Abstract
Comparing within-species variations of traits can be used in testing ecological theories. In these comparisons, it is useful to remove the effect of the difference in mean trait values, therefore measures of relative variation, most often the coefficient of variation (CV), are used. The studied traits are often calculated as the ratio of the size or mass of two organs: e.g. specific leaf area (SLA) is the ratio of leaf size and leaf mass. Often the inverse of these ratios is also meaningful; for example, the inverse of SLA is often referred to as LMA (leaf mass per area). Relative variation of a trait and its inverse should not considerably differ. However, it is illustrated that using the coefficient of variation may result in differences that could influence the interpretation, especially if there are outlier trait values. The alternative way for estimating CV from the standard deviation of log-transformed data assuming log-normal distribution and Kirkwood’s geometric coefficient of variation free from this problem, but they proved to be sensitive to outlier values. Quartile coefficient of variation performed best in the tests: it gives the same value for a trait and its inverse and it is not sensitive to outliers.
Subject terms: Ecology, Ecological modelling
Introduction
Values of qualitative traits can considerably vary among and within species1,2. The structure of variation can be explored by partitioning total variance into components related to different sources (e.g. variation between species, between sites within species, between individuals within site, and within individuals). The calculated variance components express the relative contribution of sources in percentage, therefore their values can be compared between sites, species, or traits. However, some ecological hypotheses are related to the extent of trait variation1,2. For example, it is hypothesized that the extent of intraspecific trait variation (ITV) is higher in generalists than in specialist species3,4, and it may change along environmental and species richness gradients5–7.
The coefficient of variation (CV), the standard deviation divided by the arithmetic mean, is the most widely used measure of the extent of trait variation e.g.8–11. CV has two advantages: it is a dimensionless measure of relative variation2. The extent of trait variation can be compared among traits only if it is measured in the same units. For example, the standard deviation of height, measured in cm, and SLA, measured in g cm−2 cannot be compared, while their CV is comparable because it is dimensionless. Comparing absolute variation of the same trait between species also can be misleading when the difference between means is large. Ten centimeters departure from the mean height of the species is large for a short forb but small for a tall tree. That is why better to use relative measures, such as coefficient of variation, for among-species comparisons too.
Several papers called attention to cases where CV should not be used e.g.12,13. The most important restrictions are that the domain of the variable has to be non-negative (otherwise, its arithmetic mean could be zero preventing calculation CV) and it has to be measured in ratio or log-interval scale, where the meaning of “zero” value is unarbitrary. It cannot be calculated for nominal or ordinal scale data, where the mean and standard deviation is undefined. CV also should not be calculated for interval and difference scale (i.e. for log-transformed ratio-scale variables14), where changing of unit influences the mean value. Brendel15 pointed out that the CV of standardized stable isotope ratios depends on the applied reference isotope ratio. The aims of this paper are (1) calling attention to another problem: swapping nominator and denominator of ratio type traits results in an altered CV value; and (2) suggesting to use of quartile coefficient of variation that is free from this problem.
Ratios of size or mass of plant organs are widely used as functional traits, such as the ratio of leaf area and leaf dry mass (specific leaf area, SLA or leaf mass per area, LMA), the ratio of root length and root dry mass (specific root length, SRL) or ratio of the shoot and root mass16. In these ratios, nominator and denominator are often interchangeable without loss of meaning; for example, instead of the specific leaf area (SLA) often its inverse, the leaf mass ratio (LMA) is calculated17. We would expect the relative variation of the two forms of ratio (e.g. SLA and LMA) to be the same. Note that some ratios can be transformed into proportions. For example, instead of shoot mass: root mass ratio, we can use proportion of shoot mass, i.e. shoot mass/(shoot mass + root mass). In case of proportions, relative variation of their complement is considered.
