Abstract
Constantly mutating SARS-CoV-2 is a global concern resulting in COVID-19 infectious waves from time to time in different regions, challenging present-day diagnostics and therapeutics. Early-stage point-of-care diagnostic (POC) biosensors are a crucial vector for the timely management of morbidity and mortalities caused due to COVID-19. The state-of-the-art SARS-CoV-2 biosensors depend upon developing a single platform for its diverse variants/biomarkers, enabling precise detection and monitoring. Nanophotonic-enabled biosensors have emerged as ‘one platform’ to diagnose COVID-19, addressing the concern of constant viral mutation. This review assesses the evolution of current and future variants of the SARS-CoV-2 and critically summarizes the current state of biosensor approaches for detecting SARS-CoV-2 variants/biomarkers employing nanophotonic-enabled diagnostics. It discusses the integration of modern-age technologies, including artificial intelligence, machine learning and 5G communication with nanophotonic biosensors for intelligent COVID-19 monitoring and management. It also highlights the challenges and potential opportunities for developing intelligent biosensors for diagnosing future SARS-CoV-2 variants. This review will guide future research and development on nano-enabled intelligent photonic-biosensor strategies for early-stage diagnosing of highly infectious diseases to prevent repeated outbreaks and save associated human mortalities.
Keywords: SARS-COV-2 variants, Biosensors, Nanophononics, Artificial intelligence, Mutations evolution
Graphical abstract

1. Introduction
In early 2023, there had been >758,390,564 confirmed cases of the new coronavirus disease (COVID-19) caused by SARS-CoV-2, including 6,859,093 deaths, according to the World Health Organization (WHO). Considering the rapid spread of the virus, a total of 13,226,873,459 vaccine doses were administered. Additionally, variants of SARS-CoV-2 that have greater transmissibility, pathogenicity, and potential for immune evasion challenge existing diagnostic methods (Harapan et al., 2020; Kasibhatla et al., 2020). Strict rules were implemented to control the spread of the disease. Frequent hand washing and maintaining social distance by avoiding close contact with others, especially in crowded places, were encouraged. Many businesses and educational institutions were forced to close or drastically reduce in-person interactions and shift to a remote workforce. The inclusion of SARS-CoV-2 would have prevented these catastrophic outcomes. The main challenge encountered in stopping the spread of COVID-19 was the inaccessibility of fast virus tests (Huang et al., 2020; Ruiz-Hitzky et al., 2020; Baloch et al., 2020).
The development of dependable and swift diagnosis procedures is vital, as scale-based diagnostic abilities are crucial in controlling infections and reducing mortality rates (Chaudhary et al., 2022a). If no immunizations or treatments can efficiently contain the spread of the disease, the most effective way to manage it is by isolating all infected individuals, whether they display symptoms or not (Rai et al., 2021; Ahmed et al., 2022; Taha, 2021; Taha et al., 2021). Biosensors are highly beneficial in many essential areas, such as medicine, food safety, environmental monitoring, security, pharmaceuticals, and forensics (Chaudhary et al., 2022c, Chaudhary et al., 2023a, Chaudhary et al., 2023b; Sonu, 2022). It is estimated that around 70 % of healthcare decisions are supported by diagnostic technology, making this market a major biosensor market (Ngo et al., 2017).
The revelation of the molecular profile of a patient could play a crucial role in the advancement of precision medicine. Wearable biosensors that are affordable and easy to use are likely to be useful as health tracking devices for individuals. The integration of automated and autonomous biosensors into public infrastructures, such as transportation nodes, educational institutions, workplaces, etc., is planned to increase public safety by warning the public about biological risks. Connected via the “internet of things,” these devices have the potential to provide massive spatio-temporal information at the level of a whole population (Shrivastava et al., 2020; Ho et al., 2020). In addition, the use of nanobiotechnology for the detection of the SARS-COV-2 virus has been explored, including components of biosensing, nanomedicine, and nanovaccine (Bidram et al., 2021). In the same manner, there has been an increase in expectations placed on microbiology laboratories located within hospitals to produce accurate and reliable test results for their patients. Several factors have contributed to this stress, including demands from physicians for same-day test results, an increase in the number of samples, and the increasing organism resistance, etc. (Keighley et al., 2021). Rapid diagnostics on site make patient identity and contact tracking more efficient. However, such diagnostic tools do not yet exist. Although immunogenic lateral flow tests are quick, portable, and inexpensive, they are not ideal for detecting early-stage viruses (Sheridan, 2020). Developing a test method with good selectivity requires understanding the structure and function of the virus. The “virions” comprise the entire viral particle, including the genetic material (nucleic acids) and its protective outer coating. DNA and RNA are the genetic materials of most viruses, and nucleic acids can be single- or double-stranded (van Belkum et al., 2019; Taha et al., 2019). In this review, the existing literature on emerging modern variants of SARS-CoV-2 is reviewed, discussing the state-of-the-art and recent advances in biosensor techniques for accurately detecting SARS-CoV-2 biomarkers and offering a critical analysis of both, along with the difficulties and prospects of creating biosensors for diagnosing COVID-19 and other viruses. The taxonomy of current research on SARS-COV-2 variants evaluation, structures, and biosensor diagnosis is depicted in Fig. 1 .
Fig. 1.
Taxonomy of studies for evaluation, structures, and biosensors diagnosis of SARS-COV-2 variants.
2. SARS-CoV-2: a highly infectious and evolving virus
This section describes the SARS-CoV-2 virus evolutions and the appearance of new variations. It highlights the necessity of early discovery and monitoring novel variations to restrict viral propagation. SARS-CoV-2 is a highly infectious respiratory virus that was first identified in December 2019 in Wuhan, China, and has since spread worldwide, leading to a global pandemic. It is mainly transmitted by infected people's respiratory drops and aerosols and can cause severe respiratory diseases and potentially fatal complications in some people (Azuma et al., 2020). The virus evolves over time and new variants have emerged that may be more transmitted, virulent or resistant to existing vaccines and treatments. These concern variants include Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1.529), which are associated with increased transmission and severity (Aleem et al., 2021).
2.1. Current state of SARS-CoV-2 and its variants
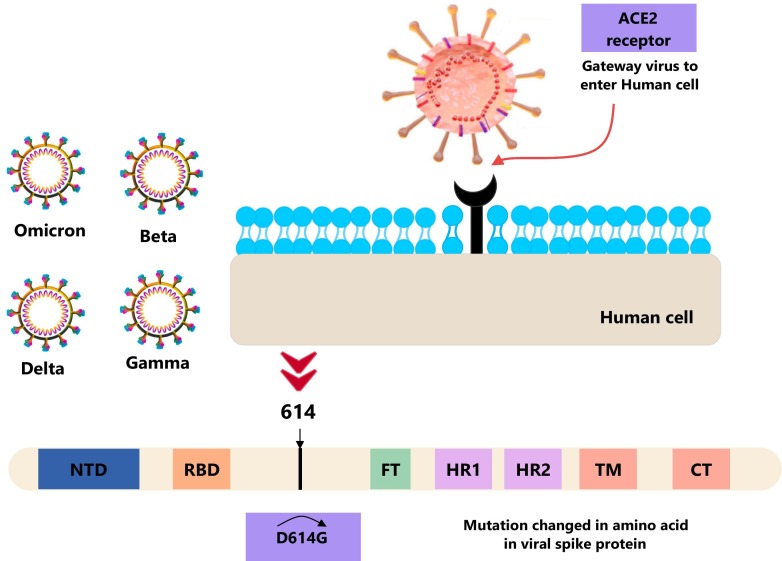
In September 2020, several new variants of SARS-CoV-2 suddenly appeared in locations without a previous connection. The B.1.1.7 mutation was initially identified in Britain (Tang et al., 2021). Another mutation that emerged in South Africa was Beta (B.1.351), Delta variant (B.1.617) in India, and Coronavirus Gamma variant (P.1) in Brazil (Samarasekera, 2021; Thye et al., 2021). Recent studies have revealed several variants of SARS-CoV-2, including variants Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and recently discovered Omicron (B.1.1.529) variants. The Omicron mutation is the most mutated strain of SARS-CoV-2, with 50 mutations in its genome, including 30 in the spherical protein (S). This increase in mutations has increased transmission and resistance to partial immunity caused by current COVID-19 vaccines and antibody-based treatments (Tang et al., 2021; Samarasekera, 2021; Thye et al., 2021). The spike protein, which allows viruses to enter host cells and is the target of modern vaccines, differs for each virus type. Delta and omicron SARS-CoV-2 strains have mutations in the spike protein, making them more contagious. These changes may affect the spikes protein receptor-binding domain (RBD) and its interaction with angiotensin-converting enzyme 2 (ACE2), which could alter cell entry, viral immune evasion, and immunogenicity (Watanabe et al., 2020). Omicron is notable because it has three different sub-lineages (BA.1, BA.2, and BA.3) found almost simultaneously, even though they are similar to Alpha, Beta, Gamma, and Delta. Scientists have recently looked at the ability of the Omicron BA.4 and BA.5 sub-lineages of Omicron BA.1-infected people. It found that the potential of vaccinated people to neutralize BA.4 and BA.5 had a three-fold reduction (Haseltine, 2022; Tallei et al., 2022), and unprotected people had a seven-fold decrease. Thus, future Omicron sub-lineages are unlikely to be protected against previous infections, and these variations BA.4 and BA.5 might cause new ones (Lewnard et al., 2022; Khan et al., 2022). Overall, Omicron is thought to have significantly more mutations in its receptor binding area (RBD) and family chylomicronemia syndrome (FCS), resulting in higher infection compared to the original SARS-CoV-2 strain and subsequent variants such as Delta (Zahradník et al., 2021). Fig. 2 illustrates a comparison of various SARS-CoV-2 variants in terms of infection rates, symptomatic cases, hospitalizations, and cumulative deaths.
Fig. 2.
Compares the population (healthy, infected, symptomatic) across the six variants of SARS-CoV-2 strains per day, with data obtained from https://ourworldindata.org.
2.2. Variants of concern (VOC) and spike mutations
A mutation refers to a single alteration in the genetic code or genome of a virus. While mutations are common, they typically do not significantly impact the properties of the virus. However, several mutations in SARS-CoV-2 have been found to affect the virus's properties and response to the pandemic. Initial analyses have suggested that at least one mutation shared by three concerning variants could potentially enhance the virus transmissions efficiency (Ünlü et al., 2021). The first variant had the D614G mutation, which made it more infectious, while the second variant had the N501Y mutation, which appears to make it more transmissible (see Fig. 3 ). Both variants have been detected in many countries worldwide, causing concern among public health officials. In the BA.1 lineage, the Omicron variant has >60 mutations. Of these, 38 are in the spike protein (S), two are in the membrane protein (M), one is in the envelope protein (E), and six are in the nucleocapsid protein (N). Also, the BA.2 lineage has 57 mutations, 31 of which are in the S protein. BA.2 has a different N-terminus than BA.1. The S protein is essential when attaching to host receptors like ACE2 (Garcia-Beltran et al., 2021; Wang et al., 2022; Araf et al., 2022). The B.1.1.298 lineage, like the second N439K lineage, B.1.258, is characterized by deletions in the amino-terminal domain (NTD) [69, 70]. This deletion has occurred multiple times in the global SARS-CoV-2 population. Deletions in 69–70 are associated with increased infectivity and are believed to impact the configuration of a prominent NTD loop (Meng et al., 2021). Understanding the dynamics of mutation and infection requires examining virus sizes, shapes, and structures. Most investigations on SARS-CoV-2 in the literature have focused on mutations in the spike protein, which is critical in binding to receptors and fusing with cell membranes to infect a human host. The spike protein is separated into two parts, N-terminal S1 and C-terminal S2, which have distinct but complementary activities and are present in the viral envelope (Abraham, 2020; Li, 2015; Gui et al., 2017). Several tests have shown that mutations in the spike glycoproteins of SARS-CoV-2 variants improve the fitness of the spike protein. The structure of the Gamma variant's N-terminal domain has changed a lot, which may explain why convalescent sera don't work as well to neutralize it. These spike mutations tell us a lot about how weak spike proteins are built, how they work, and how they react with antibodies (Resende, 2021; Resende et al., 2021; Zhu et al., 2019; Mannar et al., 2022).
