Summary
Skeletal muscle development and regeneration requires the activities of myogenic and non-myogenic muscle stem cell populations. Non-myogenic muscle stem cells, such as fibro-adipogenic progenitors (FAPs), play important roles in muscle regeneration after injury. Activated FAPs promote myogenic muscle stem cell differentiation and contribute to the restoration of muscle architecture. In pathological conditions, FAPs can differentiate into adipocytes or fibroblasts, causing fatty infiltrations or muscle fibrosis, respectively. Here, we identified the extracellular matrix protein ADAMTS-like 2 (ADAMTSL2) as a regulator of adipogenic and fibrogenic FAP differentiation. In the context of fibrogenic FAP differentiation, ADAMTSL2 inhibited the differentiation of primary mouse and human FAPs into fibroblasts in a transforming growth factor β (TGF-β)-dependent manner. Together with our previous data, a model emerges where ADAMTSL2 has a dual role in skeletal muscle biology, a pro-myogenic role, where ADAMTSL2 promotes myogenic muscle stem cell differentiation, and a TGF-β-dependent anti-fibrotic role where ADAMTSL2 attenuates FAP-to-fibroblast differentiation.
Subject areas: Biochemistry, Cell biology, Specialized functions of cells
Graphical abstract

Highlights
-
•
ADAMTS-like 2 regulates fibro-adipogenic progenitor (FAP) differentiation
-
•
ADAMTSL2 promotes FAP differentiation into adipocytes
-
•
ADAMTSL2 attenuates FAP differentiation into fibroblasts in a TGF-β dependent manner
Biochemistry; Cell biology; Specialized functions of cells
Introduction
Adult skeletal muscle has a remarkable intrinsic capacity to regenerate minor injuries caused by daily physical activity or moderate exercise.1,2 This regeneration program requires the coordinated differentiation and activity of myogenic and non-myogenic muscle stem cell populations. PAX7-positive myogenic muscle stem cells, i.e., satellite cells, become activated after muscle injury, proliferate, and differentiate into myoblasts and subsequently into myocytes.3,4,5 Myocytes then fuse with existing multinucleated myofibers or with each other to form new myofibers.6 This process ultimately restores the contractile function of skeletal muscle after injury. Mesenchymal PDGFRα-positive fibro-adipogenic progenitors (FAPs) represent an additional, non-myogenic muscle-resident stem cell population.7,8 Similar to satellite cells, quiescent FAPs become activated after muscle injury and expand through proliferation. Activated FAPs promote satellite cell and myoblast differentiation and are key players in the restoration of muscle extracellular matrix (ECM) and overall muscle architecture.9,10 During the resolution phase of the muscle injury response, activated FAPs are eliminated by tumor necrosis factor α (TNFα)-induced apoptosis.11 However, under pathological conditions, FAPs can differentiate into adipocytes or fibroblasts. Adipogenic FAP differentiation results in fatty infiltrates, which characterize diseased rotator cuff muscles following rotator cuff tears and are indicators of poor clinical outcomes.12,13,14 Fibrogenic FAP differentiation drives muscle fibrosis, a sequelae of muscular dystrophies and traumatic muscle injuries such as volumetric muscle loss.15,16,17 Muscle fibrosis is a key contributor to poor functional recovery after traumatic muscle injuries.18,19
Activation, elimination, and pathological differentiation of FAPs are regulated by several signaling pathways. Triggered by muscle injury, FAPs are activated by a combination of biomechanical cues, platelet-derived growth factor signaling, and transforming growth factor β (TGF-β) signaling.11,20,21 TGF-β ligands are secreted in a latent form bound to latent TGF-β binding proteins (LTBPs) and upon their activation transmit signals through TGF-β type I and type II receptors.22 Multiple cell sources secrete TGF-β isoforms after muscle injury, including infiltrating macrophages, regenerating myofibers, and FAPs themselves.11,23,24,25 When FAPs fail to be eliminated by TNFα-induced apoptosis, continued TGF-β signaling promotes the transition of activated FAPs into myofibroblasts, which deposit excessive collagen type-I-rich ECMs; the key signature of muscle fibrosis.16,21,26 At the same time, TGF-β signaling inhibits adipogenic differentiation of FAPs.21 Canonical wingless-related integration site (WNT) signaling through the WNT3A ligand also promotes the differentiation of activated FAPs into pro-fibrotic fibroblasts and regulates ECM gene expression, while canonical and non-canonical WNT signaling through WNT5A and WNT7A inhibits adipogenesis.27,28 Finally, bone-morphogenetic protein (BMP)-induced osteogenic differentiation of FAPs contributes to heterotopic ossification in the genetic disorder fibrodysplasia ossificans.29,30 Collectively, the integration of multiple signaling pathways determines the activation and injury- or disease-specific differentiation trajectory of FAPs into adipocytes, fibroblasts, or bone-forming cells.
TGF-β and WNT signaling are complex signaling pathways that frequently crosstalk with each other and elicit cell type-specific pleiotropic effects on growth arrest, cell proliferation, or the magnitude of ECM deposition.31,32,33 In addition to direct transcriptional regulation of TGF-β and WNT ligands and their respective receptors, both signaling pathways are also regulated by cell surface-bound or secreted ECM proteins.22,34,35 One example of such a regulatory ECM protein is the matricellular protein A disintegrin and metalloprotease with thrombospondin type I motif-like 2 (ADAMTSL2), which was shown to negatively regulate TGF-β signaling in skin and cardiac fibroblasts and to promote WNT signaling in myoblasts.36,37,38,39 Despite their homology to ADAMTS proteases, ADAMTS-like proteins, including ADAMTSL2, lack a protease domain and therefore fulfill other roles in the ECM.40 Recessive pathogenic variants in ADAMTSL2 cause geleophysic dysplasia, a syndromic connective tissue disorder characterized by short stature, delayed bone age, notably muscle hypertrophy, and severe cardiac and pulmonary anomalies.37 Pathogenic ADAMTSL2 variants have also been identified in the neonatal lethal Al-Gazali skeletal dysplasia and a dominant connective tissue disorder with similarities to dermatosparaxis Ehlers-Danlos syndrome.41,42 Global Adamtsl2 knockout mice do not survive past birth.43
In the current study, we identified a role for FAP-derived ADAMTSL2 in regulating adipogenic and fibrogenic FAP differentiation. Using primary mouse and human FAPs, we show that ADAMTSL2 attenuates fibrogenic FAP cell differentiation into pro-fibrotic fibroblasts in a TGF-β-dependent manner. Together with our previously published data, the findings presented here support a bifunctional role for ADAMTSL2 in skeletal muscle, where ADAMTSL2 promotes myogenic differentiation in a WNT-dependent manner and regulates FAP cell differentiation in a TGF-β-dependent manner.39
Results
ADAMTSL2 depletion in FAPs does not alter overall skeletal muscle architecture
We recently demonstrated a role for myoblast-derived ADAMTSL2 in skeletal muscle as a regulator of myogenic muscle stem cell differentiation.39 However, deletion of ADAMTSL2 in myogenic precursor cells reduced the amount of ADAMTSL2 mRNA and protein in skeletal muscle by 50–60%, suggesting additional cellular sources for ADAMTSL2.39 In an effort to identify other ADAMTSL2-expressing cell types in muscle, we mined two published single cell transcriptomic datasets. In the Tabula muris dataset, we identified a subset of non-myogenic mesenchymal stem cells, which include FAPs, as positive for Adamtsl2 expression (Figure 1A). Adamtsl2-positive FAPs represented 17% of the total mesenchymal stem cell population and significantly exceeded the proportion of Adamtsl2-positive cells in other muscle-resident cell populations under homeostatic conditions.44 In a second dataset, which demonstrated heterogeneity within skeletal muscle fibroblast populations, we identified Adamtsl2 expression in several but not all fibroblast sub-clusters (Figures 1B and 1C).45 Adamtsl2 was notably expressed in three of the five endomysial sub-clusters and in paramysial cells. Paramysial cells are described as a distinct cell population located between the perimysium and the endomysium characterized by high expression of Thbs4, Col1a1, and Postn, all encode regulatory (Thbs4 and Postn) or structural (Col1a1) ECM proteins, and low expression of Thbs1.45
Figure 1.
ADAMTSL2 is expressed in FAPs but does not affect skeletal muscle development or postnatal growth
(A) Single-cell transcriptomics data extracted from the Tabula muris dataset (https://tabula-muris.ds.czbiohub.org/) showing the percentage of Adamtsl2-expressing FAPs in adult mice.44 Total number of analyzed cells is indicated. T, T cells; End, endothelial cells; Sat, satellite cells; MΦ, macrophages; B, B cells.
(B) Bar plot showing the expression of Adamtsl2 in individual cells of distinct endomysial and paramysial skeletal muscle fibroblast sub-populations. Sub-clusters based on location are indicated and numbered according to Muhl et al.45 (https://betsholtzlab.org/Publications/FibroblastMural/database.html). SMC, smooth muscle cells.
(C) Percentage of Adamtsl2-positive cells with a count >1 from (B). Fibroblast sub-clusters with significant Adamtsl2 expression are highlighted in blue and the total number of analyzed cells per sub-cluster is indicated. Sub-cluster are numbered according to the legend in (B).
(D) Schematic showing the generation of mice with muscle connective tissue cell-specific inactivation of Adamtsl2. Note that Prx1-Cre deletes ADAMTSL2 also in other limb bud-derived tissues, such as bone and cartilage but not in myogenic progenitor cells.
(E) Mid-belly cross-sections of the TA from Ctrl and CKO-Prx mice stained with an ADAMTSL2 peptide antibody. Nuclei were counterstained with DAPI.
(F) Wet weight of Ctrl and CKO-Prx extensor digitorum longus (EDL), gastrocnemius (GM), and TA muscle normalized to body weight (n = 7 mice, 2 fields of view).
(G) Mid-belly cross-sections of the TA from Ctrl and CKO-Prx mice stained for the basement membrane component laminin, which outlines myofibers. Nuclei were counterstained with DAPI.
(H) Violin plot of myofiber cross-sectional areas (CSA) from (G). n = 768 (Ctrl) and n = 708 (CKO-Prx) myofibers from n = 3 mice were analyzed. Scale bars in (E) and (H) represent 50 μm.
Data are represented as mean ± SD. Statistical significance in (F), (G), and (I), was calculated with a two-sided Student’s t test, ∗∗p < 0.01 and ∗∗∗p < 0.001.