Theory
The coefficient of variation is defined as the ratio of standard deviation and mean of the distribution:
Regarding the ratio of two random variates to bivariate function allows approximating its mean () and standard deviation () by Taylor series expansion (see Supplementary Appendix A for the derivation of formulas):
If CVs of x/y and y/x equal:
therefore,
This equation should be—at least approximately – hold to approximate means and standard deviations, but:
Since the equation does not hold for the approximate value, we can expect that CVs of a ratio and its inverse may differ. A real example will be shown in the Results section to illustrate that the difference could be important.
However, there is an important exception, when the ratio follows log-normal distribution. If x/y is log-normally distributed, its logarithm follows normal distribution, with mean and standard deviation
where and are the mean and standard deviation of the log-transformed ratio, respectively. The mean and standard deviation of the log-normal distribution are
Therefore, CV depends on only, and it is independent from 18:
Since
The logarithm of the inverse ratio is also normally distributed with the same standard deviation:
Thus in this case CV is the same for the ratio and its inverse.
CV can be estimated by replacing standard deviation (σ) and means (μ) with their estimates (s and m, respectively):
If x/y follows lognormal distribution, there is another estimator of CV:
where , is the arithmetic means of log-transformed ratios and n is the sample size. can be used as a descriptive statistic even if the ratio does not follow log-normal distribution.
Kirkwood19proposed another descriptive statistic the so-called geometric coefficient of variation:
GCV is not an estimate of CV, even if z follows log-normal distribution.
The logic of calculating CV is that dividing the measure of dispersion (standard deviation in CV) by the measure of location (mean in CV) removes the effect of differences in dispersion due to different locations, and if both are measured in the same units results in a dimensionless measure. Following this logic, several alternatives to CV were developed. The main motivation was to develop more robust (i.e. less sensitive to outlier values) alternatives to CV20 and references therein. Unfortunately, most of the proposed robust relative variation measures are also sensitive to swapping nominator and denominator in ratio type traits. An exception is the quartile coefficient of variation (CVQ):
where and are the first and third quartiles of variable x20,21.
For proving that we will use the equation . Therefore, first, this equation has to be proved. Let us start from the definition of first
and third quartile
From the definition of the first quartile of x
thus
If then , therefore
Since for a continuous variable, the probability of any possible value is zero, on the right side the “less than or equal to” can be replaced by “less than”
and this equation holds only if
Now, we can turn back to the proof of equality.
Note that finite sample estimates of and may slightly differ.
Finally, let us shortly overview the relative variation of proportions. The standard deviation of a proportion and its complement is the same: . But their mean is different, , therefor their c.v. also will be different. First and third quartile of a proportion and its complement is related:
The interquartile range is the same for both y/(x + y) and x/(x + y), but the sum of the two quartiles and therefore the quartile coefficient of variation is different. The absolute variation (i.e. standard deviation or interquartile range) of proportions and their complement is the same, but their relative variation is different. We have to keep in our mind that a proportion and this complement are interchangeable when absolute variation is studied, but they have different meaning when relative variation is calculated.
Results
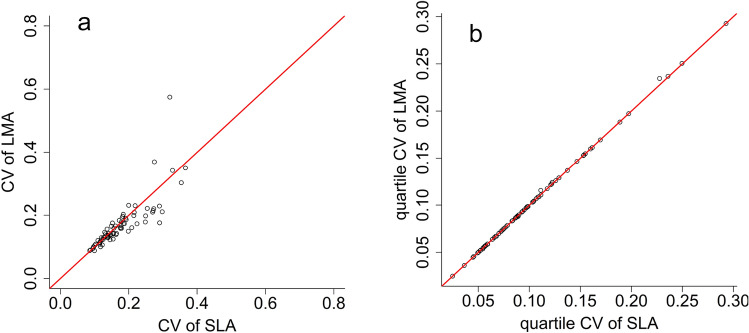
As expected, the differences between SLA and LMA in and GCV came only from rounding errors: the order of largest difference was 10–16. In the quartile coefficient of variation, the highest difference was 0.007 (Fig. 1a). However, differences hardly influenced the ranking of species according to the amount of intraspecific trait variation: the largest difference in ranks was 1, and 67 of 79 species the rank was the same for both traits. However, the amount of intraspecific trait variation (ITV) of SLA and LMA measured by (i.e. estimated standard deviation divided by sample mean) differed considerably (Fig. 1b): the largest difference was 1.07. Although the rank of species based on SLA and LMA was strongly correlated even if ITV was measured by (Fig. 2), the position of some species was strongly influenced: the largest difference in ranks between the two traits (SLA and LMA) was 21, and only 4 of 79 species remained ranks the same.
