuncu O, Ravindranathan R, Mukkavalli S, Martignetti DB, Nguyen H, et al. Single-stranded DNA gap accumulation is a functional biomarker for USP1 inhibitor sensitivity. Cancer Res 2024;84:3435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002;419:135–41. [DOI] [PubMed] [Google Scholar]
- 14. Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 2003;425:188–91. [DOI] [PubMed] [Google Scholar]
- 15. Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 1998;8:397–403. [DOI] [PubMed] [Google Scholar]
- 16. Tanizawa A, Fujimori A, Fujimori Y, Pommier Y. Comparison of topoisomerase I inhibition, DNA damage, and cytotoxicity of camptothecin derivatives presently in clinical trials. J Natl Cancer Inst 1994;86:836–42. [DOI] [PubMed] [Google Scholar]
- 17. Roulston A, Zimmermann M, Papp R, Skeldon A, Pellerin C, Dumas-Bérube É, et al. RP-3500: a novel, potent and selective ATR inhibitor that is effective in preclinical models as a monotherapy and in combination with PARP inhibitors. Mol Cancer Ther 2022;21:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Illuzzi G, Staniszewska AD, Gill SJ, Pike A, McWilliams L, Critchlow SE, et al. Preclinical characterization of AZD5305, a next generation, highly selective PARP1 inhibitor and trapper. Clin Cancer Res 2022;28:4724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zatreanu D, Robinson HMR, Alkhatib O, Boursier M, Finch H, Geo L, et al. Polθ inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat Commun 2021;12:3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buckmelter AJ, Ioannidis S, Follows B, Gustafson G, Wang M, Caravella JA, et al. Purinones as ubiquitin-specific protease 1 inhibitors. patent WO/2017/087837. 2017. [Google Scholar]
- 21. Arkinson C, Chaugule VK, Toth R, Walden H. Specificity for deubiquitination of monoubiquitinated FANCD2 is driven by the N-terminus of USP1. Life Sci Alliance 2018;1:e201800162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rennie ML, Arkinson C, Chaugule VK, Toth R, Walden H. Structural basis of FANCD2 deubiquitination by USP1−UAF1. Nat Struct Mol Biol 2021;28:356–64. [DOI] [PubMed] [Google Scholar]
- 23. Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 2017;14:290–6. [DOI] [PubMed] [Google Scholar]
- 24. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- 25. Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 2019;75:861–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood C, Burnley T, Patwardhan A, Scheres S, Topf M, Roseman A, et al. Collaborative computational project for electron cryo-microscopy. Acta Crystallogr D Biol Crystallogr 2015;71:123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michlits G, Hubmann M, Wu S-H, Vainorius G, Budusan E, Zhuk S, et al. CRISPR-UMI: single-cell lineage tracing of pooled CRISPR–Cas9 screens. Nat Methods 2017;14:1191–7. [DOI] [PubMed] [Google Scholar]
- 28. Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer 2015;15:166–80. [DOI] [PubMed] [Google Scholar]
- 29. Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, et al. A genetic map of the response to DNA damage in human cells. Cell 2020;182:481–96.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michlits G, Jude J, Hinterndorfer M, de Almeida M, Vainorius G, Hubmann M, et al. Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat Methods 2020;17:708–16. [DOI] [PubMed] [Google Scholar]
- 31. Zheng S, Wang W, Aldahdooh J, Malyutina A, Shadbahr T, Tanoli Z, et al. SynergyFinder plus: toward better interpretation and annotation of drug combination screening datasets. Proteom Bioinform 2022;20:587–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rennie ML, Arkinson C, Chaugule VK, Walden H. Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site. Sci Adv 2022;8:eabq6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coleman KE, Yin Y, Lui SKL, Keegan S, Fenyo D, Smith DJ, et al. USP1-trapping lesions as a source of DNA replication stress and genomic instability. Nat Commun 2022;13:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 2009;284:5343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sztupinszki Z, Diossy M, Krzystanek M, Reiniger L, Csabai I, Favero F, et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer 2018;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta R, Somyajit K, Narita T, Maskey E, Stanlie A, Kremer M, et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 2018;173:972–88.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dev H, Chiang T-WW, Lescale C, de Krijger I, Martin AG, Pilger D, et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat Cell Biol 2018;20:954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noordermeer SM, Adam S, Setiaputra D, Barazas M, Pettitt SJ, Ling AK, et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 2018;560:117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gogola E, Duarte AA, de Ruiter JR, Wiegant WW, Schmid JA, de Bruijn R, et al. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 2018;33:1078–93.e12. [DOI] [PubMed] [Google Scholar]
- 41. Pettitt SJ, Krastev DB, Brandsma I, Dréan A, Song F, Aleksandrov R, et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun 2018;9:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cadzow L, Brenneman J, Tobin E, Sullivan P, Nayak S, Ali JA, et al. The USP1 inhibitor KSQ-4279 overcomes PARP inhibitor resistance in homologous recombination–deficient tumors. Cancer Res 2024;84:3419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lohse I, Borgida A, Cao P, Cheung M, Pintilie M, Bianco T, et al. BRCA1 and BRCA2 mutations sensitize to chemotherapy in patient-derived pancreatic cancer xenografts. Br J Cancer 2015;113:425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Al Abo M, Sasanuma H, Liu X, Rajapakse VN, Huang S-YN, Kiselev E, et al. TDP1 is critical for the repair of DNA breaks induced by sapacitabine, a nucleoside also targeting ATM- and BRCA-deficient tumors. Mol Cancer Ther 2017;16:2543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 2015;518:254–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yazinski SA, Comaills V, Buisson R, Genois M-M, Nguyen HD, Ho CK, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 2017;31:318–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee JD, Ryu W-J, Han HJ, Kim TY, Kim MH, Sohn J. Molecular characterization of BRCA1 c.5339T>C missense mutation in DNA damage response of triple-negative breast cancer. Cancers 2022;14:2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nacson J, Krais JJ, Bernhardy AJ, Clausen E, Feng W, Wang Y, et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep 2018;24:3513–27.