To investigate if ADAMTSL2 expression in FAPs is required for skeletal muscle development, we crossbred mice with a previously described conditional Adamtsl2 flox allele with mice where the expression of Cre recombinase was controlled by the Prx1 promoter (Prx1-Cre), resulting in CKO-Prx mice (Figure 1D).46,47 Prx1-Cre inactivates Adamtsl2 in the limb bud starting at E9.5 prior to the influx of myogenic precursor cells and therefore does not inactivate Adamtsl2 in satellite cells or myofibers. This approach allows to distinguish between contributions of FAP-derived ADAMTSL2 versus myogenic progenitor cell-derived ADAMTSL2 to muscle development and homeostasis.47,48 We first analyzed ADAMTSL2 protein deposition in tibialis anterior (TA) muscle cross-sections using a custom-made ADAMTSL2 peptide antibody. We observed localization of ADAMTSL2 in the endomysium, which surrounds laminin-positive myofibers, in the Ctrl but to a lesser extent in CKO-Prx muscles (Figure 1E). The residual amount of ADAMTSL2 is likely contributed by myogenic muscle stem cells, which expressed ADAMTSL2 during differentiation.39 Despite a role for FAPs in muscle development and the significant reduction of ADAMTSL2 protein levels, we did not observe an apparent gross muscle phenotype under homeostatic conditions (Figures 1F–1H). The normalized wet weights of CKO-Prx extensor digitorum longus (EDL), gastrocnemius (GM), and TA muscles were not changed compared to Adamtsl2-fl/fl control (Ctrl) mice (Figure 1F). Muscle architecture as assessed by measuring the cross-sectional area (CSA) of individual myofibers was also not changed in the TA and the number of nuclei per field of view was similar (Figures 1G and 1H).
ADAMTSL2-deficiency in FAPs attenuates muscle regeneration after volumetric muscle loss
We previously showed delayed muscle regeneration after BaCl2 injury when ADAMTSL2 was deleted in myogenic muscle stem cells.49 However, muscle regeneration after BaCl2 injury in CKO-Prx TA was not impaired (data not shown). Therefore, we pivoted to the more severe and clinically relevant volumetric muscle loss (VML) injury model and followed muscle regeneration after VML surgery in CKO-Prx and Ctrl mice.50,51 After VML, muscle stem cells and FAPs migrate into the defect site from uninjured muscle tissue but are unable to restore muscle architecture and function.51 To induce VML, we excised a full-thickness 2 mm diameter punch biopsy from the mid-belly section of the TA in Ctrl and CKO-Prx mice and collected muscle tissue at 7 dpi and 28 dpi for histological analyses (Figures 2A and 2B).52 To validate the VML injury model, we first visualized changes in muscle architecture by laminin staining and regenerating myofibers by eMyHC staining in cross-sections through the TA after VML surgery compared to uninjured TA (Figure 2C). Compared to the well-organized myofibers in the uninjured TA, VML surgery resulted in the presence of disjointed myofibers with large spacing in-between. These myofibers stained positive for eMyHC, which indicates regenerating myofibers. In addition, a high cellularity shown by the increased number of DAPI-positive nuclei in between myofibers suggested a robust influx of immune cells, which is also a characteristic response to VML. We then compared the injury response to VML in Ctrl and CKO-Prx mice, where ADAMTSL2 was depleted in FAPs (Figures 2D–2H). At 7 dpi, we observed a significant decrease in eMyHC staining in injured CKO-Prx TA compared to injured Ctrl TA, suggesting delayed muscle regeneration after VML in the absence of ADAMTSL2 (Figures 2D and 2E). Laminin staining around injured and regenerating myofibers was disrupted in both genotypes. At 28 dpi, we observed a partial restoration of muscle architecture in both genotypes as indicated by groups of connected, laminin-positive myofibers with spaces in between (Figure 2F). However, the myofiber CSA in injured CKO-Prx TA was significantly reduced compared to injured Ctrl TA muscles (Figure 2G). Concomitant with the reduction in myofiber CSA in injured CKO-Prx TA, we observed an increase in the percentage of myofibers with centralized nuclei, which permanently label regenerated myofibers (Figure 2H). These observations are consistent with delayed and impaired muscle regeneration after VML in ADAMTSL2-deficient CKO-Prx TA.
Figure 2.
ADAMTSL2 in FAPs impacts muscle regeneration after volumetric muscle loss (VML)
(A) Surgical approach using a 2 mm biopsy punch resulting in full-thickness VML to the mid-belly section of the TA.
(B) Schematic depicting analysis time points post-VML injury.
(C) Laminin and embryonic myosin heavy chain (eMyHC) staining at 7 dpi in VML-injured TA compared to contralateral uninjured TA of WT mice showing significant muscle regeneration activity in the injured TA only.
(D) Laminin and eMyHC staining of VML-injured CKO-Prx and Ctrl TA at 7 dpi. Nuclei were counterstained with DAPI.
(E) Quantification of eMyHC signal intensity from (D) (n = 4 mice, 2 fields of view).
(F) Laminin of VML-injured CKO-Prx and Ctrl TA at 28 dpi. Nuclei were counterstained with DAPI.
(G) Quantification of myofiber cross-sectional area (CSA) from F (n = 456 (Ctrl) and n = 724 (KO-Prx) myofibers were analyzed).
(H) Quantification of the percentage of myofibers with centrally located nuclei, indicating regenerated myofibers (n = 3 mice, 2 fields of view). Scale bars in (C), (D), and (F) represent 50 μm.
Data are represented as mean ± SD. Statistical significance in (E), (G), and (H) was calculated with a two-sided Student’s t test, ∗p < 0.05 and ∗∗∗p < 0.001.
FAP-derived ADAMTSL2 regulates cell proliferation and can compensate for ADAMTSL2-deficiency in myoblasts
To compare cellular phenotypes of Ctrl and CKO-Prx-derived ADAMTSL2-deficient FAPs in vitro, we adapted a cell surface antigen-mediated magnetic activated cell sorting (MACS) protocol to isolate FAPs from mouse hindlimb muscles (Figure 3A).27,53 In a negative selection step, muscle cell suspensions were first depleted of CD31+ endothelial cells, CD45+ hematopoietic cells, and integrin α7+ satellite cells. FAPs were then enriched using the mouse-specific SCA1 antigen and cultured on laminin- and collagen type I-coated cell culture plates. To determine the purity of the isolated FAPs, we used immunostaining for PDGFRα, a cell-surface epitope that is expressed by most FAP sub-populations (Figure 3B).7,10 Close to 100% of MACS-purified FAPs stained positive for PDGFRα demonstrating the successful isolation of pure FAP populations, which were then used in subsequent experiments.
Figure 3.
FAP isolation and phenotypes of ADAMTSL2-deficient FAPs
(A) Schematic of the magnetic activated cell sorting (MACS)-based FAP purification protocol.
(B) PDGFRα immunostaining showing purity of FAP preparation from wild-type mice. Nuclei were counterstained with DAPI.
(C) Adamtsl2 mRNA quantification in Ctrl and CKO-Prx FAPs after 4 days of fibrogenic differentiation. Adamtsl2 mRNA levels were normalized to Gapdh and Ctrl.
(D) Western blot analysis of ADAMTSL2 protein in cell lysates from Ctrl and CKO-Prx FAPs.
(E) Quantification of ADAMTSL2 band intensity in D normalized to GAPDH and Ctrl (n = 3 independent FAP preparations).
(F) Western blot analysis of ADAMTSL2 protein in cell lysates from primary Ctrl and CKO-Prx myoblasts isolated via pre-plating.
(G) Ki67 immunostaining identifies proliferating FAPs isolated from Ctrl or CKO-Prx. Nuclei were counterstained with DAPI.
(H) Quantification of percentage of proliferating Ki67+ cells from (G). n = 6 fields of view from n = 1 FAP isolate.
(I) Schematic depicting the setup of the FAP/C2C12 myoblast co-culture system.
(J) Visualization of multinucleated myotubes by immunostaining for myosin heavy chain (MyHC) and DAPI staining for nuclei. sh-Adamtsl2-expressing C2C12 myoblasts were co-cultured with Ctrl or CKO-Prx FAPs. sh-Ctrl and sh-Adamtsl2 treated C2C12 myoblasts in single cultures served as controls for myotube formation and the effect of ADAMTSL2-depletion in myoblast differentiation as reported previously.39
(K) Quantification of fusion index, i.e., the percentage of nuclei in multinucleated myotubes, from (J). n = 3 co-cultures. Scale bars in (B), (G), and (J) represent 50 μm.
Data are represented as mean ± SD. Statistical significance in (C), (E), and (H) was calculated with a two-sided Student’s t test and in (K) with a one-way ANOVA with posthoc-Tukey test. ∗p < 0.05 and ∗∗∗p < 0.001.
Next, we isolated FAPs from Ctrl or CKO-Prx muscles using MACS and quantified the reduction of ADAMTSL2 mRNA by quantitative real-time PCR (qPCR) and ADAMTSL2 protein by western blot analysis (Figures 3C–3E). We observed a significant reduction of ADAMTSL2 mRNA and protein levels in CKO-Prx FAPs with some batch-to-batch variability. Cre-mediated deletion of exon 5 of Adamtsl2 results in a reading frameshift and a premature stop codon, which likely triggers nonsense-mediated mRNA decay.46 The residual amount of ADAMTSL2 could be explained by incomplete targeting of FAPs by Prx1-Cre, co-purification of PDGFRα-positive cell types other than FAPs that express residual amounts of ADAMTSL2, or co-purification of ADAMTSL2 protein secreted by other muscle cell types, such as differentiating myoblasts that adheres to the surface of FAPs. As a control, we also isolated muscle stem cell-derived myoblasts from Ctrl and CKO-Prx muscles with a pre-plating method and determined ADAMTSL2 protein quantities by western blot analysis.54 As expected, ADAMTSL2 protein levels were not reduced in CKO-Prx myoblasts since CRE under the control of the Prx1 promoter is not expressed in myogenic precursor cells or their derivatives (Figure 3F). This demonstrates that in CKO-Prx muscles ADAMTSL2 is selectively depleted in non-myogenic muscle cells, including FAPs but not in the myogenic muscle stem cell populations. When culturing CKO-Prx FAPs, we noticed increased cell numbers compared to Ctrl FAPs. To determine if increased Prx-CKO FAP cell numbers were due to increased proliferation, we quantified the percentage of Ki67-positive cells in Ctrl and CKO-Prx FAPs (Figures 3G and 3H). ADAMTSL2-deficient FAPs indeed showed a significant increase in the proliferation index, i.e., percentage of Ki67-positive cells, compared to Ctrl FAPs.
Since FAPs can promote the differentiation of myogenic muscle stem cells into multinucleated myotubes, we examined if ADAMTSL2 from FAPs can rescue the attenuated differentiation of ADAMTSL2-deficient myoblasts that we have observed previously.10,39 For that, we stably depleted ADAMTSL2 in C2C12 myoblasts using shRNA, co-cultured the ADAMTSL2-deficient myoblasts with Ctrl or CKO-Prx FAPs, and quantified myotube formation via the fusion index, i.e., the percentage of nuclei within multinucleated, myosin heavy chain (MyHC)-positive myotubes (Figure 3I). As shown previously, the fusion index in differentiating ADAMTSL2-deficient C2C12 myoblasts was strongly reduced and multinucleated MyHC-positive myotubes were largely absent (Figures 3J and 3K).39 When these ADAMTSL2-deficient myoblasts were co-cultured with Ctrl FAPs that express ADAMTSL2, myotube formation was restored, suggesting a paracrine contribution of ADAMTSL2 that promoted myoblast differentiation. Conversely, co-culture of ADAMTSL2-deficient myoblasts with ADAMTSL2-deficient CKO-Prx FAPs did not rescue myotube formation as indicated by the near absence of MyHC-positive myotubes, suggesting that this effect was indeed mediated by ADAMTSL2.