Fig. 3.
Illustrates the impact of the D614G mutation on the amino acid composition of the viral spike protein at position 614, which has been changed from aspartate (D) to glycine (G).
Ultimately, it emphasizes the need for monitoring and understanding SARS-CoV-2 development, mainly introducing new variations and spike mutations, to successfully manage the virus's propagation and create viable therapies. The mutation from aspartate (D) to glycine (G) at 614 is particularly significant because it is located in the receptor binding domain (RBD) of the spike protein and plays an important role in the virus' ability to enter human cells. Changes in amino acid composition resulting from these mutations can increase the infectivity and transmission of viruses by stabilizing the open structure of spike proteins as shown in Fig. 3.
3. Biosensors for SARS-CoV-2 detection
3.1. Optical biosensors and their potential for SARS-CoV-2 detection
Optical affinity biosensors use biomolecular receptors that can interact specifically with an analyte and an optical readout mechanism to turn this interaction into a measurable output. Spectroscopic biosensors, like Raman, infrared, and chiral, get their output directly from the optical properties of the analyte (Novotny and Hulst, 2011; Li et al., 2019; Ruiz-Vega et al., 2021). Photonic biosensors make use of evanescent fields to isolate optoelectronic components from liquid samples, making them useful for analyzing analytes. The evanescent field has an exponential decline, with a decay length on the order of hundreds of nanometers perpendicular to the sensor surface. Surface confinement is a crucial component that enhances light-matter interaction and provides precise spatial control of measurements (Altug et al., 2022; Liedberg et al., 1995; Homola and Piliarik, 2006). Two strategies, direct detection of SARS-CoV-2 viruses for early diagnosis and population screening and identification of PCR-free viral RNA, are used by biosensors based on bimodal waveguide (BiMW) interferometric technology. A photonic nanosensor that targets the SARS-CoV-2 RBD region using bioengineered nanobodies (Nb) has been developed and bimodal waveguide (BiMW) interferometric technology is used for rapid and direct measurement of the RBD of the SARS-CoV-2 virus (Ruiz-Vega et al., 2022). A microfluidic device that can directly see the levels of SARS-CoV-2 antibodies in a way that doesn't require any instruments (Cherusseri et al., 2022). Magnetic and polystyrene microparticles are used to bind to SARS-CoV-2 antibodies simultaneously, which reduces the incidence of free particles escaping from magnetic separation and shortens the accumulation length at a particle dam. The results show a LOD of 13.3 ng/ml at 70 min and a rapid mode of 57.8 ng/ml at 20 min (Wu et al., 2022). In Payne et al., it was demonstrated that the unique vibration characteristics of the surface modified by the S-protein binding peptide can be identified and quantified by the SARS-CoV-2 SERS sensor, which is based on ACE2 as a viral protein capture probe. The LOD was reported to be 300 nM (Payne et al., 2021). Sensitivity detection of the SARS-CoV-2 nucleocapsid protein (N protein) is achieved using surface plasmon resonance (SPR) methods that are boosted by nanoparticles. The method uses AuNPs with diameters of 100 nm and this enhancement of SPR provided by these large nanoparticles results in the lowest limit of detection (LOD) of the SARS-CoV-2 N protein (85 fM) ever obtained by SPR as per Yano et al. (2022). Researchers proposed using hyperspectral imaging based on optical imaging spectroscopy to detect SARS-CoV-2 spike samples of 5 μl of fluid at the pixel, droplet, and patient levels using spectral characteristic descriptors and partial least squares discriminant analysis in the visible and near infrared, achieving 100 % sensitivity as per Gomez-Gonzalez et al. (2022). COVID-19 Low-cost optical diagnostic for rapid testing is a colorimetric biosensor manufactured on cotton swabs and modified with an enzyme of angiotensin-converting enzyme 2 (ACE2) that detects SARS-CoV-2 in 5 min. It is manufactured from micro-sized gold nanoparticles and changed with human ACE2. The COLOR method saw very low viral particle loads at LOD 0.154 pg ml−1 (Ferreira et al., 2021). Here, a single pot RT-LAMP based on a multifluid probe assay was reported to detect fragments of the SARS-CoV-2 ORF1ab and N gene. The results were a sensitivity of 20 copies/l for ORF1ab gene fragments and a sensitivity of 2 copies/l for N gene fragments using two primer sets and two molecular beacon probes (MB), each labeled with a different fluorophore (Talap et al., 2022). A study shows an integrated silicon photonic crystal microarray structure for multiplexed sensing and the multimode interference coupler architecture for COVID-19 testing (Asghari et al., 2021). Fig. 4 shows four different state-of-the-art optical sensors for detecting SARS-CoV-2 variants.
Fig. 4.
Showcases the latest advancements in optical sensors for detecting SARS-CoV-2 variants. These include: A. a SERS biosensor, reprinted with permission of Payne et al. (2021) Copyright 2021, ACS sensors. B. a silicon photonic crystal microarray structure for multiplexed sensing, reprinted with permission of Asghari et al. (2021) Copyright 2021, Applied Physics Reviews. C. a microfluidic particle dam for directly visualizing SARS-CoV-2 antibody levels in COVID-19 vaccinees, reprinted with permission of Wu et al. (2022) Copyright 2022, Science Advances, and D. a standard SPR-based biosensor configuration. Reprinted with permission of Asghari et al. (2021) Copyright 2021, Applied Physics Reviews.
3.2. Field effect transistors (FET) electronic biosensors
A field-effect transistor (FET) electron sensor is only one of many potentiometric techniques. Separately from the biological recognition element, an insulator layer (like SiO2) functions as a selective transducer for the target molecule. Electrostatic surface potential variations in semiconductors are used to identify nucleic acids and proteins by correlating them with changes in the charge distribution on the surface (Kaisti, 2017). The study discusses the use of a field effect transistor (FET) biosensing unit to detect the SARS-CoV-2 virus in clinical samples using a transistor biosensing unit. As sensor, a graphene sheet field effect transistor (FET) immobilized with an antibody specific for the SARS-CoV-2 spike protein was used. Sensor output was determined using an antigen protein cultured virus and a nasopharyngeal swab from a patient with COVID-19. Detection in medical tests was possible at a concentration of 2.42 102 copies/ml 62. Researchers show analysis of nucleic acids directly on graphene field effect transistors (g-FETs) using dual Y-shaped DNA probes.
The SARS-CoV-2 nucleic acid is targeted by adapted Y-dual probes on g-FET, allowing for a high recognition ratio and a detection limit (0.03 copies l−1) 1–2 orders of magnitude lower than current nucleic acid assays. The assay is the first to perform a direct 5-in-1 pooled nucleic acid test in <1 min (Kong et al., 2021). Develop a biosensor based on viral receptors for rapid screening of COVID-19 that is sensitive and portable. A dual-gate field effect transistor was used to boost the performance of a viral receptor-based biosensor, which is notoriously insensitive. The optimized biosensor was created using a synthetic virus that resembled SARS-CoV-2 in size, structure, and composition. The new biosensor recognized SARS-CoV-2 with a sensitivity of 165 copies/ml, which is on par with molecular diagnostic assays, and did so in just 20 min (Park et al., 2022). These sensors are also inexpensive and user-friendly.
However, coronavirus samples require extensive pretreatment and filtering prior to a final diagnosis. A digital platform can achieve a binary classification based on artificial intelligence at the level of identifying a single marker/virus in 0.1 ml. Validated by a total of 240 tests, including a small-scale clinical study, diagnostic sensitivity, specificity, and precision are 99.2 %. Saliva, blood serum, and swabs can be tested with the flexible immunometric system, identifying the SARS-CoV-2 virus, spike S1 and antigen proteins.
BioScreen is compact, since it consists of a disposable cartridge and a portable electronic reader that can be synced with a smart device. Suitable molecular handling and a quick test time of 21 min are two of the many benefits of this method (Macchia et al., 2022). Fig. 5 describes the steps taken to develop a portable electrical biosensor that uses virus receptors, such as SARS-CoV-2 replication and infection, the synthesis of the synthetic virus using microfluidics and the portable electrical biosensor.
Fig. 5.
Represented A portable electrical biosensor is built using virus receptors, such as replication and infection of SARS-CoV-2, and synthesis of the synthetic virus using microfluidics and the portable electrical biosensor. A synthetic SARS-COV-2 with the SP1 spike protein bound to the avidin molecule is available for cryo-electron microscopy. Reprinted with permission of Park et al. (2022), Copyright 2022, Nano Letters.
3.3. Electrochemical biosensors
Electrochemical biosensors are evidence biochemicals that convert a voltage or current into an analytically valuable signal (Chaudhary et al., 2022c; Cherusseri et al., 2022). An electrochemical biosensor device (eCovSens) was designed to detect COVID-19 spike protein antigens in spit samples to detect the sensitive COVID-19 antibody. A gold nanoparticle was deposited on a fluorine-doped tin oxide surface and measured electrical conductivity. The results show the sensitivity levels of the assay of (90 and 120) fM (Mahari et al., 2020). Electrochemical biosensors have been reported to detect MERS-CoV. This experiment aims to demonstrate the successful use of a variety of gold nanoparticles that have been modified for use with carbon electrodes. It used an antibody specific for MERS-CoV during the screening process to recognize the virus spike protein (S1). The virus was detected 20 min after being isolated and purified (Layqah and Eissa, 2019). Developed a low-cost biosensor composed of pencil graphite electrodes that was able to detect the presence of SARS-CoV-2 on a minute scale (de Lima et al., 2021). Similarly, a study shows that an electrochemical impedance spectroscopy biosensor can be used at the point of care to detect SARS-CoV-2 and variant B.1.1.7 in real time (Torres et al., 2021). Another study reported developing an electrochemical sensor chip based on a paper that was <5 min, inexpensive, easy to apply and quantitative detection of SARS-CoV-2. Methods have been used using AuNP coated with ssDNA encoding a phosphoprotein found in viral nucleocapsids (N-gene) (Alafeef et al., 2020). Fig. 6 shows the electrochemical sensor instrument for the SARS-COV-2 virus and describes its underlying working principle. The electrochemical sensor has a significant amount of potential to convert a biochemical signal to an electrical one because of the many benefits it offers in the evaluation of a biological sample. On the other hand, isolating and purifying the selection might be a time-consuming process.
Fig. 6.
Illustrate of the SARS-COV-2 variant electrochemical sensors instrument: A. Infected samples are collected from nasal swabs or patient saliva under observation, B. SARS-CoV-2 RNA is extracted, C. Viral RNA is added to graphene-ssDNA-AuNP, D. Incubation is allowed for 5 min, and E. Electrochemical output is recorded digitally. Reprinted with permission of Alafeef et al. (2020), Copyright 2020, ACS Nano.