Figure 1.
Within-species relative variation of specific leaf area (SLA) and leaf mass per area (LMA) calculated by (a) CV (coefficient of variation, standard deviation divided by mean) and (b) quartile coefficient of variation (see formula in the main text). Red line is the 1:1 line.
Figure 2.
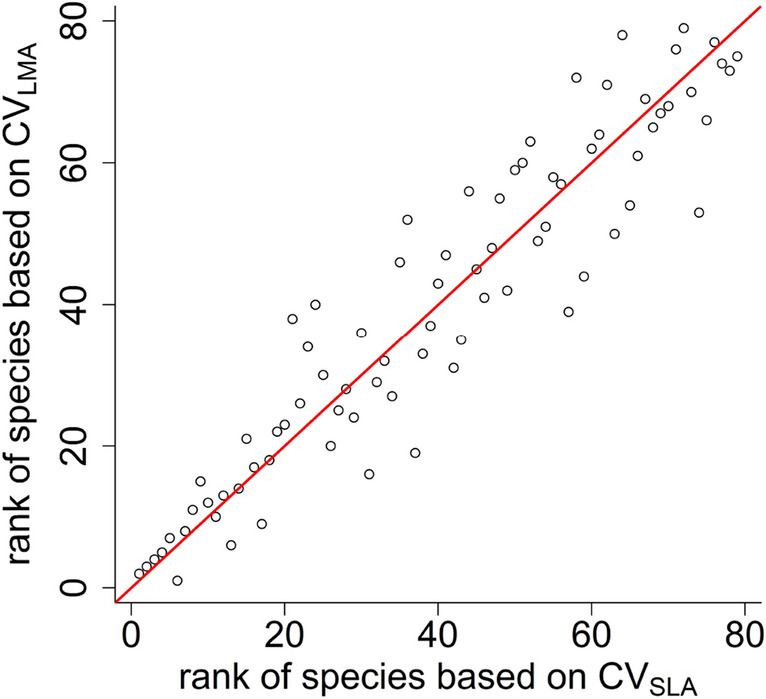
Rank of species based on their within-species relative variation of specific leaf area (SLA) and leaf mass per area (LMA) calculated by CV (coefficient of variation, standard deviation divided by mean). Red line is the 1:1 line.
The differences in between SLA and LMA were mainly caused by outlier values. After species-wise excluding outlier SLA values, the highest difference reduced to 0.25, but the difference between ranks of species according to ITV of SLA and LMA remained large: the highest rank difference was 24 (even larger than without excluding outliers), and only for 14 of 79 species were the two ranks the same.
Excluding outlier values had a negligible effect on ITV measured by quadratic CV, the correlation between values estimated with and without excluding outliers was 0.99. The same correlation of was 0.84. Surprisingly, the correlations between ITV calculated with or without excluding species-wise outliers were even smaller for and GCV (0.67 and 0.65, respectively).
All of the four measures of ITV indicate almost the same property of species (Table 1): the lowest linear correlation was 0.61, while the lowest Spearman’s rank correlation was 0.72. Quartile coefficient of variation was the most different from the other three measures because it depends only on the central part of trait distribution, and therefore it is fully insensitive to outlier values.
Table 1.