ADAMTSL2-deficient FAPs show altered differentiation into adipocytes and fibroblasts
Since FAPs can differentiate into adipocytes or fibroblasts, we investigated if ADAMTSL2 is regulating these processes. We first assessed the capacity of ADAMTSL2-deficient FAPs to differentiate into adipocytes by culturing FAPs in the presence of an adipogenic cocktail consisting of 3-isobutyl-1-methylxanthine, dexamethasone, and insulin (Figure 4A). Under these adipogenic differentiation conditions, ADAMTSL2-deficient CKO-Prx FAPs showed reduced perilipin staining, a marker protein that is associated with intracellular lipid droplets in adipocytes suggesting that ADAMTSL2 facilitates adipogenic differentiation of FAPs (Figures 4B and 4C). Next, we determined if ADAMTSL2 regulates TGF-β2-induced fibrogenic FAP differentiation (Figure 4D). Under these fibrogenic differentiation conditions, we observed an increased propensity of CKO-Prx FAPs to differentiate into fibroblasts when compared to Ctrl FAPs indicated by the increased signal intensity for α-smooth muscle actin (αSMA) (Figures 4E and 4F). These results were confirmed by western blot analysis, where the amount of αSMA protein in differentiated CKO-Prx FAPs was significantly increased compared to differentiated Ctrl FAPs (Figures 4G and 4H). Conversely, when Ctrl FAPs were differentiated in the presence of 100 μg/mL recombinant ADAMTSL2 protein, mRNA levels of Acta2 (encoding αSMA) and the fibrosis marker Col1a1 (encoding the alpha chain of collagen type I) were significantly decreased (Figure 4I).
Figure 4.
ADAMTSL2 limits fibrogenic FAP differentiation in a TGF-β signaling-dependent manner
(A) Diagram of adipogenic FAP differentiation.
(B) Perilipin immunostaining of differentiated Ctrl and CKO-Prx FAPs. Nuclei were counterstained with DAPI.
(C) Quantification of perilipin mean fluorescence intensity (MFI) from (B). n = 3 independent FAP isolates.
(D) Diagram of TGF-β2-induced fibrogenic FAP differentiation.
(E) Α-smooth muscle actin (αSMA) immunostaining of differentiated Ctrl and CKO-Prx FAPs. Nuclei were counterstained with DAPI.
(F) Quantification of αSMA MFI from (E). n = 3 independent FAP isolates.
(G) Western blot analysis of αSMA protein in cell lysates from Ctrl and CKO-Prx FAPs.
(H) Quantification of αSMA band intensity from G normalized to GAPDH and Ctrl (n = 5).
(I) Acta2 and Col1a1 mRNA quantification in Ctrl FAPs after 4 days of fibrogenic differentiation in the presence of 100 mg/mL recombinant ADAMTSL2 (rL2). mRNA levels were normalized to Gapdh and PBS treated Ctrl FAPs.
(J) Diagram of spontaneous fibrogenic FAP differentiation.
(K) αSMA immunostaining of differentiated Ctrl and CKO-Prx FAPs. Nuclei were counterstained with DAPI.
(L) Quantification of αSMA MFI from (K). n = 3 independent FAP isolates.
(M) Acta2 mRNA quantification in Ctrl and CKO-Prx FAPs after 4 days of spontaneous fibrogenic differentiation. mRNA levels were normalized to Gapdh and Ctrl FAPs.
(N) Western blot analysis of phosphorylated SMAD2 protein (pSMAD2) in cell lysates from Ctrl and CKO-Prx FAPs undergoing spontaneous fibrogenic differentiation.
(O) Quantification of pSMAD2 band intensity from N normalized to GAPDH and Ctrl (n = 3).
(P) αSMA immunostaining of spontaneously differentiated Ctrl and CKO-Prx FAPs after treatment with vehicle (DMSO) or 5 μm SB431542, a potent TGF-β type I receptor inhibitor. Nuclei were counterstained with DAPI.
(Q) Quantification of αSMA MFI from (P). n = 3 independent FAP isolates.
(R) Collagen type I immunostaining of spontaneously differentiated Ctrl and CKO-Prx FAPs. Nuclei were counterstained with DAPI.
(S) Quantification of collagen type I MFI from (R). n = 3–4 independent FAP isolates.
(T) Col1a1 mRNA quantification in Ctrl and CKO-Prx FAPs after 4 days of spontaneous fibrogenic differentiation. mRNA levels were normalized to Gapdh and Ctrl FAPs. Scale bars in (B), (E), (K), (P), and (R) represent 100 μm.
Data are represented as mean ± SD. Statistical significance in (C), (F), (H), (I), (L), (M), (O), (S), and (T) was calculated with a two-sided Student’s t test and in (Q) with a one-way ANOVA with posthoc-Tukey test. ∗∗p < 0.01 and ∗∗∗p < 0.001.
Since ADAMTSL2 was described as a negative regulator of TGF-β signaling and TGF-β signaling promotes fibrogenic FAP differentiation, we next examined if ADAMTSL2-deficient FAPs showed a similar increased propensity toward fibrogenic differentiation when cultured in the absence of recombinant TGF-β2 ligand (Figure 4J).36,37,38 Under these spontaneous fibrogenic differentiation conditions, we also observed a strong increase in αSMA-positive fibroblasts originating from CKO-Prx FAPs compared to Ctrl FAPs (Figures 4K and 4L). This was confirmed by qPCR analysis of Acta2 mRNA levels, which robustly increased in CKO-Prx FAPs (Figure 4M). This suggested that the absence of ADAMTSL2 resulted in increased FAP proliferation and fibrogenic differentiation; processes that are regulated by TGF-β signaling.21 To directly test, if canonical TGF-β signaling is dysregulated in CKO-Prx FAPs, we analyzed SMAD2 phosphorylation by western blot analysis during spontaneous FAP differentiation (Figures 4N and 4O). We observed a significant increase in the amount of phosphorylated SMAD2 protein (pSMAD2) in ADAMTSL2-deficient CKO-Prx FAPs compared to Ctrl FAPs. This suggested aberrant activation of canonical TGF-β signaling in ADAMTSL2-deficient FAPs. To further probe if the ADAMTSL2-TGF-β axis regulates fibrogenic FAP differentiation, we allowed for spontaneous CKO-Prx FAP differentiation in the presence of the compound SB431542, a frequently used, highly specific small molecule TGF-β type I receptor inhibitor.55 In the presence of SB431542, we observed a significant reduction in αSMA-positive fibroblasts originated from ADAMTSL2-deficient CKO-Prx FAPs, reaching the level of untreated Ctrl FAPs (Figures 4P and 4Q). A hallmark of pro-fibrotic FAP-derived fibroblasts is their capacity to deposit collagen type I. By immunostaining and qPCR, we detected increased collagen type I deposition in the ECM of spontaneously differentiating CKO-Prx FAPs compared to Ctrl FAPs and increased Col1a1 mRNA levels, respectively (Figures 4R–4T).
ADAMTSL2 promotes adipogenic and attenuates fibrogenic differentiation of human FAPs
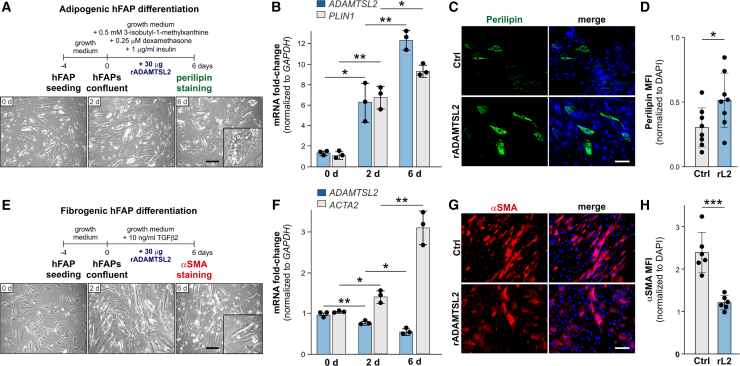
To determine if ADAMTSL2 is also involved in regulating the differentiation of human FAPs (hFAPs), we cultured primary vastus lateralis muscle-derived hFAPs under adipogenic and fibrogenic conditions and quantified ADAMTSL2 mRNA. Under adipogenic differentiation conditions, ADAMTSL2 mRNA was strongly induced at 2 days and 6 days post-differentiation coinciding with the induction of perilipin (PLIN) mRNA (Figures 5A and 5B). When hFAPs were differentiated in the presence of recombinant ADAMTSL2, adipogenesis increased as indicated by increased perilipin immunostaining (Figures 5C and 5D). These findings are consistent with reduced adipogenesis observed in ADAMTSL2-deficient primary mouse CKO-Prx FAPs (Figures 4A–4C). When fibrogenic differentiation of hFAPs was induced with recombinant TGF-β2, ADAMTSL2 mRNA decreased somewhat unexpectedly during the time course of differentiation by ∼50%, while the fibroblast marker gene encoding αSMA (ACTA2) increased (Figures 5E and 5F). When exogenous recombinant ADAMTSL2 protein was added, fibrogenic hFAP differentiation was reduced, as indicated by reduced αSMA immunostaining (Figures 5G and 5H). These data were also consistent with the increased fibrogenic differentiation observed in ADAMTSL2-deficient primary mouse CKO-Prx FAPs (Figures 5D–5H).
Figure 5.
Adipogenic and fibrogenic differentiation of human FAPs (hFAPs) is regulated by recombinant ADAMTSL2
(A) Top: diagram of adipogenic hFAP differentiation. Bottom: brightfield images of hFAPs undergoing adipogenic differentiation. Inset shows an adipocyte with lipid droplets.
(B) Relative quantification of ADAMTSL2 and perilipin (PLIN1) mRNA isolated from hFAPs undergoing adipogenic differentiation. The fold-change after normalization to GAPDH expression is shown (n = 3).
(C) Perilipin immunostaining of hFAPs differentiated in the presence or absence of 30 μg (100 μg/mL) exogenous recombinant ADAMTSL2 (rL2) protein. Nuclei were counterstained with DAPI.
(D) Quantification of perilipin MFI from (C). 2 fields of view from n = 4 wells per FAP isolate were analyzed.
(E) Top: diagram of fibrogenic hFAP differentiation. Bottom: brightfield images of hFAPs undergoing fibrogenic differentiation. Inset shows elongated fibroblast-like cell morphology.
(F) Relative quantification of ADAMTSL2 and ACTA2 (aSMA) mRNA isolated from hFAPs undergoing fibrogenic differentiation. The fold-change after normalization to GAPDH expression is shown (n = 3).
(G) aSMA immunostaining of hFAPs differentiated in the presence or absence of 30 μg (100 μg/mL) exogenous recombinant ADAMTSL2 (rL2) protein. Nuclei were counterstained with DAPI.
(H) Quantification of αSMA MFI from (G). 2 fields of view from n = 4 wells per FAP isolate were analyzed. Scale bars in (A), (C), (E), and (G) represent 100 μm.