3.4. CRISPR sensors
Genome editing and nucleic acids can be detected with the use of CRISPR technology combined with the many Cas enzymes associated with CRISPR. It is necessary to isolate the nucleic acid SARS-CoV-2 to confirm the presence of COVID-19 (Rahman et al., 2021). A study shows that CRISPR-Cas13d provides a broad-spectrum antiviral (BSA) that inhibits >99 % of viral titers from various human coronaviruses and variants of SARS-CoV-2 (Zeng et al., 2022). Recent advances in CRISPR-Cas13 transcription amplification methods have facilitated the investigation that allows the detection of SARS-CoV-2 and its mutant versions. The target-activated ribonuclease activity of CRISPR-Cas13a helps to produce sensitive amplification signals, and this is made possible by the aptamer of light-up RNA. In addition to influenza viruses such as H1N1, H7N9, and H9N2, the RNA viral test can detect Coronavirus, SARS-CoV-2, Middle East respiratory sickness (MERS) and SARS viruses. Furthermore, the detection of SARS-CoV-2 was at levels as low as 82 copies with this modified test. In particular, it made it possible to precisely differentiate a crucial mutation of the SARS-CoV-2 variation called D614G, which can cause a greater risk of epidemic and pathogenetic spread (Wang et al., 2021). Present an amplification-free CRISPR-Cas13aassay to directly detect SARS-CoV-2 from nasal swab RNA using a mobile phone microscope. The test achieved a sensitivity of 100 copies/ml in <30 min after identifying reextracted RNA from a panel of positive clinical samples in <5 min. The method combines crRNAs targeting SARS-CoV-2 RNA to increase sensitivity and specificity by measuring viral load using enzyme kinetics (Fozouni et al., 2021). Researchers have developed a fully automated CRISPR-LAMP platform that can manipulate droplets and perform combined reactions with high sensitivity and specificity, detecting SARS-CoV-2 mutations with a detection limit of 102 copies/μl (Zhang et al., 2022). Fig. 7 shows the compact CRISPR-Cas13a system integrated into mobile phone microscopes, as well as the design of mobile phone microscopes that detect fluorescence. CRISPR-Cas13a is a gene editing tool for the detection of RNA. The compact system consists of Cas13a enzymes and guide RNAs that specifically target and separate the interesting RNA sequences, such as SARS-CoV-2. The system is designed to generate fluorescent signals when targeting RNA is cut, and can be detected with the help of mobile phone microscopes (Table 1).
Fig. 7.
Detection of SARS-COV-2 variants based on CRISPR: A. Mobile phone-based microscope for fluorescence detection and images of CRISPR-Cas13a-based mobile phone microscopy. Reprinted with permission of Fozouni et al. (2021), Copyright 2021, Cell. B. CRISPR-LAMP platform that is automated, Reprinted with permission of Zhang et al. (2022), Copyright 2022, Analytical Chemistry.
Table 1.
Analysis and summary of the latest innovations techniques for detecting the SARS-CoV-2 virus and its variants.
| Instruments/year | Target | Variant detection | LOD | Duration | Sample | Sensitivity | Application | Ref |
|---|---|---|---|---|---|---|---|---|
| Photonics nanosensor (2022) | SARS-CoV-2 | ✓ | 598 FFU ml−1 | ˂20 min | RBD | 97 % | Hospitals and laboratories | (Ruiz-Vega et al., 2022) |
| Microfluidic (2022) | SARS-CoV-2 | ✓ | 57.8 ng/ml | 20 min | Antibody levels | 46.67 % | POC test | (Wu et al., 2022) |
| SERS (2021) | SARS-CoV-2 | ✓ | 300 nM | Na | ACE2 and RBD | NA | On-site and POC test | (Payne et al., 2021) |
| SPR with large gold (AuNPs) (2022) | SARS-CoV-2 | ✓ | 85 f. (4 pg/ml) | 15 min. | Nucleocapsid protein | NA | POC test | (Yano et al., 2022) |
| Optical imaging spectroscopy (2022) | SARS-CoV-2 | ✓ | 102–106 copies ml−1 | 54 patients in 300 S | Spike protein | 100 % | POC test | (Gomez-Gonzalez et al., 2022) |
| Colorimetric (2021) | SARS-CoV-2 | ✓ | 0.154 pg ml−1 | 5 min | ACE2 | 96 % | POC test | (Ferreira et al., 2021) |
| RT-LAMP assay with multi fluorescent probes (2022) | SARS-CoV-2 | ✓ | 20 and 2 copies/μl | NA | RNA protein, ORF1ab, and N gene | 0.9915 and 0.9956 | Clinical settings in resource-limited regions | (Talap et al., 2022) |
| Graphene FET sensor (2021) | Screening of COVID-19 | ✓ | 0.03 copy μl−1 | ∼1 min | RNA protein, ORF1ab, and N gene | ∼1.3 × 10−4 and ∼3.4 × 10−5 mV | On-site and POC test | (Kong et al., 2021) |
| FET sensor (2022) | SARS-CoV-2 | ✓ | 100 pg ml−1 | 20 min | ACE2, Spike, and antibody | ∼165 copies/ml | Screening of infectious diseases | (Park et al., 2022) |
| Electronic sensor/BioScreen (2022) | SARS-CoV-2 | ✓ | 0.1 ml | 21 min | Spike S1, G antigen, blood serum, and swab | 99.2 % | Point of care | (Macchia et al., 2022) |
| Electrochemical (2021) | SARS-CoV-2 | ✓ | 229 fg ml−1 | 6.5 min | ACE2 | 100.0 % | POC test and population surveillance | (de Lima et al., 2021) |
| Electrochemical (2021) | SARS-CoV-2 | ✓ | 2.8 fg ml−1 | 4 min | ACE2 | 100 % | POC test | (Torres et al., 2021) |
| Electrochemical chip (2020) | SARS-CoV-2 | ✓ | 6.9 copies/μl | ˃5 min | RNA and N-gene | 231 (copies μl−1)−1 | POC test | (Alafeef et al., 2020) |
| CRISPR-Cas13 (2021) | MERS, SARS-CoV and SARS-CoV-2 | ✓ | 82 copies | 20 min | RNA and D614G | NA | Monitoring Clinical | (Wang et al., 2021) |
| CRISPR-Cas13 with mobile phone (2021) | SARS-CoV-2 | ✓ | 100 nM | ˃5 min | RNA | ∼100 copies/μml | POC test | (Fozouni et al., 2021) |
| CRISPR-LAMP platform (2021) | SARS-CoV-2 | ✓ | 102 copies/μl | 1 h | Spike, T478K, D614G, P681R, and P681H | 100 % | Clinical diagnostic | (Zhang et al., 2022) |
| Smart – phone miSHERLOCK (2022) | SARS-CoV-2 | ✓ | 100,000 cp/ml | 55 min | RNA protein | 2–20-Fold | Test at home and POC | (De Puig et al., 2021) |
| Harmony COVID-19 KIT with smartphone (2021) | SARS-CoV-2 | ✕ | 0.5–2.5 copies/μl | 17 min | RNA protein | 95 % | Test at home and POC | (Panpradist et al., 2021) |
| Smartphone-based LAMP (2022) | SARS-CoV-2/influenza | ✓ | 103 copies/ml | 25 min | variant gRNAs | 100 % | POC | (Heithoff et al., 2022) |
| Smartphone using Quantum dot (2021) | SARS-CoV-2 | ✓ | 1.99 & 0.11 pM | Real-time | RBD and N-gene | 90 % | Healthcare system | (Zhang et al., 2021) |
| RT-LAMP based on smartphone (2022) | SARS-CoV-2 | ✓ | 6 copies/μl | 45 min | Nucleic acid, sequence | NA | Test at home | (Tang et al., 2022) |
3.5. Smartphone-based biosensors for at-home testing
This section discusses the development of smartphone-compatible biosensors. These biosensors utilize the smartphone's camera and other sensors to perform various tests, such as identifying the presence of certain chemicals or pathogens in a sample highlights the most recent innovations in smartphone-based biosensors and their potential for detection SARS-CoV-2 variants. A diagnostic device called SHERLOCK-minimally instrumented (miSHERLOCK) was developed by scientists at the Harvard University Wyss Institute for Biologically Inspired Engineering, the Massachusetts Institute of Technology. Diagnostic tool for treating patients using natural saliva to extract, purify, and concentrate viral RNA. Performs amplifier and detection reactions, delivers fluorescent visual output in 1 h, and requires only three user actions (Madrid et al., 2022). Furthermore, it can be quickly adjusted to identify different viruses and SARS-CoV-2 and mutations associated with variants B.1.1.7, B.1.351 and P.1 by multiplication of compassion. An optional smartphone app allows for output quantification, automatic interpretation, and the potential for remote, dispersed result reporting, producing data that can be read and validated by an accompanying smartphone application in a matter of hours (De Puig et al., 2021), as shown in Fig. 8-A. A study provides home testing, and a POC called “Harmony COVID-19” was created using low-cost consumables, prefilled reagents, and straightforward equipment. The method develops a multiplexed reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay that can report results in as little as 17 min for samples with a high viral load (5000 copies/ml) using a nasal matrix or saliva sample containing 0.5 virus particles/l; Harmony was able to detect 97 or 83 % of the simulated models (Panpradist et al., 2021). A smartphone uses a loop-mediated isotopic amplification assay to detect SARS-CoV-2 and influenza. The method tested two groups of 20 individuals with symptoms in positive symptomatic SARS-CoV-2, and the SARS-CoV-2 screening tests at admission were negative for 30 asymptomatic members of the same population. However, this study had some limitations that required more participants (Heithoff et al., 2022). COVID-19 patient monitoring system using quantum barcodes on smartphones have also been evaluated. Compared to lateral flow tests, its technology was 90 % more sensitive and 100 % more specific for SARS-CoV-2. The corresponding values for the lateral flow assays were 34 % and 100 %, respectively, (Zhang et al., 2021), as shown in Fig. 8-B. Biosensors and characterization methods have tremendous potential due to their wide use and educational benefits. However, biological and chemical sensors face scientific challenges before providing reliable, accurate, and early disease detection in four primary categories of split virus detection: First, direct viral diagnosis: A perfect virus can only be detected by biosensors or, more generally, by cell culture methods (Caygill et al., 2010). Establish a saliva-based SARS-CoV-2 reverse transcription loop-mediated isothermal amplification (RT-LAMP) system. A platform handles everything from preparing viral particles by thermal lysis to sample dispensing, from target sequence RT-LAMP amplification to the result from reporting and communication. The results can identify in LoD 5 copies/μl of saliva with a turnaround time of <45 min (Tang et al., 2022).
Fig. 8.
Portable variants of the SARS-COV-2 virus tested based on the smart phone: A. MiSHERLOCK schematic showing the integration of instrument-free viral RNA extraction and concentration from raw saliva, reactions that identify SARS-CoV-2 and variations, fluorescence output and an auxiliary mobile phone app for automatic result interpretation. Reprinted with permission of De Puig et al. (2021), Copyright 2021, Science Advances. B. Smartphone-based imaging device for quantum dot barcodes scan immunoassay. Reprinted with permission from Zhang et al. (2021), Copyright 2021, ACS Nano Letters.
4. Challenges and opportunities
In its early stages, COVID-19 diagnosis is based on clinical symptoms and interactions with potentially infected individuals. However, the clinical symptoms and signs are inconclusive, thus requiring further diagnostic and serological tests (Filipić et al., 2020; Liu et al., 2020). Low- and middle-income nations bear the majority of the disease burden due to a lack of laboratory capacity for diagnosis, treatment, and care of current diseases and new infections (Bandawe et al., 2022; Vitoria et al., 2009). Developed molecular diagnostic tests based on RT-qPCR for detecting SARS-CoV-2 are available, but their usage in resource-poor areas is challenging due to the need for specialized personnel and equipment. SARS-CoV-2 detection methods are becoming increasingly sophisticated as research progresses, and new mutant viruses require periodic validation of testing procedures for both sensitivity and specificity (Wei et al., 2023; World Health Organization, 2020). Further information on these new infections and their control is expected to become available as viral genome sequencing and research progress, along with innovative vaccination techniques (Ramachandran et al., 2020).