Correlations between within-species relative variation of SLA with (upper half-matrix) and without (lower half-matrix) excluding outliers.
| GCV | ||||
|---|---|---|---|---|
| 0.982 | 0.982 | 0.679 | ||
| 0.819 | 1.000 | 0.718 | ||
| GCV | 0.797 | 0.999 | 0.718 | |
| 0.765 | 0.629 | 0.609 |
Discussion
Presented results illustrate that ratio of sample standard deviation and sample mean () is sensitive both to outlier values and choosing a ratio-trait or its inverse (for example SLA or LMA). Three alternatives to this measure were evaluated in this paper. Both and GCV gave the same value for a trait and its inverse, but they are more sensitive to outlier values than . Quartile CV proved to be the most robust measure of ITV, it was hardly influenced by either excluding outliers and choosing a trait or its inverse. Therefore, I suggest that in studies testing hypotheses related to the amount of intra-specific trait variation, the quartile coefficient of variation should be used, especially if the inverse of the studied trait (i.e. 1/trait) is also meaningful.
Materials and methods
An R function for calculating two estimates of CV ( and ), geometric coefficient of variation (GCV), and quartile coefficient of variation () were developed (Supplementary Appendix B). All analyses were done in R environment, and the script and data will be available in a public repository.
For illustrating purposes, the dataset of Gyalus et al.22 was used that contains plot level measurement of leaf traits. In this paper, only specific leaf (SLA, leaf area in cm2 per leaf dry mass in g) data were used. Leaf mass per area (LMA) was calculated as 1/SLA. Four indices of relative variation of SLA and LMA were calculated for each species with at least 10 SLA data. Then the absolute differences between SLA and LMA in relative within-species variation and species rank according to within-species variation were calculated. Since could be more sensitive to outlier values than other measures, all analyses were repeated after excluding outlier values.
Supplementary Information
Acknowledgements
This research was supported by the NKFIH-K124671 grant.
Author contributions
Z.B.-D. conceived, designed, and executed this study and wrote the manuscript. No other person is entitled to authorship.
Funding
Open access funding provided by ELKH Centre for Ecological Research.
Data availability
Data and code available from Zenodo https://doi.org/10.5281/zenodo.6907699.
Competing interests
The author declares no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-31711-8.
References
- 1.Albert CH, et al. Intraspecific functional variability: Extent, structure and sources of variation. J. Ecol. 2010;98:604–613. doi: 10.1111/j.1365-2745.2010.01651.x. [DOI] [Google Scholar]
- 2.Albert CH, Grassein F, Schurr FM, Vieilledent G, Violle C. When and how should intraspecific variability be considered in trait-based plant ecology? Perspect. Plant Ecol. Evol. Syst. 2011;13:217–225. doi: 10.1016/j.ppees.2011.04.003. [DOI] [Google Scholar]
- 3.Sides CB, et al. Revisiting Darwin’s hypothesis: Does greater intraspecific variability increase species’ ecological breadth? Am. J. Bot. 2014;101:56–62. doi: 10.3732/ajb.1300284. [DOI] [PubMed] [Google Scholar]
- 4.Wellstein C, et al. Intraspecific phenotypic variability of plant functional traits in contrasting mountain grasslands habitats. Biodivers. Conserv. 2013;22:2353–2374. doi: 10.1007/s10531-013-0484-6. [DOI] [Google Scholar]