Data are represented as mean ± SD. Statistical significance in (B) and (F) was calculated with a one-way ANOVA with posthoc-Tukey test and in (D) and (H) with a two-sided Student’s t test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Discussion
The coordinated differentiation of myogenic and non-myogenic muscle stem cell populations is critical for muscle development, homeostasis, and regeneration.48,56 In particular, dysregulated FAP cell differentiation can contribute to muscle pathologies such as fatty infiltrates or muscle fibrosis.11,21,57,58 Secreted regulatory ECM proteins, such as ADAMTSL2 can influence the differentiation trajectories of these muscle stem cell populations and thus contribute to muscle homeostasis and attenuate or augment muscle pathologies. In the context of muscle development, we showed previously that ADAMTSL2 promoted the differentiation of myoblasts by augmenting canonical WNT signaling.39 Since ADAMTSL2 is also expressed in non-myogenic mesenchymal-derived muscle stem cells, i.e., FAPs, we here examined its role during FAP differentiation. We identified ADAMTSL2 as a positive regulator of adipogenic FAP differentiation and a negative regulator of fibrogenic FAP differentiation. The latter effect was attributed to the TGF-β regulatory activity of ADAMTSL2. Despite aberrant differentiation of FAPs in the absence of ADAMTSL2, in vivo inactivation of Adamtsl2 did not cause apparent muscle phenotypes under homeostatic conditions. However, the response to severe muscle injury, i.e., VML, was delayed in the absence of ADAMTSL2.
The fact that we did not identify an apparent muscle phenotype after ADAMTSL2 inactivation in FAPs in vivo under homeostatic conditions was somewhat unexpected, given the prominent roles of FAPs in muscle development and homeostasis and the potent signaling regulator activities of ADAMTSL2. For example, depletion of FAPs from skeletal muscle resulted in loss of muscle mass and muscle strength, reduced myofiber CSA, and impairment of neuromuscular junctions.7,59 The lack of a muscle phenotype in CKO-Prx mice is in contrast to mice, where ADAMTSL2 was deleted in myogenic progenitor cells.39 Here we observed increased muscle weight and myofiber CSA in the TA and EDL, but not the GM. This suggests that FAP-derived ADAMTSL2 cannot compensate for the absence of ADAMTSL2 in myogenic progenitor cells in vivo. An additional explanation for the lack of a gross muscle phenotype in the CKO-Prx muscle under homeostatic conditions is a potential compensation for ADAMTSL2-deficiency in FAPs by ADAMTSL2 from differentiating myoblasts or other cellular sources. The lack of a phenotype in CKO-Prx muscle could indicate that ADAMTSL2 originating from differentiating myoblasts compensates for the lack of ADAMTSL2 in FAPs but not vice versa. However, we still observed reduced ADAMTSL2 deposition in CKO-Prx muscle tissue. A combined myogenic progenitor- and FAP-specific ADAMTSL2 deletion would resolve this question. In our co-culture experiments, we showed that ADAMTSL2 from FAPs could rescue the differentiation of ADAMTSL2-deficient myoblasts. That suggests that, in principle, ADAMTSL2 from FAPs can promote myogenesis and crosstalk with differentiating myoblasts. However, in uninjured muscle in vivo, FAPs and satellite cells are separated by a basement membrane, and it is unknown if ADAMTSL2 can cross this barrier. If ADAMTSL2 cannot cross the muscle basement membrane, then there may be a biological need for two independent pools of ADAMTSL2, a FAP-derived ADAMTSL2 pool and a differentiating myoblast-derived ADAMTSL2 pool, both fulfilling largely cell autonomous roles.
Since FAPs typically do not differentiate into adipocytes or fibroblasts under homeostatic conditions, we believe ADAMTSL2 may play a more important role in regulating FAP differentiation after acute muscle injuries. When we used a mild muscle injury model (BaCl2 injection), we did not observe aberrant muscle regeneration in CKO-Prx TA muscles. This was in contrast to the delayed muscle regeneration that we have observed using the same injury model after ADAMTSL2 ablation in myogenic muscle stem cells.49 This suggests that ADAMTSL2 from FAPs does not play a prominent role during muscle regeneration of smaller injuries or that myoblast-derived ADAMTSL2 can fully compensate for the lack of ADAMTSL2 from FAPs. In contrast, we did observe delayed and aberrant muscle regeneration after VML in CKO-Prx mice. VML induces a much more severe muscle injury, including a strong inflammatory response and the loss of muscle ECM and muscle-resident stem cells.51 Delayed muscle regeneration in CKO-Prx mice was indicated by reduced eMyHC staining at 7 dpi and reduced myofiber CSA at 28 dpi. These results suggested that ADAMTSL2 in FAPs is required to promote muscle regeneration after VML. This could be achieved through a supportive role for ADAMTSL2, where FAP-derived ADAMTSL2 promotes the migration and differentiation of myoblasts into the defect site or in a more active role, where FAPs-derived ADAMTSL2 contributes to the restoration of muscle architecture through regulation of TGF-β signaling or other signaling pathways. The fact that ADAMTSL2 in FAPs is required for muscle regeneration after VML, but apparently dispensable for muscle regeneration after BaCl2 injury could also be explained by quantitative requirements depending on the severity of muscle injuries. In such a scenario, ADAMTSL2 from myoblasts is sufficient to promote complete muscle regeneration after less severe injuries, but ADAMTSL2 from both cell sources, myogenic muscle stem cells and FAPs, is required to promote regeneration of larger muscle injuries, such as VML. In line with this hypothesis, we also observed delayed muscle regeneration after VML when ADAMTSL2 was depleted in myogenic muscle stem cells (data not shown).
One approach to determine the role of ADAMTSL2 during FAP differentiation is to resort to purified FAPs in vitro. By using in vitro FAP differentiation assays, we identified a role for ADAMTSL2 in regulating adipogenic and fibrogenic differentiation of FAPs. In the absence of ADAMTSL2, FAPs had a higher propensity to differentiate into αSMA-positive fibroblasts. This was linked to elevated TGF-β signaling as evidenced by increased SMAD2 phosphorylation in cell lysates from CKO-Prx FAP-derived fibroblasts and, more importantly, by the attenuation of fibrogenic differentiation of CKO-Prx FAPs after treatment with a small molecule TGF-β receptor type I inhibitor. These findings are in line with previous reports on the role of ADAMTSL2 as a negative regulator of TGF-β signaling in skin and cardiac fibroblasts. TGF-β ligand and therefore, canonical TGF-β signaling, was increased in skin fibroblasts isolated from patients with geleophysic dysplasia due to mutations in ADAMTSL2.37,38 In cardiac fibroblasts, overexpression of ADAMTSL2 decreased TGF-β signaling possibly by limiting the amount of latent TGF-β-binding protein 1 (LTBP1) secretion and/or its ECM incorporation.36 This attenuated the differentiation of cardiac fibroblasts into pro-fibrotic myofibroblasts and could play a role in the regulation of cardiac fibrosis. In this system, ADAMTSL2 did not appear to directly interfere with active TGF-β signaling for example by sequestering active TGF-β ligand or by limiting the TGF-β ligand-TGF-β receptor interaction. However, the TGF-β-regulatory role of ADAMTSL2 may also be cell-type or tissue specific. For example, we showed that ADAMTSL2 augmented WNT signaling during myoblast differentiation by engaging with WNT ligands and the WNT receptor.39 The ADAMTSL2-mediated promotion of myoblast differentiation was independent of TGF-β signaling. We also showed that a TGF-β neutralizing antibody treatment could not rescue bronchial epithelial dysplasia in Adamtsl2 knockout mice, despite normalization of TGF-β signaling within the bronchial epithelium.43 Finally, elevated TGF-β signaling was not observed in two disease-specific Adamtsl2 knock-in mouse models that recapitulated key features of geleophysic dysplasia nor was TGF-β signaling elevated in patient-derived fibroblasts from a patient with Al-Gazali skeletal dysplasia due to mutations in ADAMTSL2, which represents the most severe spectrum of ADAMTSL2-related disorders.41,60 Due to the severity of Al-Gazali skeletal dysplasia, which includes a high incidence of neonatal lethality, one would have predicted that TGF-β would be highly dysregulated. Therefore, it remains unclear if TGF-β dysregulation is disease-driving in ADAMTSL2-related connective tissue disorders and if targeting aberrant TGF-β signaling would represent a viable therapeutic option for treating affected patients.
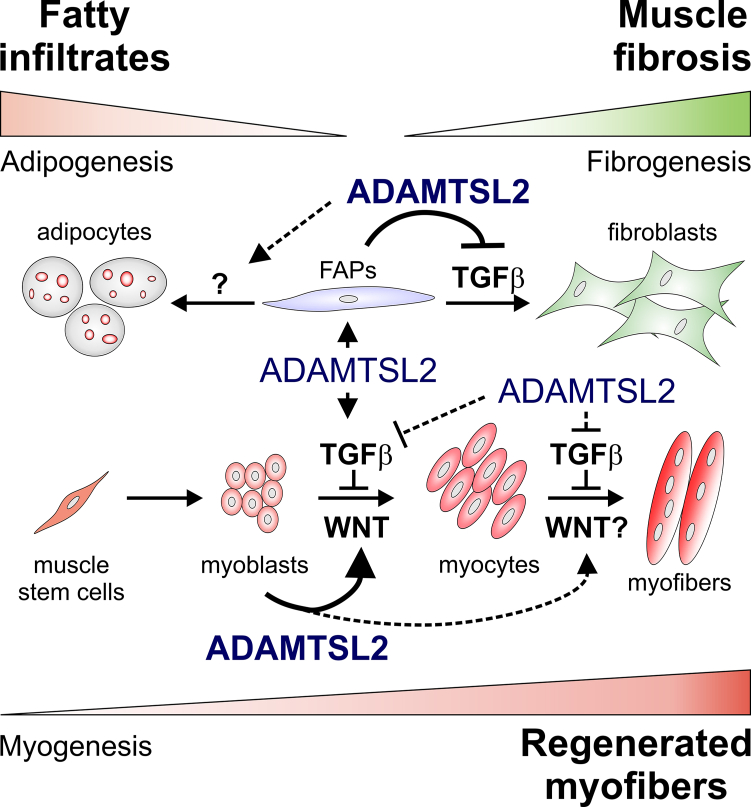
Collectively, our data support a model, where ADAMTSL2 has a dual role in skeletal muscle (Figure 6). In one role, FAP-derived ADAMTSL2 is required to attenuate FAP differentiation into fibroblasts and regulate adipogenic FAP cell differentiation. These roles are likely relevant to regenerate microinjuries due to daily activity and moderate exercise and to prevent the accumulation of fibrosis in skeletal muscle. In another role, myoblast-derived ADAMTSL2 was not only required for normal muscle development and homeostasis but also promoted muscle regeneration after injury.39,49 Future research will examine to what extent ADAMTSL2 coordinates FAP and muscle stem cell differentiation during regeneration either fully cell autonomously or by mediating the crosstalk between both muscle stem cell populations.
Figure 6.
Conceptual model for the dual role of ADAMTSL2 in skeletal muscle
Bottom: by promoting the differentiation of myogenic muscle stem cells in a WNT-dependent manner, ADAMTSL2 promotes the formation of myofibers during muscle development and muscle regeneration after injury.39,49 Top: by attenuating fibrogenic differentiation of FAPs in a TGF-β-dependent manner, ADAMTSL2 prevents muscle fibrosis. The relevance of ADAMTSL2 for adipogenic FAP differentiation and the regulated signaling pathways are currently unclear. Experimentally validated ADAMTSL2 regulatory mechanisms are indicated with solid arrows/lines and proposed ADAMTSL2 regulatory mechanisms are indicated with dashed lines.