Many systemic challenges associated with laboratory systems exist in low- and middle-income countries, including a lack of laboratory supplies, equipment, skilled personnel, educators, and training programs, insufficient logistic support, a de-emphasis on laboratory testing, inadequate testing quality monitoring, decentralization of laboratory facilities, and a lack of government laboratory standard (Petti et al., 2006; Shen et al., 2021; Arya et al., 2019). The efficiency and accuracy of detection methods depend heavily on the quality of samples used, such as throat swabs, nasal swabs, deep throat saliva, and sputum samples collected from the upper and lower respiratory systems (Oh et al., 2021; Luka et al., 2015). The logistics of collecting and transporting samples to a lab are crucial to making an accurate diagnosis, and misidentification due to sample contamination, poor sample collection, handling, transportation, or storage, insufficient samples, or the presence of interfering chemicals may contribute to diagnostic errors. Preparing the workforce for new responsibilities could lower the number of required workers and redirect the focus on activities that add value, such as quality evaluation and the implementation of new diagnostic tests (Lippi and Da Rin, 2019; Yu et al., 2020; Lin et al., 2020) Therefore, A sophisticated network of nanophotonic biosensors can monitor virus mutations to enhance healthcare quality. Limitations of biosensors in detecting SARS-COV-2 mutations are presented in Table 2 .
Table 2.
A summary of biosensor challenges for the detection of SARS-COV-2 variants.
| Biosensor categories | Challenges |
|---|---|
| Optical biosensor |
|
| |
| |
| Electronic biosensors |
|
| Electrochemical biosensors |
|
| CRISPR biosensors |
|
| Smartphone-based biosensors |
|
5. Advancements in nanophotonic-based biosensors for SARS-CoV-2 variants detection
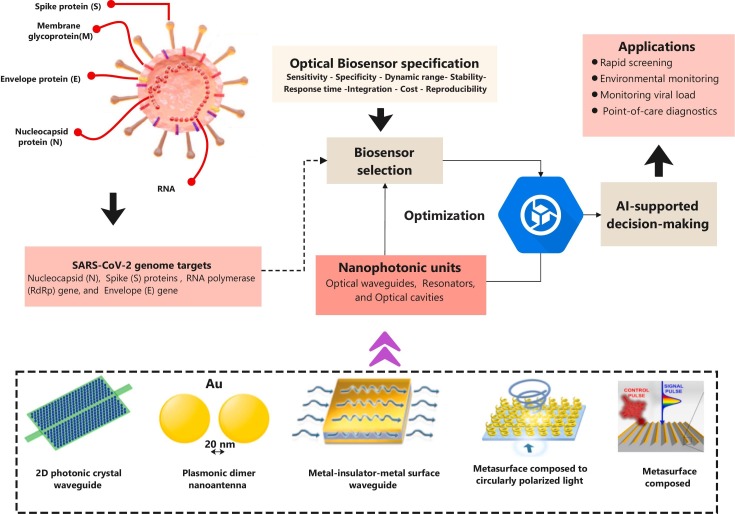
Today, nanophotonic biosensors are capable of a wide detection range of pathogens/diseases (Chaudhary et al., 2022c, Chaudhary et al., 2023c). Typically, a suitable capture reagent is all that is required. Although larger molecules, such as antibodies and enzymes, are recognized by these sensors, smaller molecules, such as metabolites, are difficult to detect (Yoo and Lee, 2016; Chaudhary et al., 2022b). However, this variation still allows for a great deal of versatility in use. Even though it is still in its infancy, many scientists are already planning for the promising future of the field of point-of-care nanophotonic. Real-time tracking of essential biomarkers is being investigated for its potential with implanted and wearable nanophotonic devices (Guo et al., 2021). Additionally, there are still significant obstacles to overcome, since light delivery within the body is problematic and the body has a natural predisposition to reject alien objects (Kwon, 2022). The Mexican Optical Research Center is developing wearable sensors incorporated in nanophotonic materials, including gold (Gao et al., 2019). This paper proposes a new family of 3D nanophotonic structures inspired by general relativity, in which light is subjected to the curvature of the medium as it evolves. Wave packets traveling through our curved-space structure can have their trajectories, diffraction characteristics, phase, and group velocities manipulated to our liking (Bekenstein et al., 2017). Nanophotonic biosensors with improved compactness, portability, and throughput are enabled by integrated photonic circuits. Integration strategies can be classified as vertical or planar. Nanostructures can be used in vertical integration to reduce optical coupling requirements, eliminating the need for external optical couplers such as prisms, gratings, or waveguides. The use of collinear light can efficiently miniaturize and multiplex nanostructures, resulting in high multiplexing limited only by the diffraction limits of light (Lopez et al., 2017; Zanchetta et al., 2017; Taha et al., 2022). Optical waveguides can be arranged in a 1D array and functionalized separately for multiplexed detection in planar integration approaches. Sensor chips can be further miniaturized by hybridizing the optoelectronic components and the electronics layer. Commercially viable silicon-based integrated sensors using conventional waveguides and optical microresonators such as micro rings and photonic crystal cavities are used in most planar sensors (Hu et al., 2019; Lee et al., 2019). However, to analyze extremely absorptive or turbid solutions, one must consider the effects of optical loss, interference, and scattering, which might occur because light must travel through the sample during vertical integration. Individual sensing components often use nanostructure arrays to provide a more excellent output signal than individual particles in practice. This set of parts may be configured in 2D arrays for multiplexed detection (Wang et al., 2020; Mudumba et al., 2017; Chaudhary et al., 2023c). Future work on nanophotonic biosensors for SARS-CoV-2 detection can improve sensor sensitivity and specificity, and increase their multiplexing capabilities. Researchers can also explore the use of different types of nanostructures and materials for biosensor applications and new integration strategies for smaller and more portable devices (Vaisocherová et al., 2015; Maan et al., 2020; Yoo et al., 2015; Galloway et al., 2013).
5.1. Nanophononics units
Nanophotonic units are subwavelength optical devices that can operate light and manipulate light at the nanoscale, which typically involves structures with dimensions smaller than the wavelength of light. These units consist of various nanoscale components, such as waveguides, resonances, and optical cavities, which limit and control light diffusion. Controls the properties of light, such as its intensity, polarization, phase, and propagation direction, via nanoscale materials and electronics (Tran et al., 2022). Nanophotonics can be utilized to increase the sensitivity and selectivity of optical biosensors by taking advantage of the small interaction between light and biomolecules and can accomplish this by creating nanophotonic structures capable of confining and amplifying the electromagnetic field around the sensor region, enabling more efficient biomolecular interaction detection (Prasad, 2004; Koenderink et al., 2015). A 2D photonic crystal waveguide, coupled to resonant cavities, can serve as a wavelength division multiplexer in optical communications. This waveguide is made of dielectric materials with different refractive indices, creating a photonic bandgap that restricts specific wavelengths of light. As a result, light with certain wavelengths can be trapped in resonant cavities and sent through a waveguide to different output ports. This technology has potential applications in high-speed optical communications and optical sensing (Akahane et al., 2003). A plasmonic dimer nanoantenna coupled to an optical emitter is a structure that can produce directional light emission. It consists of two closely spaced metallic nanoparticles supporting plasmon resonances and an optical emitter emitting light when excited. The plasmon resonances can enhance the emission of light from the emitter and direct it in the desired direction. This technology has potential applications in imaging, sensing, and optical communication systems. However, careful design and experimental characterization are necessary for optimal performance due to various factors affecting the structure (Netherton et al., 2022). A metasurface made of chiral antennas selectively interacts with circularly polarized light by modifying the phase and amplitude of its left- and right-circularly polarized components. Furthermore, this creates a chiral filter discriminating between the two polarizations with optical communication, spectroscopy, and biological imaging applications. Chiral metasurface design and fabrication can be challenging, but recent nanofabrication advances have opened up new research possibilities (Lee et al., 2014). The metal-insulator-metal (MIM) surface plasmon polariton (SPP) waveguide can confine light and reduce its wavelength at the nanoscale. It comprises a metal layer between two insulator layers with a nanoscale gap. When light is coupled into the waveguide, surface plasmon polaritons can be excited, leading to strong confinement of the light and a reduction of its wavelength. This technology has potential applications in nanophotonics, sensing, and optical communication systems that must consider limitations such as propagation length and losses (Bidault et al., 2019).
5.2. Nanophotonics enabling artificial neural networks
The unique properties of photons could solve some of the problems electronics face. For example, photons are helpful as information carriers because they have high speeds of propagation, low chances of interference, and many parallelisms. In addition, nanophotonics enable signal multiplexing over time, space, polarization, angular momentum, and wavelength. As a result, optical waveguides or fibers have replaced copper connections in computer processors and data centers, due to the development of optical interconnect technology (Deng and Liu, 2014; Ren et al., 2016; X. Li et al., 2015; Deng et al., 2015). Consequently, artificial neural networks (ANNs) are computational systems that are inspired by the structure and function of biological neural networks in the human brain. ANNs are made up of interconnected nodes that can process and transmit information. Recently, researchers have started exploring the use of nanophotonics to enable ANNs with improved performance and energy efficiency (Appeltant et al., 2011). Nanophotonics also allows for the development of compact and efficient ANNs. By using nanoscale photonic components, such as waveguides and photonic crystals, ANNs can be integrated into small form factors, making them ideal for use in portable and wearable devices. One of the main advantages of using nanophotonics in ANN is the ability to perform parallel calculations. In traditional electronic ANNs, the computation is performed sequentially, which can cause bottlenecks and delays in processing. With nanophotonics, parallel computations can significantly improve processing speed and reduce energy consumption (Mesaritakis et al., 2016; Kaikhah and Loochan, 2001; Lin et al., 2018). Overall, the use of nanophotonics in ANNs holds great promise for the development of efficient and high-performance computing systems. However, many challenges remain to be overcome, such as the integration of nanophotonic components with existing electronic systems and the development of new fabrication techniques for nanophotonic devices. (Jones et al., 2019; M. Li et al., 2015; Owoicho et al., 2021)
5.3. Benefits of AI-enabled nano-photonic optical biosensors
The development of optical biosensors based on nanophotonics and artificial intelligence can be very useful in detecting SARS-CoV-2 variants. Some potential applications are: Rapid screening of SARS-CoV-2 variants:
-
•
Rapid screening of SARS-CoV-2 variants: Optical biosensors can be used to detect specific mutations or SARS-CoV-2 variants with specific probes or antibodies. Using nanophotonic structures improves the sensitivity and specificity of biosensors, and AI algorithms help detect and classify different variations.
-
•
Monitoring of viral loads and replication: optical biosensors can be used to monitor the viral load and replication of SARS-CoV-2 in patient samples and provide feedback on the effectiveness of antiviral treatment in real time. Nanophotonic structures can detect low viral loads and artificial intelligence algorithms can help analyze and interpret sensor data.
-
•
Environmental monitoring of SARS-CoV-2 variants: Optical biosensors can be used to monitor SARS-CoV-2 variation in the environment, such as wastewater and air samples. Nanophotonic structures can improve the sensitivity of biosensors, while artificial intelligence algorithms can help detect and track different variants.
-
•
SARS-CoV-2 Point of Care Diagnostics: Optical biosensors can be designed as portable and easy-to-use devices for SARS-CoV-2 Point of Care Diagnostics. The use of nanophotonic structures can allow biosensors to be miniaturized and integrated, while artificial intelligence algorithms can analyze and interpret sensor data in real time. Overall, the combination of nanophotonic structures and AI algorithms may lead to the development of highly sensitive, selective and efficient optical biosensors for the detection of SARS-CoV-2 variants and have potential applications in the field of clinical, environmental and public health.