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharma Saha S, Gentles L, Bradbury A, Brecht D, Robinson R, O’Donnell R, et al. Genomic, transcriptomic, and functional alterations in DNA damage response pathways as putative biomarkers of chemotherapy response in ovarian cancer. Cancers 2021;13:1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma J, Shah R, Bell AC, McDermott N, Pei X, Selenica P, et al. Increased synthetic cytotoxicity of combinatorial chemo-radiotherapy in homologous recombination deficient tumors. Int J Radiat Oncol Biol Phys 2024 Jul 10. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A 2009;106:3166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 2009;28:2461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cong K, Peng M, Kousholt AN, Lee WTC, Lee S, Nayak S, et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol Cell 2021;81:3128–44.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moore CE, Yalcindag SE, Czeladko H, Ravindranathan R, Wijesekara Hanthi Y, Levy JC, et al. RFWD3 promotes ZRANB3 recruitment to regulate the remodeling of stalled replication forks. J Cell Biol 2023;222:e202106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sims AE, Spiteri E, Sims RJ, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 2007;14:564–7. [DOI] [PubMed] [Google Scholar]
- 56. Nijman SMB, Huang TT, Dirac AMG, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 2005;17:331–9. [DOI] [PubMed] [Google Scholar]
- 57. Leung W, Baxley RM, Traband E, Chang Y-C, Rogers CB, Wang L, et al. FANCD2-dependent mitotic DNA synthesis relies on PCNA K164 ubiquitination. Cell Rep 2023;42:113523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood 2011;117:5078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, Gutiérrez-Enríquez S, Ducy M, Ibrahim YH, et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med 2018;10:EMMM201809172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yap TA, Lakhani NJ, Patnaik A, Lee EK, Gutierrez M, Moore KN, et al. First-in-human phase I trial of the oral first-in-class ubiquitin specific peptidase 1 (USP1) inhibitor KSQ-4279 (KSQi), given as single agent (SA) and in combination with olaparib (OLA) or carboplatin (CARBO) in patients (pts) with advanced solid tumors, enriched for deleterious homologous recombination repair (HRR) mutations. J Clin Oncol 2024;42:3005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods include the chemical synthesis procedure of TNG348 and additional biochemical assay protocols not included in the main text.
Supplementary Figure S1 shows additional biochemical and in vivo exposure data to support Figure 1
Supplementary Figure S2 shows additional selectivity data from Eurofins KinomeScan and DiscoverX Safety Scan to support Figure 2. It additionally shows western blot controls for USP1 KO and cDNA expression, as well as the impact of the USP1 V156 K mutation on USP1 substrate and other mutations on the cell growth.
Supplementary Figure S3 shows CRISPR screening data suggesting that loss of the BRCA1-A complex sensitizes cells to TNG348, but not olaparib
Supplementary Figure S4 shows additional data supporting Figure 4. This includes an alternative representation for the combination cell line panels shown in Figure 4D-E, examples of combination between TNG348 and saruparib, olaparib, cisplatin or SN38, and the viability effects in MCF10A of TNG348, RP3500 and ART558 alone or in combination with olaparib.
Supplementary Figure S5 shows additional data supporting Figure 5 including mouse body weight following TNG348, PARPi or combination treatment, additional in vivo experiments with lower TNG348 doses, an in vivo experiment in a BRCA1/2wt PDX model, and plasma concentration comparisons between TNG348 or PARPi alone versus their combination.
Supplementary Table S1 shows the list of cell lines and culture conditions used in this study
Supplementary Table S2 shows the list of antibodies used in this study
Supplementary Table S3 shows information on Cryo-EM data collection, refinement and validation
Supplementary Table S4 lists the area under the curve values for each cell line in the TNG348 single agent cell line panel shown in Figure 3A-B.
Supplementary Table S5 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the UWB1.289 cell line from Figure 3C
Supplementary Table S6 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the MDA-MB-436 cell line from Figure 3D
Supplementary Table S7 lists the wtheta and p-values for each gene in the olaparib vs DMSO CRISPR screen in the UWB1.289 cell line from Figure 3E
Supplementary Table S8 lists the wtheta and p-values for each gene in the olaparib vs DMSO CRISPR screen in the MDA-MB-436 cell line from Figure 3F
Supplementary Table S9 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the COV362 cell line from Figure 4A
Supplementary Table S10 lists the wtheta and p-values for each gene in the TNG348 vs DMSO CRISPR screen in the HCC1395 cell line from Figure 4B
Supplementary Table S11 lists the bliss synergy score values for each cell line in the TNG348 and olaparib combination cell line panel shown in Figure 4D.
Supplementary Table S12 lists the bliss synergy score values for each cell line in the TNG348 and saruparib combination cell line panel shown in Figure 4E.
Supplementary Table S13 shows the statistical analysis of the different treatment groups from the different in vivo PDX studies
Supplementary Table S14 shows additional information on the PDX used in this study, including model histology, clinical subtype, relevant mutation and treatment history
Data Availability Statement
The cryo-EM data generated in this study are publicly available in RCSB protein data bank at PDB ID: 9EBS. CRISPR screening data generated in this study are publicly available in Gene Expression Omnibus at GSE284239. All other data are available upon request.