Limitations of the study
One limitation of our study is that we did not determine if ADAMTSL2 regulates FAP differentiation after muscle injury in vivo. While glycerol injury elicits adipogenic FAP differentiation, models to elicit fibrogenic FAP differentiation leading to muscle fibrosis, are more challenging and may require repeat acute injuries, chronic muscle injuries, or co-injection of pro-fibrotic agents, such as TGF-β1.61,62,63,64 Another limitation is the use of Prx1-Cre to delete ADAMTSL2 since it also inactivates ADAMTSL2 in other musculoskeletal tissues.46 Therefore, we cannot rule out that the phenotype of Prx-CKO FAPs could be directly or indirectly altered due to changes during embryonic skeletal muscle development or by the lack of ADAMTSL2 in other skeletal muscle cell types, respectively, or that non-Prx-lineage cells contribute to the FAP population in the TA muscle.
Resource availability
Lead contact
Requests for resources and reagents should be directed to the lead contact, Dirk Hubmacher (dirk.hubmacher@mssm.edu).
Materials availability
The Adamtsl2-flox mouse will be made available upon request according to institutional and NIH guidelines. The original “knock-out” first allele (C57BL/6N-Adamtsl2tm1a(KOMP)Wtsi/MbpMmucd, #046490-UCD), generated by KOMP is available from Mutant Mouse Resource & Research Centers (MMRRC). The ADAMTSL2 antibody will be made available upon request.
Data and code availability
All data are contained within the manuscript or referenced. Any additional information required to reanalyze the data will be shared by the lead contact upon request. No original code was reported in this paper.
Acknowledgments
This work was in part supported by a grant from the National Institutes of Health (R01AR070748 to D.H. and R01AR080616 to W.M.H.). We thank Drs. Muhl and Betsholz (Karolinska Institute, Solna, Sweden) for the extraction of the ADAMTSL2 expression data in skeletal muscle fibroblasts.45 We thank Ms. Zerina Balic for technical support.
Author contributions
C.R. and N.T. performed the experiments, analyzed the data, and drafted the manuscript and figures. J.H.C. and K.J.H. isolated hFAPs and established their differentiation protocols. B.C.-Y., K.O., and W.M.H. provided expertise and reagents for mouse FAP isolation and established the mouse FAP differentiation protocols. D.H. conceived the study, analyzed data, edited the manuscript and figures. All authors revised the manuscript and approved the final version.
Declaration of interests
J.H.C. and K.J.H. are employees of Cook MyoSite Inc.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD31 | BioLegend | 102503; RRID: AB_312910 |
| Anti-CD45 | BioLegend | 103103; RRID: AB_312968 |
| Biotinylated anti-integrin α7 | Miltenyl Biotech | 130-128-938; RRID: AB_2905296 |
| Anti-SCA1 | BioLegend | 108103; RRID: AB_313340 |
| Anti-myosin heavy chain | DSHB | MF-20; RRID: AB_2147781 |
| Anti-PDGFRα | Thermo Fisher Scientific | 14-1401-82; RRID: AB_467491 |
| Ki67 | Cell Signaling Technology | 9129; RRID: AB_2687446 |
| Anti-perilipin | Cell Signaling Technology | 9349; RRID: AB_1082991 |
| Anti-αSMA | eBioscience | 14-9760-82; RRID: AB_2572996 |
| Collagen type I | Abcam | Ab34710; RRID: AB_731684 |
| Rhodamine Red™-X (RRX) AffiniPure™ Goat Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch | 111-295-144 |
| Rhodamine Red™-X (RRX) AffiniPure™ Goat Anti-Mouse IgG (H + L) | Jackson ImmunoResearch | 115-295-146 |
| Alexa Fluor® 488 AffiniPure™ Goat Anti-Mouse IgG (H + L) | Jackson ImmunoResearch | 115-545-146 |
| Alexa Fluor® 488 AffiniPure™ Goat Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch | 111-545-144 |
| Anti-ADAMTSL2 peptide antibody | Hubmacher Laboratory39 | |
| Anti-pSMAD2 | Cell Signaling Technology | 18338; RRID: AB_2798798 |
| Anti-GAPDH | EMD Millipore | MAB374; RRID: AB_2107445 |
| IR Dye 800CW Goat anti mouse IgG | LI-COR Biosciences | 926-32210; RRID: AB_621842 |
| IR Dye 800CW Goat anti rabbit IgG | LI-COR Biosciences | 926-32211; RRID: AB_621843 |
| IR Dye 680 RD Goat anti mouse IgG | LI-COR Biosciences | 926-68070; RRID: AB_10956588 |
| IR Dye 680 RD Goat anti rabbit IgG | LI-COR Biosciences | 926-68071; RRID: AB_10956166 |
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase type 4 | Worthington Biochemical Corporation | LS004188 |
| Dispase II | Gibco | 17105-041 |
| Streptavidin nanobeads | BioLegend | 480015 |
| MojoSort Buffer | BioLegend | 480017 |
| Laminin for coating | Gibco | 23017-015 |
| Collagen type 1 for coating | Gibco | A10483-01 |
| Recombinant FGF-2 | R&D | 3718-FB |
| Recombinant TGFβ2 | Thermo Fisher Scientific | PV6122 |
| 3-isobutyl-1-methylxanthine | Millipore Sigma | I5879 |
| dexamethasone | Millipore Sigma | D4902 |
| Insulin | Millipore Sigma | I1882 |
| SB431542 | Selleckchem | S1067 |
| Polyethylenimine | Polysciences | 23966 |
| paraformaldehyde | MP Biomedicals | 150146 |
| 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium | Abcam | AB104139 |
| Fluoromount-G containing DAPI | Abcam | AB104139 |
| Tragacanth | Thermo Scientific | A18502.36 |
| Methyl butane | Avantor | Q223-08 |
| Pierce Protease Inhibitor Mini Tablets | Thermo Fisher Scientific | A32955 |
| PhosSTOP | Millipore Sigma | 4906845001 |
| Experimental models: Cell lines | ||
| Human fibro-adipogenic progenitor cells | Cook MyoSite | T01743 |
| Experimental models: Organisms/strains | ||
| Adamtsl2-flox mice generated from C57BL/6N-Adamtsl2tm1a(KOMP)Wtsi/MbpMmucd | KOMP/Hubmacher laboratory43,46 | #046490-UCD |
| Prrx1Cre (Prx1-Cre) transgenic mice | The Jackson Laboratory | #033173 |
| C2C12 myoblasts | ATCC | CRL-1772 |
| HEK293T | ATCC | CRL-3216 |
| Oligonucleotides | ||
| ACTA2-f (5′-CTATGAGGGCTATGCCTTGCC-3′) | Thermo Fisher Scientific | n.a. |
| ACTA2-r (5′-GCTCAGCAGTAGTAACGAAGGA-3′) | Thermo Fisher Scientific | n.a. |
| ADAMTSL2-f (5′-GTGACGGGGTGCTTTTCTC-3′) | Thermo Fisher Scientific | n.a. |
| ADAMTSL2-r (5′-ATTCCCCTTGCGATAGTTGCC-3′) | Thermo Fisher Scientific | n.a. |
| PLIN1-f (5′-CCATGTCCCTATCAGATGCCC-3′) | Thermo Fisher Scientific | n.a. |
| PLIN1-r (5′-CTGGTGGGTTGTCGATGTC-3′) | Thermo Fisher Scientific | n.a. |
| GAPDH-f (5′-ACAACTTTGGTATCGTGGAAGG’-3′) | Thermo Fisher Scientific | n.a. |
| GAPDH-r (5′-GCCATCACGCCACAGTTTC-3′) | Thermo Fisher Scientific | n.a. |
| Acta2-f (5′-GACGTACAACTGGTATTGTG-3′) | Thermo Fisher Scientific | n.a. |
| Acta2-r (5′-TCAGGATCTTCATGAGGTAG-3′ | Thermo Fisher Scientific | n.a. |
| Adamtsl2-f (5′-CTTCAACTCCCGTGTGTATGAC-3′ | Thermo Fisher Scientific | n.a. |
| Adamtsl2-r (5′-TGTAGGTCGCATGGCTTACTG-3′) | Thermo Fisher Scientific | n.a. |
| Col1a1-f (5′-GCTCCTCTTAGGGGCCACT-3′) | Thermo Fisher Scientific | n.a. |
| Col1a1-r (5′-CCACGTCTCACCATTGGGG-3′) | Thermo Fisher Scientific | n.a. |
| Gapdh-f (5′-AGCTTCGGCACATATTTCATCTG-3′) | Thermo Fisher Scientific | n.a. |
| Gapdh-r (5′-CGTTCACTCCCATGACAAACA-3′ | Thermo Fisher Scientific | n.a. |
| Recombinant DNA | ||
| pcDNAMycHis3.1-ADAMTSL2 | Apte Laboratory64 | n.a |
| Software and algorithms | ||
| ImageJ/Fiji | Open Access | n.a |
| OriginPro 2019 | Origin Lab | n.a |
| CorelDraw Graphics Suite 2018 | Corel Draw | n.a |
Experimental model and study participant details
FAP-specific Adamtsl2 knockout
The generation and characterization of the conditional Adamtsl2 allele in C57BL/6 mice has been described previously.46 The floxed exon 5 of Adamtsl2 was excised using Prx1-Cre (#033173, Jackson Laboratory), which removes the sequences between the floxed regions in muscle connective tissue cells, including FAPs, but not in myogenic muscle stem cells.47 Removal of exon 5 results in a reading frameshift and a premature stop codon, which triggers nonsense-mediated mRNA decay. The muscle phenotype of CKO-Prx mice was analyzed between 6 and 8 weeks of age in male and female mice. Mice were fed a normal diet and kept under a 12 h light-dark cycle. At the time of tissue collection, mice were euthanized by CO2 asphyxiation followed by cervical dislocation. 6-10 week-old male and female mice were used. Animal experimentation was overseen by the Center for Comparative Medicine and Surgery at the Icahn School of Medicine at Mount Sinai and followed all federal regulations and the guidelines contained in the National Research Council Guide for the Care and Use of Laboratory Animals. Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai (#PROTO202000259).
Volumetric muscle loss surgery
VML surgery was performed as previously described in skeletally mature male and female mice between 3-6 months of age.52 After injecting 0.1 mg/kg buprenorphine hydrochloride intraperitoneally, Ctrl and CKO-Prx mice were anesthetized with 3% isoflurane inhalation using a nose cone. The front of the hind limb was shaved and the surgical site was disinfected with betadine and 70% isopropyl alcohol. A longitudinal skin incision was made to expose the TA muscle and the tip of a metal spatula was inserted posterior to the TA to prevent unintended damage to the tibia. Then, a circular, 2 mm diameter full-thickness defect was created in the mid-belly section of the TA using a disposable biopsy punch (Miltex). After hemostasis was achieved the wound was closed with 7 mm wound clips and mice were allowed to recover on a heating pad prior to returning them to their home cages. Mice were subsequently monitored for signs of distress, weight loss, and ambulation until euthanasia and tissue harvest at 7 dpi and 28 dpi.