Fig. 9 shows integrated nanophotonic biosensors that are the future of artificial intelligence for multiattribute decision support systems to detect analytes in different environments. This integration represents a promising avenue for future applications of nanophotonics biosensors in analyte detection.
Fig. 9.
Showcases the potential of integrated nanophotonics biosensors and artificial intelligence (AI) for creating multi-attribute decision support systems capable of detecting analytes in diverse environments.
6. Conclusions
The outbreak of coronavirus diseases has had a significant impact on human life and economic development. SARS-CoV-2 continues to mutate during transmission, making epidemic control and prevention more complex and potentially reducing the effectiveness of the vaccine. The severity of the disease has decreased due to vaccinations and virus mutations and may coexist with humans in the future. Clinicians have faced significant challenges in quickly identifying microorganisms and pathogens, but validated technologies such as optical biosensors and photonics have been developed in microbiology laboratories to decrease the time required to detect viruses in a few minutes. This advancement in technology also addresses the decreasing number of experienced laboratory personnel and makes patient care more cost-effective and efficient by automating tasks. Technician responsibilities will shift from manual handling to handling sophisticated instruments in the future. Automation also helps to alleviate the strain caused by the growing backlog of test samples and the shrinking workforce of microbiologists. By combining cutting-edge biological technologies with intelligent image analysis, non-productive workflow disruptions can be eliminated and the microbiologist's time can be freed up for more valuable tasks like result interpretation and client counseling. In summary, AI-powered laser-assisted diagnosis can improve food safety inspections, air virus density analysis, and environmental monitoring. An intelligent optical network of medical biosensors can also improve healthcare quality by collecting information for virus detection using an optical network linked to a multi-intelligent microbiology laboratory.
Abbreviations
- COVID-19
coronavirus disease
- SARS-CoV-2
Coronavirus Severe Acute Respiratory Syndrome 2
- TEM
Transmission Electron Microscopy
- ACE2
angiotensin-converting enzyme 2
- HR
Heptad Repeat
- SLIC
Simple Linear Iterative Clustering
- SARS-COV
Severe Acute Respiratory Syndrome Coronavirus
- RSIs
Receptor-Specific Interactions
- hACE2
higher angiotensin-converting enzyme 2
- RBD
receptor-binding domain
- BNNs
biological neural networks
- λ
wavelength
CRediT authorship contribution statement
B.A.T., wrote the initial manuscript draft, Conceptualization and methodology, Y.A.M., investigation, Conceptualization and methodology, Q.A., Writing - Review & Editing., N.A., supervision and validation., Formal analysis and resources., Y.L., Validation., Z.C. & S.R., Writing - Review & Editing., and V.C., Writing - Review & Editing. All authors have reviewed and accepted the published version of the manuscript.
Funding
The authors would like to express their gratitude to the Ministry of Higher Education, Malaysia for its generous grant (FRGS/1/2021/TK0/UKM/02/17), which provided essential support for the successful completion of this study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors gratefully acknowledge the financial support of the Department of Electrical, Electronic and Systems Engineering/Faculty of Engineering and Built Environment/Universiti Kebangsaan Malaysia (UKM) for their encouragement. VC acknowledges British Council for providing grants and support under Going Global Partnerships grants (Exploratory Grant: ECOBUSS project and Industrial Academia Collaborative Grant 2022-23: Unique ID :7 of project titled: Design and development of bio/chemical sensors to address antibiotics abuse in India). VC also acknowledge Vice Chancellor, University of Delhi, India for continous support.
Editor: Warish Ahmed
Data availability
Data availability is available on GISAID: (https://www.gisaid.org/) and (https://ourworldindata.org/).
References
- Abraham J. Passive antibody therapy in COVID-19. Nat. Rev. Immunol. 2020;20:401–403. doi: 10.1038/s41577-020-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed B., Qussay T., Jubouri A., Al Y., Mohd M., Dzulkefly S., Zan B., Ashrif A., Mahmoud A.B., Fadhel M., Arsad N. Photonics enabled intelligence system to identify SARS - CoV 2 mutations. Appl. Microbiol. Biotechnol. 2022 doi: 10.1007/s00253-022-11930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahane Y., Asano T., Song B.S., Noda S. High-Q photonic nanocavity in a two-dimensional photonic crystal. Nature. 2003;425:944–947. doi: 10.1038/nature02063. [DOI] [PubMed] [Google Scholar]
- Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor Chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleem A., Samad A.B.Akbar, Slenker A.K. StatPearls. StatPearls Publishing; 2021. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19)http://www.ncbi.nlm.nih.gov/pubmed/3403334 [PubMed] [Google Scholar]
- Altug H., Oh S.H., Maier S.A., Homola J. Advances and applications of nanophotonic biosensors. Nat. Nanotechnol. 2022;17:5–16. doi: 10.1038/s41565-021-01045-5. [DOI] [PubMed] [Google Scholar]
- Appeltant L., Soriano M.C., Van Der Sande G., Danckaert J., Massar S., Dambre J., Schrauwen B., Mirasso C.R., Fischer I. Information processing using a single dynamical node as complex system. Nat. Commun. 2011;2:1–6. doi: 10.1038/ncomms1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araf Y., Akter F., Dong Tang Y., Fatemi R., Parvez M.S.A., Zheng C., Hossain M.G. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022;94:1825–1832. doi: 10.1002/jmv.27588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya A., Gangwar A., Kumar A. Elsevier Inc.; 2019. Biosensors in Animal Biotechnology. [DOI] [Google Scholar]
- Asghari A., Wang C., Yoo K.M., Rostamian A., Xu X., Shin J.D., Dalir H., Chen R.T. Fast, accurate, point-of-care COVID-19 pandemic diagnosis enabled through advanced lab-on-chip optical biosensors: opportunities and challenges. Appl. Phys. Rev. 2021;8 doi: 10.1063/5.0022211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma K., Yanagi U., Kagi N., Kim H., Ogata M., Hayashi M. Environmental factors involved in SARS-CoV-2 transmission: effect and role of indoor environmental quality in the strategy for COVID-19 infection control. Environ. HealthPrev. Med. 2020;25:1–16. doi: 10.1186/s12199-020-00904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloch Z., Ma Z., Ji Y., Ghanbari M., Pan Q., Aljabr W. Unique challenges to control the spread of COVID-19 in the Middle East. J. Infect. Public Health. 2020;13:1247–1250. doi: 10.1016/j.jiph.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandawe G., Chitenje M., Bitiliyu-Bangoh J., Kampira E. Approaches to deployment of molecular testing for SARS-CoV-2 in resource-limited settings. Clin. Lab. Med. 2022;42:283–298. doi: 10.1016/j.cll.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekenstein R., Kabessa Y., Sharabi Y., Tal O., Engheta N., Eisenstein G., Agranat A.J., Segev M. Control of light by curved space in nanophotonic structures. Nat. Photonics. 2017;11:664–670. doi: 10.1038/s41566-017-0008-0. [DOI] [Google Scholar]
- Bidault S., Mivelle M., Bonod N. Dielectric nanoantennas to manipulate solid-state light emission. J. Appl. Phys. 2019;126 doi: 10.1063/1.5108641. [DOI] [Google Scholar]
- Bidram E., Esmaeili Y., Amini A., Sartorius R., Tay F.R., Shariati L., Makvandi P. Nanobased platforms for diagnosis and treatment of COVID-19: from benchtop to bedside. ACS Biomater. Sci. Eng. 2021;7:2150–2176. doi: 10.1021/acsbiomaterials.1c00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill R.L., Blair G.E., Millner P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta. 2010;681:8–15. doi: 10.1016/j.aca.2010.09.038. [DOI] [PubMed] [Google Scholar]
- Chaudhary V., Bhadola P., Kaushik A., Khalid M., Furukawa H., Khosla A. Assessing temporal correlation in environmental risk factors to design efficient area-specific COVID-19 regulations: Delhi based case study. Sci Rep. 2022;12:12949. doi: 10.1038/s41598-022-16781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary V., Gautam A., Silotia P., Malik S., de Oliveira Hansen R., Khalid M., Khosla A., Kaushik A., Mishra Y.K. Internet-of-nano-things (IoNT) driven intelligent face masks to combat airborne health hazard. Mater. Today. 2022;60:201–226. doi: 10.1016/j.mattod.2022.08.019. [DOI] [Google Scholar]
- Chaudhary V., Kaushik A., Furukawa H., Khosla A. Review—Towards 5th Generation AI and IoT Driven Sustainable Intelligent Sensors Based on 2D MXenes and Borophene. ECS Sens. Plus. 2022;1(013601) doi: 10.1149/2754-2726/ac5ac6. [DOI] [Google Scholar]
- Chaudhary V., Awan H.T.A., Khalid M., Bhadola P., Tandon R., Khosla A. Progress in engineering interlayer space modulated MXenes to architect next-generation airborne pollutant sensors. Sensors Actuators B Chem. 2023;379:133225. doi: 10.1016/j.snb.2022.133225. [DOI] [Google Scholar]
- Chaudhary V., Talreja R.K., Khalid M., Rustagi S., Khosla A. Polypyrrole-Ag/AgCl Nanocomposite-Enabled Exhaled Breath-Acetone Monitoring Non-Invasive Biosensor for Diabetes Diagnostics. ECS J. Solid State Sci. Technol. 2023;12(037003) doi: 10.1149/2162-8777/acc2e4. [DOI] [Google Scholar]
- Chaudhary V., Khanna V., Ahmed Awan H.T., Singh K., Khalid M., Mishra Y.K., Bhansali S., Li C.Z., Kaushik A. Towards hospital-on-chip supported by 2D MXenes-based 5th generation intelligent biosensors. Biosens. Bioelectron. 2023;220 doi: 10.1016/j.bios.2022.114847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherusseri J., Savio C.M., Khalid M., Chaudhary V., Numan A., Varma S.J., Menon A., Kaushik A. SARS-CoV-2-on-Chip for Long COVID Management. Biosensors. 2022;12:890. doi: 10.3390/bios12100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima L.F., Ferreira A.L., Torres M.D.T., de Araujo W.R., de la Fuente-Nunez C. Minute-scale detection of SARS-CoV-2 using a low-cost biosensor composed of pencil graphite electrodes. Proc. Natl. Acad. Sci. U. S. A. 2021;118:1–9. doi: 10.1073/pnas.2106724118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Puig H., Lee R.A., Najjar D., Tan X., Soekensen L.R., Angenent-Mari N.M., Donghia N.M., Weckman N.E., Ory A., Ng C.F., Nguyen P.Q., Mao A.S., Ferrante T.C., Lansberry G., Sallum H., Niemi J., Collins J.J. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021;7:23–26. doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R., Liu X. Optical multiplexing: tunable lifetime nanocrystals. Nat. Photonics. 2014;8:10–12. doi: 10.1038/nphoton.2013.353. [DOI] [Google Scholar]