- 5.Helsen K, et al. Biotic and abiotic drivers of intraspecific trait variation within plant populations of three herbaceous plant species along a latitudinal gradient. BMC Ecol. 2017;17:38. doi: 10.1186/s12898-017-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuppler J, et al. Global gradients in intraspecific variation in vegetative and floral traits are partially associated with climate and species richness. Glob. Ecol. Biogeogr. 2020;29:992–1007. doi: 10.1111/geb.13077. [DOI] [Google Scholar]
- 7.Lemke IH, et al. Patterns of phenotypic trait variation in two temperate forest herbs along a broad climatic gradient. Plant Ecol. 2015;216:1523–1536. doi: 10.1007/s11258-015-0534-0. [DOI] [Google Scholar]
- 8.Cheng J, Chu P, Chen D, Bai Y. Functional correlations between specific leaf area and specific root length along a regional environmental gradient in inner Mongolia grasslands. Funct. Ecol. 2016;30:985–997. doi: 10.1111/1365-2435.12569. [DOI] [Google Scholar]
- 9.Li S, et al. Leaf functional traits of dominant desert plants in the Hexi Corridor, Northwestern China: Trade-off relationships and adversity strategies. Glob. Ecol. Conserv. 2021;28:e01666. doi: 10.1016/j.gecco.2021.e01666. [DOI] [Google Scholar]
- 10.Roscher C, et al. Trait means, trait plasticity and trait differences to other species jointly explain species performances in grasslands of varying diversity. Oikos. 2018;127:865–865. doi: 10.1111/oik.04815. [DOI] [Google Scholar]
- 11.Roscher C, et al. Functional groups differ in trait means, but not in trait plasticity to species richness in local grassland communities. Ecology. 2018;99:2295–2307. doi: 10.1002/ecy.2447. [DOI] [PubMed] [Google Scholar]
- 12.Livers JJ. Some limitations to use of coefficient of variation. J. Farm Econ. 1942;24:892. doi: 10.2307/1232009. [DOI] [Google Scholar]
- 13.Pélabon C, Hilde CH, Einum S, Gamelon M. On the use of the coefficient of variation to quantify and compare trait variation. Evol. Lett. 2020;4:180–188. doi: 10.1002/evl3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houle D, Pélabon C, Wagner GP, Hansen TF. Measurement and meaning in biology. Q. Rev. Biol. 2011;86:3–34. doi: 10.1086/658408. [DOI] [PubMed] [Google Scholar]
- 15.Brendel O. Is the coefficient of variation a valid measure for variability of stable isotope abundances in biological materials?: Is CV a valid measure for isotopic compositions? Rapid Commun. Mass Spectrom. 2014;28:370–376. doi: 10.1002/rcm.6791. [DOI] [PubMed] [Google Scholar]
- 16.Pérez-Harguindeguy N, et al. New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 2013;61:167–234. doi: 10.1071/BT12225. [DOI] [Google Scholar]
- 17.Poorter H, Niinemets Ü, Poorter L, Wright IJ, Villar R. Causes and consequences of variation in leaf mass per area (LMA): A meta-analysis. New Phytol. 2009;182:565–588. doi: 10.1111/j.1469-8137.2009.02830.x. [DOI] [PubMed] [Google Scholar]
- 18.Koopmans LH, Owen DB, Rosenblatt JI. Confidence intervals for the coefficient of variation for the normal and log normal distributions. Biometrika. 1964;51:25–32. doi: 10.1093/biomet/51.1-2.25. [DOI] [Google Scholar]
- 19.Kirkwood TBL. Geometric means and measures of dispersion. Biometrics. 1979;35:908–909. [Google Scholar]
- 20.Arachchige CNPG, Prendergast LA, Staudte RG. Robust analogs to the coefficient of variation. J. Appl. Stat. 2022;49:268–290. doi: 10.1080/02664763.2020.1808599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonett DG. Confidence interval for a coefficient of quartile variation. Comput. Stat. Data Anal. 2006;50:2953–2957. doi: 10.1016/j.csda.2005.05.007. [DOI] [Google Scholar]
- 22.Gyalus A, et al. Plant trait records of the Hungarian and Serbian flora and methodological description of some hard to measure plant species. Acta Bot. Hung. 2022;64:451–454. doi: 10.1556/034.64.2022.3-4.14. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code available from Zenodo https://doi.org/10.5281/zenodo.6907699.