Isolation of primary mouse FAPs
Primary mouse FAPs were isolated by magnetic activated cell sorting (MACS) as described previously.27,53 Briefly, murine hind limb muscles from two mice/isolation (6-8 weeks old, male or female) were dissected, minced, and digested with 0.2% collagenase type 4 (LS004188, Worthington Biochemical Corporation) and 2.5U/mL Dispase II (17105-041, Gibco) in 20 mL Dulbecco’s modified Eagles Medium (DMEM, 10-013-CV, Corning) in 50 mL conical tubes in an orbital shaker at 37°C for 1.5 h or until the solution was homogeneous. An FBS (10082147, Gibco)-coated 10 mL pipette was used to break up tissue chunks during this period. The cell suspension was filtered through a 70 μm cell strainer and 20 mL of DMEM was added to deactivate the collagenase. Cells were pelleted by centrifugation at 300 × g for 5 min. For negative selection by MACS separation, the cell suspension was labeled with 6 μL total of anti-CD31 (102503, BioLegend), anti-CD45 (103103, BioLegend) and 10 μL biotinylated anti-integrin α7 (130-128-938, Miltenyl Biotech) for 30 min on ice. Labeled cells were then incubated with 20 μL streptavidin nanobeads (480015, BioLegend) for 15 min on ice and run through an LS column attached to a magnet, which had been equilibrated with 2 mL of MojoSort Buffer (480017, BioLegend). The cell suspension in the flow through, predominantly FAPs, was labeled for positive selection with 5 μL anti-SCA1 (108103, BioLegend) for 30 min, followed by incubation with 20 μL streptavidin nanobeads for 15 min and separated with LS columns. After removal of the LS columns from the magnet the enriched SCA1+ cells, i.e., FAPs were eluted in 2 mL of MojoSort buffer using a plunger. FAPs attached to the nanobeads were pelleted at 300 × g for 5 min and 80,000 cells were seeded onto 10 cm cell culture plates coated with 10 μg/mL laminin (23017-015, Gibco) and 5 μg/mL collagen type I (A10483-01, Gibco) (passage 0). FAPs were cultured in complete DMEM (DMEM, 10% fetal bovine serum, 1% penicillin/streptomycin (15140-122, Gibco)) and expanded once they reached ∼70-80% confluency. Primary FAPs were used in passage 1 - 3 since their differentiation potential declines with further passaging.
Isolation of primary human FAPs
Frozen primary human FAPs (hFAPs, FP-1111, F01743-63F.0.2.P2) were provided by Cook MyoSite. hFAPs were isolated from the vastus lateralis muscle using a proprietary process capitalizing on their surface expression of the PDGFRα antigen. hFAPs once grown were checked for purity utilizing anti-desmin to detect myogenic cell contaminants. Fibrogenic differentiation ability was confirmed by growing hFAP in the presence of 2 ng/mL of TGFβ2, 10% FBS and 10% horse serum for seven days. Fibrogenic differentiation capacity was confirmed by expression of αSMA. Adipogenic differentiation ability was confirmed by growing hFAP in the presence of 10% FBS, 1 μg/mL insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine for 14 days. Adipogenic differentiation capacity was confirmed by the presence of perilipin-positive cells and the visual appearance of lipid droplets. All tissues were acquired from deceased donors and obtained in accordance with the United States Uniform Anatomical Gift Act.
Method details
Differentiation of FAPs into fibroblasts and adipocytes and treatments
Freshly isolated mouse FAPs were expanded for 2 days, seeded onto chamber slides, and cultured to near confluency in growth medium (complete DMEM supplemented with 2.5 ng/mL FGF-2 (3718-FB, R&D)). Fibrogenic differentiation was induced by switching the culture medium to complete DMEM supplemented with 10 ng/mL TGFβ2 (PV6122, Thermo Scientific). Medium was changed every 24 h for 4 days. For adipogenic differentiation, cells were cultured in complete DMEM supplemented with 0.5 mM 3-isobutyl-1-methylxanthine, 0.25 μM dexamethasone, and 1 μg/mL insulin for 4 days.
Frozen hFAPs, obtained from Cook MyoSite, were thawed in a water bath at 37°C and transferred into a 15 mL falcon tube containing 6 mL complete growth media. hFAPs were pelleted at 160 × g for 3 min and the pellet was resuspended in 10 mL fresh complete growth medium. 2 × 106 hFAPs were then cultured in a 10 cm cell culture plate. Fibrogenic and adipogenic differentiation was induced similar to the primary mouse FAPs, except that the media was changed every 48 h or 72 h, respectively, for 6 days. To investigate the effect of recombinant ADAMTSL2 on hFAP differentiation into fibroblasts, hFAPs were differentiated in the presence of PBS or full-length recombinant ADAMTSL2 protein at 100 μg/mL for 2 days.
To promote the spontaneous differentiation of FAPs into fibroblasts in the absence of TGFβ2, freshly isolated mouse FAPs were cultured for 2 days to near confluency in complete growth media. Then 2.2 × 106 FAPs were seeded in a 10 cm2 dish for western blot or 50,000 FAPs/chamber were seeded in an 8-well chamber slide for immunostaining. Cells were cultured for 7 days in complete growth medium. TGFβ signaling was inhibited by treatment of FAPs with 5 μM of the TGFβ type I receptor inhibitor SB431542 (S1067, Selleckchem) for 24 h prior to differentation.55,65
Co-culture of primary myoblasts and FAPs
The generation of ADAMTSL2-deficient C2C12 cells was described previously.39 Briefly, C2C12 cells (ATTC, CRL-1772) were stably transfected with a plasmid expressing shRNA that targets ADAMTSL2 and resulted in efficient depletion of ADAMTSL2 mRNA and protein and impaired C2C12 differentiation.39 For the co-cultures, we seeded 50,000 C2C12 myoblasts with 50,000 FAPs/well onto an 8-well glass chamber slide in complete DMEM medium including 10% FBS. After 24 h, the medium was switched to DMEM containing 2% horse serum to induce differentiation of myoblasts into myotubes. After 4 days, the co-cultures were fixed and stained for myosin heavy chain (MyHC, DSHB #MF-20) and DAPI to visualize myotubes and nuclei, respectively. The fusion index was quantified from immunofluorescence micrographs as the percentage of nuclei within the boundaries of MyHC-positive multinucleated myotubes.
Recombinant ADAMTSL2 protein production
For protein production, a previously described full-length mouse ADAMTSL2 expression construct was transfected into HEK293T cells (ATCC, CRL-3216) using polyethylenimine (Polysciences #23966).66 The amino acid sequence of mouse and human ADAMTSL2 is 87% identical.66 Serum-free medium from G418-selected ADAMTSL2-expressing cells was collected at 48 h and 72 h post-confluency. The pooled medium was concentrated using a KrosFlo KR2i filtration system (Spectrum labs) and ADAMTSL2 was affinity-purified using a Ni-NTA column (GE Healthcare #17-1408-01) connected to a NGC Chromatography System (Bio-Rad). The buffer was then changed to PBS via dialysis (Dialysis membrane, Thermo Scientific #66380) and the final protein concentration was determined with a Bradford Assay. Protein purity was assessed by SDS PAGE followed by staining with SimplyBlue SafeStain (Invitrogen). Purified ADAMTSL2 protein was stored at −20°C in aliquots.
mRNA quantification with qRT-PCR
Total RNA was isolated from FAPs with Trizol reagent according to the manufacturer’s protocol. Reverse transcriptase was used to convert 1 μg of RNA into cDNA. This cDNA obtained was used to perform quantitative real time PCR (qRT-PCR) to measure mRNA levels of ACTA2 (αSMA) (forward primer: 5′-CTATGAGGGCTATGCCTTGCC-3’; reverse primer: 5′-GCTCAGCAGTAGTAACGAAGGA-3′), ADAMTSL2 (forward primer: 5′-GTGACGGGGTGCTTTTCTC-3’; reverse primer: 5′-ATTCCCCTTGCGATAGTTGCC-3′), PLIN1 (perilipin) (forward primer: 5′-CCATGTCCCTATCAGATGCCC-3’; reverse primer 5′-CTGGTGGGTTGTCGATGTC-3′) and GAPDH (forward primer: 5′-ACAACTTTGGTATCGTGGAAGG’-3’; reverse primer 5′-GCCATCACGCCACAGTTTC-3′). All primers were designed and synthesized by IDT DNA. The conditions of the PCR were 48° for 30 min, 95° degrees for 15 s and 60° for 1 min for a total of 35 cycles in a C1000 Touch Thermal Cycler (BioRad). The ΔΔCt values were used as measure of the fold change of mRNA expression.
Immunostaining of FAPs
50,000 FAPs/chamber were seeded onto laminin and collagen type I-coated 8-well chamber slides in 200 μL of complete growth medium. FAPs were cultured for 3 days until they reached 95% confluence after which the medium was changed to fibrogenic or adipogenic differentiation medium. After 4 days, the medium was aspirated, and the cells were rinsed three times with 0.5 mL PBS (D8537, Sigma Life Science). All the following steps were performed at room temperature (RT). FAPs were fixed with 0.2 mL of 4% paraformaldehyde (150146, PFA- MP Biomedicals) in PBS for 15 min. Subsequently, FAPs were rinsed three times with 0.5 mL PBS for 5 min each. To permeabilize the cell membrane, FAPs were incubated with 0.2 mL methanol (179377-4L-PB, Sigma-Aldrich) for 10 min. Non-specific antibody binding sites were blocked with 0.2 mL of 5% bovine serum albumin in PBS for 1 h. Following blocking, FAPs were rinsed three times with 0.5 mL of PBS for 5 min each. FAPs were then incubated with 0.2 mL of primary antibody, diluted in PBS for 2 h. The following primary antibodies were used: PDGFRα (14-1401-82, eBioscience, 1:50), Ki67 (9129, Cell Signaling Technology, 1:200), perilipin (9349T, Cell Signaling Technology, 1:200), αSMA (14-9760-82, eBioscience, 1:200), MyHC (DSHB, #MF 20, 1:50), and collagen type I (ab34710, Abcam, 1:200). After incubation, FAPs were rinsed three times with 0.5 mL PBS for 5 min each and incubated with 0.2 mL of the following goat-anti mouse or goat-anti rabbit secondary antibodies diluted at 1:500 in PBST for 1h at RT: Rhodamine Red (111-295-144 or 115-295-146, Jackson Immuno Research) or Alexa Fluor 488 (115-545-146 or 111-545-144, Jackson Immuno Research). FAPs were rinsed three times with 0.5 mL PBS for 5 min each and mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium (AB104139, Abcam). The slides were cover-slipped and FAPs were observed using a Zeiss AxioImager Z1 equipped with the appropriate filter sets and a CCD camera. The mean fluorescence intensities of individual channels were measured using ImageJ.
Cryosectioning of TA muscle tissue
1 g of tragacanth gum (A18502.36, Thermo Scientific) was dissolved in 10 mL of water by vortexing and the paste was scooped into disposable plastic cryo-molds. The hind limb TA muscle was dissected, and the tibialis anterior muscle was vertically embedded into the mold using fine forceps. The embedded TA muscle was immersed for 30 s in liquid nitrogen pre-cooled 2-methyl butane (Q223-08, Avantor) and then stored at −80°C until cryosectioning. Using an Avantik cryostat cooled to −23°C 7 μm sections were collected onto frosted glass slides (Vistavision Microscope Slides, VWR). Tissue sections were either used fresh for immunostaining or stored at −80°C until use.