- Deng R., Qin F., Chen R., Huang W., Hong M., Liu X. Temporal full-colour tuning through non-steady-state upconversion. Nat. Nanotechnol. 2015;10:237–242. doi: 10.1038/nnano.2014.317. [DOI] [PubMed] [Google Scholar]
- Ferreira A.L., De Lima L.F., Torres M.D.T., De Araujo W.R., De La Fuente-Nunez C. Low-cost optodiagnostic for minute-time scale detection of SARS-CoV-2. ACS Nano. 2021;15:17453–17462. doi: 10.1021/acsnano.1c03236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipić A., Gutierrez-Aguirre I., Primc G., Mozetič M., Dobnik D. Cold plasma, a new Hope in the field of virus inactivation. Trends Biotechnol. 2020;38:1278–1291. doi: 10.1016/j.tibtech.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P., Son S., de León Derby M.Díaz, Knott G.J., Gray C.N., D’Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway C.M., Kreuzer M.P., Aćimović S.S., Volpe G., Correia M., Petersen S.B., Neves-Petersen M.T., Quidant R. Plasmon-assisted delivery of single nano-objects in an optical hot spot. Nano Lett. 2013;13:4299–4304. doi: 10.1021/nl402071p. [DOI] [PubMed] [Google Scholar]
- Gao M., Li J., Bao Z., Hu M., Nian R., Feng D., An D., Li X., Xian M., Zhang H. A natural in situ fabrication method of functional bacterial cellulose using a microorganism. Nat. Commun. 2019;10:437. doi: 10.1038/s41467-018-07879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez E., Barriga-Rivera A., Fernandez-Muñoz B., Navas-Garcia J.M., Fernandez-Lizaranzu I., Munoz-Gonzalez F.J., Parrilla-Giraldez R., Requena-Lancharro D., Gil-Gamboa P., Rosell-Valle C., Gomez-Gonzalez C., Mayorga-Buiza M.J., Martin-Lopez M., Muñoz O., Gomez-Martin J.C., Relimpio-Lopez M.I., Aceituno-Castro J., Perales-Esteve M.A., Puppo-Moreno A., Garcia-Cozar F.J., Olvera-Collantes L., Gomez-Diaz R., de los Santos-Trigo S., Huguet-Carrasco M., Rey M., Gomez E., Sanchez-Pernaute R., Padillo-Ruiz J., Marquez-Rivas J. Optical imaging spectroscopy for rapid, primary screening of SARS-CoV-2: a proof of concept. Sci. Rep. 2022;12:1–18. doi: 10.1038/s41598-022-06393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhou B., Du Z., Yang C., Kong L., Xu L. Soft and plasmonic hydrogel optical probe for glucose monitoring. Nanophotonics. 2021;10:3549–3558. doi: 10.1515/nanoph-2021-0360. [DOI] [Google Scholar]
- Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H., Megawati D., Hayati Z., Wagner A.L., Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health. 2020;13:667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine W.A. Birth Of The Omicron Family: BA.1, BA.2, BA.3. Each As Different As Alpha Is From Delta, Forbes. 2022. https://www.forbes.com/sites/williamhaseltine/2022/01/26/birth-of-the-omicron-family-ba1-ba2-ba3-each-as-different-as-alpha-is-from-delta/?sh=27bfe37f3da9
- Heithoff D.M., Barnes L.V., Mahan S.P., Fox G.N., Arn K.E., Ettinger S.J., Bishop A.M., Fitzgibbons L.N., Fried J.C., Low D.A., Samuel C.E., Mahan M.J. Assessment of a smartphone-based loop-mediated isothermal amplification assay for detection of SARS-CoV-2 and influenza viruses. JAMA Netw. Open. 2022;5 doi: 10.1001/jamanetworkopen.2021.45669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D., Quake S.R., McCabe E.R.B., Chng W.J., Chow E.K., Ding X., Gelb B.D., Ginsburg G.S., Hassenstab J., Ho C.M., Mobley W.C., Nolan G.P., Rosen S.T., Tan P., Yen Y., Zarrinpar A. Enabling Technologies for Personalized and Precision Medicine. Trends Biotechnol. 2020;38:497–518. doi: 10.1016/j.tibtech.2019.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homola J., Piliarik M. Surf. Plasmon Reson. Based Sensors. Springer; 2006. Surface plasmon resonance (SPR)Sensors; pp. 45–67. [DOI] [Google Scholar]
- Hu H., Yang X., Guo X., Khaliji K., Biswas S.R., García de Abajo F.J., Low T., Sun Z., Dai Q. Gas identification with graphene plasmons. Nat. Commun. 2019;10:1–7. doi: 10.1038/s41467-019-09008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Fan C., Li M., Nie H.L., Wang F.B., Wang H., Wang R., Xia J., Zheng X., Zuo X., Huang J. COVID-19: a call for physical scientists and engineers. ACS Nano. 2020;14:3747–3754. doi: 10.1021/acsnano.0c02618. [DOI] [PubMed] [Google Scholar]
- Jones R.R., Hooper D.C., Zhang L., Wolverson D., Valev V.K. Raman techniques: fundamentals and Frontiers. Nanoscale Res. Lett. 2019;14 doi: 10.1186/s11671-019-3039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikhah K., Loochan F. Computer generated holograms for optical neural networks. Appl. Intell. 2001;14:145–160. doi: 10.1023/A:1008314025737. [DOI] [Google Scholar]
- Kaisti M. Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 2017;98:437–448. doi: 10.1016/j.bios.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Kajisa T., Ono M., Miyasaka Y., Hasegawa Y., Saito A., Otsuka K., Sakane A., Sasaki T., Yasutomo K., Hamajima R., Kanai Y., Kobayashi T., Matsuura Y., Itonaga M., Yasui T., aki Yano T. Ultrasensitive detection of SARS-CoV-2 nucleocapsid protein using large gold nanoparticle-enhanced surface plasmon resonance. Sci. Rep. 2022;12:1–8. doi: 10.1038/s41598-022-05036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasibhatla S.M., Kinikar M., Limaye S., Kale M.M., Kulkarni-Kale U. Understanding evolution of SARS-CoV-2: a perspective from analysis of genetic diversity of RdRp gene. J. Med. Virol. 2020;92:1932–1937. doi: 10.1002/jmv.25909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keighley C., Garnham K., Harch S.A.J., Robertson M., Chaw K., Teng J.C., Chen S.C.A. Candida auris: diagnostic challenges and emerging opportunities for the clinical microbiology laboratory. Curr. Fungal Infect. Rep. 2021;15:116–126. doi: 10.1007/s12281-021-00420-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., Cele S., Lustig G., Amoako D., Wolter N., Samsunder N., Sivro A., San J.E., Giandhari J., Tegally H., Pillay S., Naidoo Y., Mazibuko M., Miya Y., Ngcobo N., Manickchund N., Magula N., Karim Q.A., von Gottberg A., Abdool Karim S.S., Hanekom W., Gosnell B.I., Khoza T., Smit T., Wong E., Lessells R.J., de Oliveira T., Moosa M.Y.S., Sigal A. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022;13:1–7. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink A.F., Alù A., Polman A. Nanophotonics: shrinking light-based technology. Science(80-.) 2015;348:516–521. doi: 10.1126/science.1261243. [DOI] [PubMed] [Google Scholar]
- Kong D., Wang X., Gu C., Guo M., Wang Y., Ai Z., Zhang S., Chen Y., Liu W., Wu Y., Dai C., Guo Q., Qu D., Zhu Z., Xie Y., Liu Y., Wei D. Direct SARS-CoV-2 nucleic acid detection by Y-shaped DNA dual-probe transistor assay. J. Am. Chem. Soc. 2021;143:17004–17014. doi: 10.1021/jacs.1c06325. [DOI] [PubMed] [Google Scholar]
- Kwon D. Light-based sensors set to revolutionize on-site testing. Nature. 2022;607:834–836. doi: 10.1038/d41586-022-02043-w. [DOI] [PubMed] [Google Scholar]
- Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Tymchenko M., Argyropoulos C., Chen P.Y., Lu F., Demmerle F., Boehm G., Amann M.C., Alù A., Belkin M.A. Giant nonlinear response from plasmonic metasurfaces coupled to intersubband transitions. Nature. 2014;511:65–69. doi: 10.1038/nature13455. [DOI] [PubMed] [Google Scholar]
- Lee I.H., Yoo D., Avouris P., Low T., Oh S.H. Graphene acoustic plasmon resonator for ultrasensitive infrared spectroscopy. Nat. Nanotechnol. 2019;14:313–319. doi: 10.1038/s41565-019-0363-8. [DOI] [PubMed] [Google Scholar]
- Lewnard J.A., Hong V.X., Patel M.M., Kahn R., Lipsitch M., Tartof S.Y. Clinical outcomes associated with SARS-CoV-2 omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat. Med. 2022;28:1933–1943. doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/jvi.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ren H., Chen X., Liu J., Li Q., Li C., Xue G., Jia J., Cao L., Sahu A., Hu B., Wang Y., Jin G., Gu M. Athermally photoreduced graphene oxides for three-dimensional holographic images. Nat. Commun. 2015;6 doi: 10.1038/ncomms7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Cushing S.K., Wu N. Plasmon-enhanced optical sensors: a review. Analyst. 2015;140:386–406. doi: 10.1039/c4an01079e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Liedberg B., Nylander C., Lundström I. Biosensing with surface plasmon resonance - how it all started. Biosens. Bioelectron. 1995;10:i–ix. doi: 10.1016/0956-5663(95)96965-2. [DOI] [PubMed] [Google Scholar]
- Lin X., Rivenson Y., Yardimci N.T., Veli M., Luo Y., Jarrahi M., Ozcan A. All-optical machine learning using diffractive deep neural networks. Science (80-.) 2018;361:1004–1008. doi: 10.1126/science.aat8084. [DOI] [PubMed] [Google Scholar]
- Lin C., Lin C., Xiang J., Yan M., Li H., Huang S., Huang S., Shen C., Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19) Clin. Chem. Lab. Med. 2020;58:1089–1094. doi: 10.1515/cclm-2020-0187. [DOI] [PubMed] [Google Scholar]
- Lippi G., Da Rin G. Advantages and limitations of total laboratory automation: a personal overview. Clin. Chem. Lab. Med. 2019;57:802–811. doi: 10.1515/cclm-2018-1323. [DOI] [PubMed] [Google Scholar]
- Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., Feng Y., Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from jan to feb 2020. Clin. Chim. Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G.A., Estevez M.C., Soler M., Lechuga L.M. Recent advances in nanoplasmonic biosensors: applications and lab-on-a-chip integration. Nanophotonics. 2017;6:123–136. doi: 10.1515/nanoph-2016-0101. [DOI] [Google Scholar]