Immunostaining of fresh-frozen TA sections
Frozen muscle sections were allowed to reach RT and then fixed with 4% EM-grade PFA in PBS for 20 min at RT in the dark. Afterward, sections were rinsed 2× in PBS for 10 min each and the tissues were permeabilized with methanol for 10 min. After rinsing 2× in PBS for 5 min each the sections were immersed in 0.01M citric acid preheated to 90°C in Coplin jars and steamed for 5 min with microwaving for 10 s followed by 50 s pause. This process was repeated for an additional 5 min using a second Coplin jar. Muscle sections were allowed to cool for 30 min in the citric acid solution and then rinsed 3x in PBS for 5 min each. Non-specific antibody binding sites were blocked with 5% BSA in PBS for 1h and sections were incubated with 100-200 μL of the following primary antibodies diluted in PBST at 4°C overnight: anti-ADAMTSL2 (1:50, custom), anti-perilipin (9349T, Cell Signaling Technology, 1:200), anti-αSMA (14-9760-82, eBioscienceTM, 1:200). After overnight incubation, the slides were rinsed 2x with PBS for 10 min and incubated with the secondary antibodies listed above (1:500 dilution in PBST) for 1 h at room temperature. The slides were then rinsed 2x with PBS for 10 min and mounted with Fluoromount-G containing DAPI (Abcam, AB104139) and cover-slipped. The slides were imaged using a Zeiss AxioImager Z1 microscope. Mean fluorescence intensities of individual channels were quantified using ImageJ software.
Western blot
FAPs were collected in Eppendorf tubes in PBS using cell scrapers, pelleted at 6000 rpm for 3 min, and lysed using lysis buffer (2x the estimated pellet volume) consisting of 900 μL radio-immunoprecipitation assay (RIPA) buffer supplemented with 100 μL EDTA-free protease inhibitor (Pierce Protease Inhibitor Mini Tablets, Fisher Scientific) and 100 μL phosphatase inhibitor (PhosSTOP, Roche), to extract protein. The tubes were placed on ice and allowed to lyse for 30 min. The samples were then spun at 12,000 rpm for 10 min and the supernatant was transferred into a fresh Eppendorf tube. The protein concentration was determined using a Bradford assay and 100 μg of total protein was used for western blot analyses. Proteins were denatured at 95°C for 5 min and separated using a 10% SDS polyacrylamide gel electrophoresis gel. The proteins were transferred onto a methanol-activated PVDF membrane by applying 400 mA of current for 3 h in a Tris-glycine-methanol transfer buffer. After the transfer, the membrane was washed with TBST (Tris-buffered saline with 0.1% Tween 20) for 2 × 5 min and blocked in 5% non-fat dry milk for 1 h. For antibodies against phosphorylated proteins, 5% bovine serum albumin in TBST was used as blocking buffer. After blocking, the membrane was rinsed with TBST for 5 min and incubated with the following primary antibodies for 2 h at RT. Anti-αSMA (14-9760-82, eBioscience, 1:2000), anti-pSMAD2 (18338S, Cell Signaling Technology 1:2000), anti ADAMTSL2 (1:500, custom), or anti-GAPDH (EMD Millipore, MAB374, 1:2000). Following incubation with the primary antibody, the membrane was washed 2x with TBST for 5 min each and incubated with the following goat-anti mouse or goat-anti rabbit secondary antibodies (1:10,000 dilution in TBST) for 1 h at RT: IR Dye 800CW Goat anti mouse IgG (926-32210, LI-COR Biosciences), IR Dye 800CW Goat anti rabbit IgG (926-32211, LI-COR Biosciences,), IR Dye 680 RD Goat anti mouse IgG (926-68070, LI-COR Biosciences,), or IR Dye 680 RD Goat anti rabbit IgG (926-68071, LI-COR Biosciences). Membranes were rinsed 3 × 5 min with TBST and 1 × 5 min with TBS and imaged using an Azure Imager (C600, Azure Biosystems). Band intensities were quantified with ImageJ and normalized to the GAPDH signal and Ctrl samples.
Quantification and statistical analysis
The mean values of n = 2 groups were compared with a two-sample Student’s t-test and the mean values of n ≥ 3 groups were compared with a one-way ANOVA followed by posthoc Tukey analysis. A p-value of<0.05 was considered statistically significant. Sample sizes for the individual experiments are indicated in the figure legends. OriginPro was used for statistical calculations and data plotting. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Published: May 20, 2025
References
- 1.Sousa-Victor P., Garcia-Prat L., Munoz-Canoves P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2021;23:204–226. doi: 10.1038/s41580-021-00421-2. [DOI] [PubMed] [Google Scholar]
- 2.Helmbacher F., Stricker S. Tissue cross talks governing limb muscle development and regeneration. Semin. Cell Dev. Biol. 2020;104:14–30. doi: 10.1016/j.semcdb.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Mauro A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 5.Forcina L., Cosentino M., Musaro A. Mechanisms Regulating Muscle Regeneration: Insights into the Interrelated and Time-Dependent Phases of Tissue Healing. Cells. 2020;9 doi: 10.3390/cells9051297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leikina E., Gamage D.G., Prasad V., Goykhberg J., Crowe M., Diao J., Kozlov M.M., Chernomordik L.V., Millay D.P. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Dev. Cell. 2018;46:767–780.e7. doi: 10.1016/j.devcel.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wosczyna M.N., Konishi C.T., Perez Carbajal E.E., Wang T.T., Walsh R.A., Gan Q., Wagner M.W., Rando T.A. Mesenchymal Stromal Cells Are Required for Regeneration and Homeostatic Maintenance of Skeletal Muscle. Cell Rep. 2019;27:2029–2035.e5. doi: 10.1016/j.celrep.2019.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theret M., Rossi F.M.V., Contreras O. Evolving Roles of Muscle-Resident Fibro-Adipogenic Progenitors in Health, Regeneration, Neuromuscular Disorders, and Aging. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.673404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassari S., Duprez D., Fournier-Thibault C. Non-myogenic Contribution to Muscle Development and Homeostasis: The Role of Connective Tissues. Front. Cell Dev. Biol. 2017;5:22. doi: 10.3389/fcell.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joe A.W.B., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M.V. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemos D.R., Babaeijandaghi F., Low M., Chang C.K., Lee S.T., Fiore D., Zhang R.H., Natarajan A., Nedospasov S.A., Rossi F.M.V. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 12.Kuzel B.R., Grindel S., Papandrea R., Ziegler D. Fatty infiltration and rotator cuff atrophy. J. Am. Acad. Orthop. Surg. 2013;21:613–623. doi: 10.5435/JAAOS-21-10-613. [DOI] [PubMed] [Google Scholar]
- 13.Gerber C., Schneeberger A.G., Hoppeler H., Meyer D.C. Correlation of atrophy and fatty infiltration on strength and integrity of rotator cuff repairs: a study in thirteen patients. J. Shoulder Elb. Surg. 2007;16:691–696. doi: 10.1016/j.jse.2007.02.122. [DOI] [PubMed] [Google Scholar]
- 14.Gladstone J.N., Bishop J.Y., Lo I.K.Y., Flatow E.L. Fatty infiltration and atrophy of the rotator cuff do not improve after rotator cuff repair and correlate with poor functional outcome. Am. J. Sports Med. 2007;35:719–728. doi: 10.1177/0363546506297539. [DOI] [PubMed] [Google Scholar]
- 15.Contreras O., Rebolledo D.L., Oyarzún J.E., Olguín H.C., Brandan E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell Tissue Res. 2016;364:647–660. doi: 10.1007/s00441-015-2343-0. [DOI] [PubMed] [Google Scholar]
- 16.Davies M.R., Liu X., Lee L., Laron D., Ning A.Y., Kim H.T., Feeley B.T. TGF-beta Small Molecule Inhibitor SB431542 Reduces Rotator Cuff Muscle Fibrosis and Fatty Infiltration By Promoting Fibro/Adipogenic Progenitor Apoptosis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano A.L., Mann C.J., Vidal B., Ardite E., Perdiguero E., Muñoz-Cánoves P. Cellular and molecular mechanisms regulating fibrosis in skeletal muscle repair and disease. Curr. Top. Dev. Biol. 2011;96:167–201. doi: 10.1016/B978-0-12-385940-2.00007-3. [DOI] [PubMed] [Google Scholar]
- 18.Mahdy M.A.A. Skeletal muscle fibrosis: an overview. Cell Tissue Res. 2019;375:575–588. doi: 10.1007/s00441-018-2955-2. [DOI] [PubMed] [Google Scholar]
- 19.Cholok D., Lee E., Lisiecki J., Agarwal S., Loder S., Ranganathan K., Qureshi A.T., Davis T.A., Levi B. Traumatic muscle fibrosis: From pathway to prevention. J. Trauma Acute Care Surg. 2017;82:174–184. doi: 10.1097/TA.0000000000001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uezumi A., Fukada S.i., Yamamoto N., Takeda S., Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 21.Contreras O., Cruz-Soca M., Theret M., Soliman H., Tung L.W., Groppa E., Rossi F.M., Brandan E. Cross-talk between TGF-beta and PDGFRalpha signaling pathways regulates the fate of stromal fibro-adipogenic progenitors. J. Cell Sci. 2019;132 doi: 10.1242/jcs.232157. [DOI] [PubMed] [Google Scholar]
- 22.Robertson I.B., Horiguchi M., Zilberberg L., Dabovic B., Hadjiolova K., Rifkin D.B. Latent TGF-beta-binding proteins. Matrix Biol. 2015;47:44–53. doi: 10.1016/j.matbio.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juban G., Saclier M., Yacoub-Youssef H., Kernou A., Arnold L., Boisson C., Ben Larbi S., Magnan M., Cuvellier S., Theret M., et al. AMPK Activation Regulates LTBP4-Dependent TGF-beta1 Secretion by Pro-inflammatory Macrophages and Controls Fibrosis in Duchenne Muscular Dystrophy. Cell Rep. 2018;25:2163–2176.e6. doi: 10.1016/j.celrep.2018.10.077. [DOI] [PubMed] [Google Scholar]
- 24.Li Y., Foster W., Deasy B.M., Chan Y., Prisk V., Tang Y., Cummins J., Huard J. Transforming growth factor-beta1 induces the differentiation of myogenic cells into fibrotic cells in injured skeletal muscle: a key event in muscle fibrogenesis. Am. J. Pathol. 2004;164:1007–1019. doi: 10.1016/s0002-9440(10)63188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pessina P., Kharraz Y., Jardí M., Fukada S.i., Serrano A.L., Perdiguero E., Muñoz-Cánoves P. Fibrogenic Cell Plasticity Blunts Tissue Regeneration and Aggravates Muscular Dystrophy. Stem Cell Rep. 2015;4:1046–1060. doi: 10.1016/j.stemcr.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazala D.A., Novak J.S., Hogarth M.W., Nearing M., Adusumalli P., Tully C.B., Habib N.F., Gordish-Dressman H., Chen Y.W., Jaiswal J.K., Partridge T.A. TGF-beta-driven muscle degeneration and failed regeneration underlie disease onset in a DMD mouse model. JCI Insight. 2020;5:e135703. doi: 10.1172/jci.insight.135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu C., Chin-Young B., Park G., Guzmán-Seda M., Laudier D., Han W.M. WNT7A suppresses adipogenesis of skeletal muscle mesenchymal stem cells and fatty infiltration through the alternative Wnt-Rho-YAP/TAZ signaling axis. Stem Cell Rep. 2023;18:999–1014. doi: 10.1016/j.stemcr.2023.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reggio A., Rosina M., Palma A., Cerquone Perpetuini A., Petrilli L.L., Gargioli C., Fuoco C., Micarelli E., Giuliani G., Cerretani M., et al. Adipogenesis of skeletal muscle fibro/adipogenic progenitors is affected by the WNT5a/GSK3/beta-catenin axis. Cell Death Differ. 2020;27:2921–2941. doi: 10.1038/s41418-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wosczyna M.N., Biswas A.A., Cogswell C.A., Goldhamer D.J. Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. J. Bone Miner. Res. 2012;27:1004–1017. doi: 10.1002/jbmr.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees-Shepard J.B., Yamamoto M., Biswas A.A., Stoessel S.J., Nicholas S.A.E., Cogswell C.A., Devarakonda P.M., Schneider M.J., Jr., Cummins S.M., Legendre N.P., et al. Activin-dependent signaling in fibro/adipogenic progenitors causes fibrodysplasia ossificans progressiva. Nat. Commun. 2018;9:471. doi: 10.1038/s41467-018-02872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dzialo E., Tkacz K., Blyszczuk P. Crosstalk between the TGF-beta and WNT signalling pathways during cardiac fibrogenesis. Acta Biochim. Pol. 2018;65:341–349. doi: 10.18388/abp.2018_2635. [DOI] [PubMed] [Google Scholar]
- 32.Biressi S., Miyabara E.H., Gopinath S.D., Carlig P.M.M., Rando T.A. A Wnt-TGFbeta2 axis induces a fibrogenic program in muscle stem cells from dystrophic mice. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3008411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhmetshina A., Palumbo K., Dees C., Bergmann C., Venalis P., Zerr P., Horn A., Kireva T., Beyer C., Zwerina J., et al. Activation of canonical Wnt signalling is required for TGF-beta-mediated fibrosis. Nat. Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berendsen A.D., Fisher L.W., Kilts T.M., Owens R.T., Robey P.G., Gutkind J.S., Young M.F. Modulation of canonical Wnt signaling by the extracellular matrix component biglycan. Proc. Natl. Acad. Sci. USA. 2011;108:17022–17027. doi: 10.1073/pnas.1110629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy-Ullrich J.E., Sage E.H. Revisiting the matricellular concept. Matrix Biol. 2014;37:1–14. doi: 10.1016/j.matbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rypdal K.B., Erusappan P.M., Melleby A.O., Seifert D.E., Palmero S., Strand M.E., Tønnessen T., Dahl C.P., Almaas V., Hubmacher D., et al. The extracellular matrix glycoprotein ADAMTSL2 is increased in heart failure and inhibits TGFbeta signalling in cardiac fibroblasts. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-99032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Goff C., Morice-Picard F., Dagoneau N., Wang L.W., Perrot C., Crow Y.J., Bauer F., Flori E., Prost-Squarcioni C., Krakow D., et al. ADAMTSL2 mutations in geleophysic dysplasia demonstrate a role for ADAMTS-like proteins in TGF-beta bioavailability regulation. Nat. Genet. 2008;40:1119–1123. doi: 10.1038/ng.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piccolo P., Sabatino V., Mithbaokar P., Polishchuck E., Law S.K., Magraner-Pardo L., Pons T., Polishchuck R., Brunetti-Pierri N. Geleophysic dysplasia: novel missense variants and insights into ADAMTSL2 intracellular trafficking. Mol. Genet. Metab. Rep. 2019;21 doi: 10.1016/j.ymgmr.2019.100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taye N., Singh M., Baldock C., Hubmacher D. Secreted ADAMTS-like 2 promotes myoblast differentiation by potentiating WNT signaling. Matrix Biol. 2023;120:24–42. doi: 10.1016/j.matbio.2023.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taye N., Redhead C., Hubmacher D. Secreted ADAMTS-like proteins as regulators of connective tissue function. Am. J. Physiol. Cell Physiol. 2024;326:C756–C767. doi: 10.1152/ajpcell.00680.2023. [DOI] [PubMed] [Google Scholar]
- 41.Batkovskyte D., McKenzie F., Taylan F., Simsek-Kiper P.O., Nikkel S.M., Ohashi H., Stevenson R.E., Ha T., Cavalcanti D.P., Miyahara H., et al. Al-Gazali Skeletal Dysplasia Constitutes the Lethal End of ADAMTSL2-Related Disorders. J. Bone Miner. Res. 2023;38:692–706. doi: 10.1002/jbmr.4799. [DOI] [PubMed] [Google Scholar]
- 42.Steinle J., Hossain W.A., Lovell S., Veatch O.J., Butler M.G. ADAMTSL2 gene variant in patients with features of autosomal dominant connective tissue disorders. American journal of medical genetics. Part A. 2021;185:743–752. doi: 10.1002/ajmg.a.62030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubmacher D., Wang L.W., Mecham R.P., Reinhardt D.P., Apte S.S. Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia--a novel mouse model providing insights into geleophysic dysplasia. Dis. Model. Mech. 2015;8:487–499. doi: 10.1242/dmm.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabula Muris Consortium. Overall coordination. Logistical coordination. Organ collection and processing. Library preparation and sequencing. Computational data analysis. Cell type annotation. Writing group. Supplemental text writing group. Principal investigators Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muhl L., Genové G., Leptidis S., Liu J., He L., Mocci G., Sun Y., Gustafsson S., Buyandelger B., Chivukula I.V., et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 2020;11:3953. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubmacher D., Taye N., Balic Z., Thacker S., Adams S.M., Birk D.E., Schweitzer R., Apte S.S. Limb- and tendon-specific Adamtsl2 deletion identifies a role for ADAMTSL2 in tendon growth in a mouse model for geleophysic dysplasia. Matrix Biol. 2019;82:38–53. doi: 10.1016/j.matbio.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 48.Sefton E.M., Kardon G. Connecting muscle development, birth defects, and evolution: An essential role for muscle connective tissue. Curr. Top. Dev. Biol. 2019;132:137–176. doi: 10.1016/bs.ctdb.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taye N., Rodriguez L., Iatridis J.C., Han W.M., Hubmacher D. Myoblast-derived ADAMTS-like 2 promotes skeletal muscle regeneration after injury. NPJ Regen. Med. 2024;9:39. doi: 10.1038/s41536-024-00383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Testa S., Fornetti E., Fuoco C., Sanchez-Riera C., Rizzo F., Ciccotti M., Cannata S., Sciarra T., Gargioli C. The War after War: Volumetric Muscle Loss Incidence, Implication, Current Therapies and Emerging Reconstructive Strategies, a Comprehensive Review. Biomedicines. 2021;9 doi: 10.3390/biomedicines9050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corona B.T., Wenke J.C., Ward C.L. Pathophysiology of Volumetric Muscle Loss Injury. Cells Tissues Organs. 2016;202:180–188. doi: 10.1159/000443925. [DOI] [PubMed] [Google Scholar]
- 52.Pollot B.E., Corona B.T. Volumetric Muscle Loss. Methods Mol. Biol. 2016;1460:19–31. doi: 10.1007/978-1-4939-3810-0_2. [DOI] [PubMed] [Google Scholar]
- 53.Marinkovic M., Fuoco C., Sacco F., Cerquone Perpetuini A., Giuliani G., Micarelli E., Pavlidou T., Petrilli L.L., Reggio A., Riccio F., et al. Fibro-adipogenic progenitors of dystrophic mice are insensitive to NOTCH regulation of adipogenesis. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasut A., Jones A.E., Rudnicki M.A. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. 2013 doi: 10.3791/50074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inman G.J., Nicolás F.J., Callahan J.F., Harling J.D., Gaster L.M., Reith A.D., Laping N.J., Hill C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 56.Collins B.C., Kardon G. It takes all kinds: heterogeneity among satellite cells and fibro-adipogenic progenitors during skeletal muscle regeneration. Development. 2021;148 doi: 10.1242/dev.199861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biferali B., Proietti D., Mozzetta C., Madaro L. Fibro-Adipogenic Progenitors Cross-Talk in Skeletal Muscle: The Social Network. Front. Physiol. 2019;10:1074. doi: 10.3389/fphys.2019.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uezumi A., Fukada S., Yamamoto N., Ikemoto-Uezumi M., Nakatani M., Morita M., Yamaguchi A., Yamada H., Nishino I., Hamada Y., Tsuchida K. Identification and characterization of PDGFRalpha+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uezumi A., Ikemoto-Uezumi M., Zhou H., Kurosawa T., Yoshimoto Y., Nakatani M., Hitachi K., Yamaguchi H., Wakatsuki S., Araki T., et al. Mesenchymal Bmp3b expression maintains skeletal muscle integrity and decreases in age-related sarcopenia. J. Clin. Investig. 2021;131 doi: 10.1172/JCI139617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camarena V., Williams M.M., Morales A.A., Zafeer M.F., Kilic O.V., Kamiar A., Abad C., Rasmussen M.A., Briski L.M., Peart L., et al. ADAMTSL2 mutations determine the phenotypic severity in geleophysic dysplasia. JCI Insight. 2024;9 doi: 10.1172/jci.insight.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hardy D., Besnard A., Latil M., Jouvion G., Briand D., Thépenier C., Pascal Q., Guguin A., Gayraud-Morel B., Cavaillon J.M., et al. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Norris A.M., Fierman K.E., Campbell J., Pitale R., Shahraj M., Kopinke D. Studying intramuscular fat deposition and muscle regeneration: insights from a comparative analysis of mouse strains, injury models, and sex differences. Skelet. Muscle. 2024;14:12. doi: 10.1186/s13395-024-00344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahdy M.A.A., Warita K., Hosaka Y.Z. Effects of transforming growth factor-beta1 treatment on muscle regeneration and adipogenesis in glycerol-injured muscle. Anim. Sci. J. 2017;88:1811–1819. doi: 10.1111/asj.12845. [DOI] [PubMed] [Google Scholar]
- 64.Pessina P., Cabrera D., Morales M.G., Riquelme C.A., Gutiérrez J., Serrano A.L., Brandan E., Muñoz-Cánoves P. Novel and optimized strategies for inducing fibrosis in vivo: focus on Duchenne Muscular Dystrophy. Skelet. Muscle. 2014;4:7. doi: 10.1186/2044-5040-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laping N.J., Grygielko E., Mathur A., Butter S., Bomberger J., Tweed C., Martin W., Fornwald J., Lehr R., Harling J., et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol. Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 66.Koo B.H., Le Goff C., Jungers K.A., Vasanji A., O'Flaherty J., Weyman C.M., Apte S.S. ADAMTS-like 2 (ADAMTSL2) is a secreted glycoprotein that is widely expressed during mouse embryogenesis and is regulated during skeletal myogenesis. Matrix Biol. 2007;26:431–441. doi: 10.1016/j.matbio.2007.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript or referenced. Any additional information required to reanalyze the data will be shared by the lead contact upon request. No original code was reported in this paper.