- Luka G., Ahmadi A., Najjaran H., Alocilja E., Derosa M., Wolthers K., Malki A., Aziz H., Althani A., Hoorfar M. Microfluidics integrated biosensors: a leading technology towards lab-on-A-chip and sensing applications. Sensors (Switzerland) 2015;15:30011–30031. doi: 10.3390/s151229783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan A.M.C., Hofman A.H., de Vos W.M., Kamperman M. Recent developments and practical feasibility of polymer-based antifouling coatings. Adv. Funct. Mater. 2020;30:2000936. doi: 10.1002/adfm.202000936. [DOI] [Google Scholar]
- Macchia E., Kovács-Vajna Z.M., Loconsole D., Sarcina L., Redolfi M., Chironna M., Torricelli F., Torsi L. A handheld intelligent single-molecule binary bioelectronic system for fast and reliable immunometric point-of-care testing. Sci. Adv. 2022;8:1–14. doi: 10.1126/sciadv.abo0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid R.E., Ramallo F.A., Barraza D.E., Chaile R.E. Smartphone-based biosensor devices for healthcare: technologies, trends, and adoption by end-users. Bioengineering. 2022;9 doi: 10.3390/bioengineering9030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahari S., Roberts A., Shahdeo D., Gandhi S. Ecovsens-ultrasensitive Novel In-house Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. BioRxiv; 2020. pp. 1–20. [DOI] [Google Scholar]
- Mannar D., Saville J.W., Sun Z., Zhu X., Marti M.M., Srivastava S.S., Berezuk A.M., Zhou S., Tuttle K.S., Sobolewski M.D., Kim A., Treat B.R., Da Silva Castanha P.M., Jacobs J.L., Barratt-Boyes S.M., Mellors J.W., Dimitrov D.S., Li W., Subramaniam S. SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization. Nat. Commun. 2022;13:1–12. doi: 10.1038/s41467-022-32262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Kemp S.A., Papa G., Datir R., Ferreira I.A.T.M., Marelli S., Harvey W.T., Lytras S., Mohamed A., Gallo G., Thakur N., Collier D.A., Mlcochova P., Robson S.C., Loman N.J., Connor T.R., Golubchik T., Nunez R.T.Martinez, Ludden C., Corden S., Johnston I., Bonsall D., Smith C.P., Awan A.R., Bucca G., Torok M.E., Saeed K., Prieto J.A., Jackson D.K., Hamilton W.L., Snell L.B., Moore C., Harrison E.M., Goncalves S., Fairley D.J., Loose M.W., Watkins J., Livett R., Moses S., Amato R., Nicholls S., Bull M., Smith D.L., Barrett J., Aanensen D.M., Curran M.D., Parmar S., Aggarwal D., Shepherd J.G., Parker M.D., Glaysher S., Bashton M., Underwood A.P., Pacchiarini N., Loveson K.F., Templeton K.E., Langford C.F., Sillitoe J., de Silva T.I., Wang D., Kwiatkowski D., Rambaut A., O’Grady J., Cottrell S., Holden M.T.G., Thomson E.C., Osman H., Andersson M., Chauhan A.J., Hassan-Ibrahim M.O., Lawniczak M., Alderton A., Chand M., Constantinidou C., Unnikrishnan M., Darby A.C., Hiscox J.A., Paterson S., Martincorena I., Volz E.M., Page A.J., Pybus O.G., Bassett A.R., Ariani C.V., Chapman M.H.S., Li K.K., Shah R.N., Jesudason N.G., Taha Y., McHugh M.P., Dewar R., Jahun A.S., McMurray C., Pandey S., McKenna J.P., Nelson A., Young G.R., McCann C.M., Elliott S., Lowe H., Temperton B., Roy S., Price A., Rey S., Wyles M., Rooke S., Shaaban S., de Cesare M., Letchford L., Silveira S., Pelosi E., Wilson-Davies E., Hosmillo M., O’Toole Á., Hesketh A.R., Stark R., du Plessis L., Ruis C., Adams H., Bourgeois Y., Michell S.L., Gramatopoulos D., Edgeworth J., Breuer J., Todd J.A., Fraser C., Buck D., John M., Kay G.L., Palmer S., Peacock S.J., Heyburn D., Weldon D., Robinson E., McNally A., Muir P., Vipond I.B., Boyes J., Sivaprakasam V., Salluja T., Dervisevic S., Meader E.J., Park N.R., Oliver K., Jeffries A.R., Ott S., da Silva Filipe A., Simpson D.A., Williams C., Masoli J.A.H., Knight B.A., Jones C.R., Koshy C., Ash A., Casey A., Bosworth A., Ratcliffe L., Xu-McCrae L., Pymont H.M., Hutchings S., Berry L., Jones K., Halstead F., Davis T., Holmes C., Iturriza-Gomara M., Lucaci A.O., Randell P.A., Cox A., Madona P., Harris K.A., Brown J.R., Mahungu T.W., Irish-Tavares D., Haque T., Hart J., Witele E., Fenton M.L., Liggett S., Graham C., Swindells E., Collins J., Eltringham G., Campbell S., McClure P.C., Clark G., Sloan T.J., Jones C., Lynch J., Warne B., Leonard S., Durham J., Williams T., Haldenby S.T., Storey N., Alikhan N.F., Holmes N., Moore C., Carlile M., Perry M., Craine N., Lyons R.A., Beckett A.H., Goudarzi S., Fearn C., Cook K., Dent H., Paul H., Davies R., Blane B., Girgis S.T., Beale M.A., Bellis K.L., Dorman M.J., Drury E., Kane L., Kay S., McGuigan S., Nelson R., Prestwood L., Rajatileka S., Batra R., Williams R.J., Kristiansen M., Green A., Justice A., Mahanama A.I.K., Samaraweera B., Hadjirin N.F., Quick J., Poplawski R., Kermack L.M., Reynolds N., Hall G., Chaudhry Y., Pinckert M.L., Georgana I., Moll R.J., Thornton A., Myers R., Stockton J., Williams C.A., Yew W.C., Trotter A.J., Trebes A., MacIntyre-Cockett G., Birchley A., Adams A., Plimmer A., Gatica-Wilcox B., McKerr C., Hilvers E., Jones H., Asad H., Coombes J., Evans J.M., Fina L., Gilbert L., Graham L., Cronin M., Kumziene-Summerhayes S., Taylor S., Jones S., Groves D.C., Zhang P., Gallis M., Louka S.F., Starinskij I., Jackson C., Gourtovaia M., Tonkin-Hill G., Lewis K., Tovar-Corona J.M., James K., Baxter L., Alam M.T., Orton R.J., Hughes J., Vattipally S., Ragonnet-Cronin M., Nascimento F.F., Jorgensen D., Boyd O., Geidelberg L., Zarebski A.E., Raghwani J., Kraemer M.U.G., Southgate J., Lindsey B.B., Freeman T.M., Keatley J.P., Singer J.B., de Oliveira Martins L., Yeats C.A., Abudahab K., Taylor B.E.W., Menegazzo M., Danesh J., Hogsden W., Eldirdiri S., Kenyon A., Mason J., Robinson T.I., Holmes A., Price J., Hartley J.A., Curran T., Mather A.E., Shankar G., Jones R., Howe R., Morgan S., Wastenge E., Chapman M.R., Mookerjee S., Stanley R., Smith W., Peto T., Eyre D., Crook D., Vernet G., Kitchen C., Gulliver H., Merrick I., Guest M., Munn R., Bradley D.T., Wyatt T., Beaver C., Foulser L., Palmer S., Churcher C.M., Brooks E., Smith K.S., Galai K., McManus G.M., Bolt F., Coll F., Meadows L., Attwood S.W., Davies A., Lacy E.De, Downing F., Edwards S., Scarlett G.P., Jeremiah S., Smith N., Leek D., Sridhar S., Forrest S., Cormie C., Gill H.K., Dias J., Higginson E.E., Maes M., Young J., Wantoch M., Jamrozy D., Lo S., Patel M., Hill V., Bewshea C.M., Ellard S., Auckland C., Harrison I., Bishop C., Chalker V., Richter A., Beggs A., Best A., Percival B., Mirza J., Megram O., Mayhew M., Crawford L., Ashcroft F., Moles-Garcia E., Cumley N., Hopes R., Asamaphan P., Niebel M.O., Gunson R.N., Bradley A., Maclean A., Mollett G., Blacow R., Bird P., Helmer T., Fallon K., Tang J., Hale A.D., Macfarlane-Smith L.R., Harper K.L., Carden H., Machin N.W., Jackson K.A., Ahmad S.S.Y., George R.P., Turtle L., O’Toole E., Watts J., Breen C., Cowell A., Alcolea-Medina A., Charalampous T., Patel A., Levett L.J., Heaney J., Rowan A., Taylor G.P., Shah D., Atkinson L., Lee J.C.D., Westhorpe A.P., Jannoo R., Lowe H.L., Karamani A., Ensell L., Chatterton W., Pusok M., Dadrah A., Symmonds A., Sluga G., Molnar Z., Baker P., Bonner S., Essex S., Barton E., Padgett D., Scott G., Greenaway J., Payne B.A.I., Burton-Fanning S., Waugh S., Raviprakash V., Sheriff N., Blakey V., Williams L.A., Moore J., Stonehouse S., Smith L., Davidson R.K., Bedford L., Coupland L., Wright V., Chappell J.G., Tsoleridis T., Ball J., Khakh M., Fleming V.M., Lister M.M., Howson-Wells H.C., Berry L., Boswell T., Joseph A., Willingham I., Duckworth N., Walsh S., Wise E., Moore N., Mori M., Cortes N., Kidd S., Williams R., Gifford L., Bicknell K., Wyllie S., Lloyd A., Impey R., Malone C.S., Cogger B.J., Levene N., Monaghan L., Keeley A.J., Partridge D.G., Raza M., Evans C., Johnson K., Betteridge E., Farr B.W., Goodwin S., Quail M.A., Scott C., Shirley L., Thurston S.A.J., Rajan D., Bronner I.F., Aigrain L., Redshaw N.M., Lensing S.V., McCarthy S., Makunin A., Balcazar C.E., Gallagher M.D., Williamson K.A., Stanton T.D., Michelsen M.L., Warwick-Dugdale J., Manley R., Farbos A., Harrison J.W., Sambles C.M., Studholme D.J., Lackenby A., Mbisa T., Platt S., Miah S., Bibby D., Manso C., Hubb J., Dabrera G., Ramsay M., Bradshaw D., Schaefer U., Groves N., Gallagher E., Lee D., Williams D., Ellaby N., Hartman H., Manesis N., Patel V., Ledesma J., Twohig K.A., Allara E., Pearson C., Cheng J.K.J., Bridgewater H.E., Frost L.R., Taylor-Joyce G., Brown P.E., Tong L., Broos A., Mair D., Nichols J., Carmichael S.N., Smollett K.L., Nomikou K., Aranday-Cortes E., Johnson N., Nickbakhsh S., Vamos E.E., Hughes M., Rainbow L., Eccles R., Nelson C., Whitehead M., Gregory R., Gemmell M., Wierzbicki C., Webster H.J., Fisher C.L., Signell A.W., Betancor G., Wilson H.D., Nebbia G., Flaviani F., Cerda A.C., Merrill T.V., Wilson R.E., Cotic M., Bayzid N., Thompson T., Acheson E., Rushton S., O’Brien S., Baker D.J., Rudder S., Aydin A., Sang F., Debebe J., Francois S., Vasylyeva T.I., Zamudio M.E., Gutierrez B., Marchbank A., Maksimovic J., Spellman K., McCluggage K., Morgan M., Beer R., Afifi S., Workman T., Fuller W., Bresner C., Angyal A., Green L.R., Parsons P.J., Tucker R.M., Brown R., Whiteley M., Bonfield J., Puethe C., Whitwham A., Liddle J., Rowe W., Siveroni I., Le-Viet T., Gaskin A., Johnson R., Abnizova I., Ali M., Allen L., Anderson R., Ariani C., Austin-Guest S., Bala S., Barrett J., Bassett A., Battleday K., Beal J., Beale M., Bellany S., Bellerby T., Bellis K., Berger D., Berriman M., Bevan P., Binley S., Bishop J., Blackburn K., Boughton N., Bowker S., Brendler-Spaeth T., Bronner I., Brooklyn T., Buddenborg S.K., Bush R., Caetano C., Cagan A., Carter N., Cartwright J., Monteiro T.C., Chapman L., Chillingworth T.J., Clapham P., Clark R., Clarke A., Clarke C., Cole D., Cook E., Coppola M., Cornell L., Cornwell C., Corton C., Crackett A., Cranage A., Craven H., Craw S., Crawford M., Cutts T., Dabrowska M., Davies M., Dawson J., Day C., Densem A., Dibling T., Dockree C., Dodd D., Dogga S., Dougan G., Dougherty M., Dove A., Drummond L., Dudek M., Durrant L., Easthope E., Eckert S., Ellis P., Farr B., Fenton M., Ferrero M., Flack N., Fordham H., Forsythe G., Francis M., Fraser A., Freeman A., Galvin A., Garcia-Casado M., Gedny A., Girgis S., Glover J., Gould O., Gray A., Gray E., Griffiths C., Gu Y., Guerin F., Hamilton W., Hanks H., Harrison E., Harrott A., Harry E., Harvison J., Heath P., Hernandez-Koutoucheva A., Hobbs R., Holland D., Holmes S., Hornett G., Hough N., Huckle L., Hughes-Hallet L., Hunter A., Inglis S., Iqbal S., Jackson A., Jackson D., Verdejo C.J., Jones M., Kallepally K., Kay K., Keatley J., Keith A., King A., Kitchin L., Kleanthous M., Klimekova M., Korlevic P., Krasheninnkova K., Lane G., Langford C., Laverack A., Law K., Lensing S., Lewis-Wade A., Liddle J., Lin Q., Lindsay S., Linsdell S., Long R., Lovell J., Lovell J., Mack J., Maddison M., Makunin A., Mamun I., Mansfield J., Marriott N., Martin M., Mayho M., McClintock J., McHugh S., MapcMinn L., Meadows C., Mobley E., Moll R., Morra M., Morrow L., Murie K., Nash S., Nathwani C., Naydenova P., Neaverson A., Nerou E., Nicholson J., Nimz T., Noell G.G., O’Meara S., Ohan V., Olney C., Ormond D., Oszlanczi A., Pang Y.F., Pardubska B., Park N., Parmar A., Patel G., Payne M., Peacock S., Petersen A., Plowman D., Preston T., Quail M., Rance R., Rawlings S., Redshaw N., Reynolds J., Reynolds M., Rice S., Richardson M., Roberts C., Robinson K., Robinson M., Robinson D., Rogers H., Rojo E.M., Roopra D., Rose M., Rudd L., Sadri R., Salmon N., Saul D., Schwach F., Seekings P., Simms A., Sinnott M., Sivadasan S., Siwek B., Sizer D., Skeldon K., Skelton J., Slater-Tunstill J., Sloper L., Smerdon N., Smith C., Smith C., Smith J., Smith K., Smith M., Smith S., Smith T., Sneade L., Soria C.D., Sousa C., Souster E., Sparkes A., Spencer-Chapman M., Squares J., Stanley R., Steed C., Stickland T., Still I., Stratton M., Strickland M., Swann A., Swiatkowska A., Sycamore N., Swift E., Symons E., Szluha S., Taluy E., Tao N., Taylor K., Taylor S., Thompson S., Thompson M., Thomson M., Thomson N., Thurston S., Toombs D., Topping B., Tovar-Corona J., Ungureanu D., Uphill J., Urbanova J., Van P.J., Vancollie V., Voak P., Walker D., Walker M., Waller M., Ward G., Weatherhogg C., Webb N., Wells A., Wells E., Westwood L., Whipp T., Whiteley T., Whitton G., Widaa S., Williams M., Wilson M., Wright S., Duncan L.M., Carabelli A.M., Kenyon J.C., Lever A.M., Marco A.De, Saliba C., Culap K., Cameroni E., Matheson N.J., Piccoli L., Corti D., James L.C., Robertson D.L., Bailey D., Gupta R.K. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35:109292. doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesaritakis C., Kapsalis A., Bogris A., Syvridis D. Artificial neuron based on integrated semiconductor quantum dot mode-locked lasers. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep39317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudumba S., de Alba S., Romero R., Cherwien C., Wu A., Wang J., Gleeson M.A., Iqbal M., Burlingame R.W. Photonic ring resonance is a versatile platform for performing multiplex immunoassays in real time. J. Immunol. Methods. 2017;448:34–43. doi: 10.1016/j.jim.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Netherton A., Tao Z., Zhang X., Chen R., Bai B., Qin J. Vol. 605. 2022. Microcomb-driven Silicon Photonic Systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo A., Gandhi P., Miller W.G. Frequency that laboratory tests influence medical decisions. J. Appl. Lab. Med. 2017;1:410–414. doi: 10.1373/jalm.2016.021634. [DOI] [PubMed] [Google Scholar]
- Novotny L., Hulst N.V. Antennas for light. Nat. Photonics. 2011;5:83–90. [Google Scholar]
- Oh S.H., Altug H., Jin X., Low T., Koester S.J., Ivanov A.P., Edel J.B., Avouris P., Strano M.S. Nanophotonic biosensors harnessing van der Waals materials. Nat. Commun. 2021;12:1–18. doi: 10.1038/s41467-021-23564-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owoicho O., Olwal C.O., Quaye O. Potential of laser-induced fluorescence-light detection and ranging for future stand-off virus surveillance. Microb. Biotechnol. 2021;14:126–135. doi: 10.1111/1751-7915.13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panpradist N., Kline E.C., Atkinson R.G., Roller M., Wang Q., Hull I.T., Kotnik J.H., Oreskovic A.K., Bennett C., Leon D., Lyon V., Gilligan-Steinberg S.D., Han P.D., Drain P.K., Starita L.M., Thompson M.J., Lutz B.R. Harmony COVID-19: a ready-to-use kit, low-cost detector, and smartphone app for point-of-care SARS-CoV-2 RNA detection. Sci. Adv. 2021;7:1–18. doi: 10.1126/sciadv.abj1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim H., Woo K., Kim J.M., Jo H.J., Jeong Y., Lee K.H. SARS-CoV-2 variant screening using a virus-receptor-based electrical biosensor. Nano Lett. 2022;22:50–57. doi: 10.1021/acs.nanolett.1c03108. [DOI] [PubMed] [Google Scholar]
- Payne T.D., Klawa S.J., Jian T., Kim S.H., Papanikolas M.J., Freeman R., Schultz Z.D. Catching COVID: engineering peptide-modified surface-enhanced raman spectroscopy sensors for SARS-CoV-2. ACS Sensors. 2021;6:3436–3444. doi: 10.1021/acssensors.1c01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C.A., Polage C.R., Quinn T.C., Ronald A.R., Sande M.A. Laboratory medicine in Africa: a barrier to effective health care. Clin. Infect. Dis. 2006;42:377–382. doi: 10.1086/499363. [DOI] [PubMed] [Google Scholar]
- Prasad P.N. John Wiley & Sons; 2004. Nanophotonics. [Google Scholar]
- Rahman M.R., Hossain M.A., Mozibullah M., Al Mujib F., Afrose A., Shahed-Al-Mahmud M., Apu M.A.I. CRISPR is a useful biological tool for detecting nucleic acid of SARS-CoV-2 in human clinical samples. Biomed. Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai P., Kumar B.K., Deekshit V.K., Karunasagar I., Karunasagar I. Detection technologies and recent developments in the diagnosis.Pdf. Appl. Microbiol. Biotechnol. 2021;105:441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A., Huyke D.A., Sharma E., Sahoo M.K., Huang C., Banaei N., Pinsky B.A., Santiago J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Li X., Zhang Q., Gu M. Applied physics: on-chip noninterference angular momentum multiplexing of broadband light. Science(80-.) 2016;352:805–809. doi: 10.1126/science.aaf1112. [DOI] [PubMed] [Google Scholar]
- Resende P. SARS-CoV-2 reinfection by the new variant of concern (VOC) P.1 in Amazonas, Brazil - SARS-CoV-2 coronavirus/nCoV-2019 genomic epidemiology - virological. Virological. 2021:1–9. https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596 [Google Scholar]
- Resende P.C., Bezerra J.F., de Vasconcelos R.H.Teixeira, Arantes I., Appolinario L., Mendonça A.C., Paixao A.C., Rodrigues A.C.Duarte, Silva T., Rocha A.S., Pauvolid-Corrêa A., Motta F.Couto, Teixeira D.L.F., de Oliveira Carneiro T.F., Neto F.P.Freire, Herbster I.D., Leite A.B., Riediger I.N., do Carmo Debur M., Naveca F.G., Almeida W., Livorati M., Bello G., Siqueira M.M. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020. Virological. 2021;10:1–8. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584 [Google Scholar]
- Ruiz-Hitzky E., Darder M., Wicklein B., Ruiz-Garcia C., Martín-Sampedro R., del Real G., Aranda P. Nanotechnology responses to COVID-19. Adv. Healthc. Mater. 2020;9:2000979. doi: 10.1002/adhm.202000979. [DOI] [PubMed] [Google Scholar]
- Ruiz-Vega G., Soler M., Lechuga L.M. Nanophotonic biosensors for point-of-care COVID-19 diagnostics and coronavirus surveillance. J.Phys. Photon. 2021;3 doi: 10.1088/2515-7647/abd4ee. [DOI] [Google Scholar]
- Ruiz-Vega G., Soler M., Estevez M.C., Ramirez-Priego P., Pazos M.D., Noriega M.A., Margolles Y., Francés-Gómez C., Geller R., Matusali G., Colavita F., di Caro A., Casasnovas J.M., Fernández L.A., Lechuga L.M. Rapid and direct quantification of the SARS-CoV-2 virus with an ultrasensitive nanobody-based photonic nanosensor. Sens. Diagn. 2022;1:983–993. doi: 10.1039/D2SD00082B. [DOI] [Google Scholar]
- Samarasekera U. India grapples with second wave of COVID-19. Lancet Microbe. 2021;2 doi: 10.1016/s2666-5247(21)00123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Bin Anwar T., Mulchandani A. Current status, advances, challenges and perspectives on biosensors for COVID-19 diagnosis in resource-limited settings. Sens. Actuators Rep. 2021;3 doi: 10.1016/j.snr.2021.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020;38:515–518. doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- Shrivastava S., Trung T.Q., Lee N.E. Recent progress, challenges, and prospects of fully integrated mobile and wearable point-of-care testing systems for self-testing. Chem. Soc. Rev. 2020;49:1812–1866. doi: 10.1039/c9cs00319c. [DOI] [PubMed] [Google Scholar]
- Sonu C.V. A Paradigm of Internet-of-Nano-Things Inspired Intelligent Plant Pathogen-Diagnostic Biosensors. ECS Sens. Plus. 2022;1 doi: 10.1149/2754-2726/ac92ed. [DOI] [Google Scholar]
- Taha B.A. Perspectives of photonics technology to diagnosis COVID–19 viruses: a short review. J. Appl. Sci. Nanotechnol. 2021;1:1–6. doi: 10.53293/jasn.2021.11016. [DOI] [Google Scholar]
- Taha B.A., Al Mashhadany Y., Mokhtar M.H.H., Bin Zan M.S.D., Arsad N., An analysis review of detection coronavirus disease (Covid-19) based on biosensor application. Sensors (Switzerland). 2019;20(2020):1–29. doi: 10.3390/s20236764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha B.A., Mashhadany Y.Al, Bachok N.N., Bakar A.Ashrif A., Mokhtar M.H.Hafiz, Zan M.S.Dzulkefly Bin, Arsad N. Detection of covid-19 virus on surfaces using photonics: challenges and perspectives. Diagnostics. 2021;11:1119. doi: 10.3390/diagnostics11061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha B.A., Mashhadany Y.Al, Al-Jumaily A.H.J., Zan M.S.D.Bin, Arsad N. Multidisciplinary Digital Publishing Institute; 2022. SARS-CoV-2 Morphometry Analysis and Prediction of Real Virus Levels Based on Full Recurrent Neural Network Using TEM Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talap J., Shen M., Yu L., Zeng S., Cai S. RT-LAMP assay combining multi-fluorescent probes for SARS-CoV-2 RNA detection and variant differentiation. Talanta. 2022;248 doi: 10.1016/j.talanta.2022.123644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallei T.E., Alhumaid S., AlMusa Z., Fatimawali D., Kusumawaty A., Alynbiawi A.N., Alshukairi A.A.Rabaan. Update on the omicron sub-variants BA.4 and BA.5. Rev. Med. Virol. 2022:1–7. doi: 10.1002/rmv.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021;82:e27–e28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Cui J., Kshirsagar A., Liu T., Yon M., Kuchipudi S.V., Guan W. SLIDE: saliva-based SARS-CoV-2 self-testing with RT-LAMP in a Mobile device. ACS Sensors. 2022;7:2370–2378. doi: 10.1021/acssensors.2c01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thye A.Y.K., Law J.W.F., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H. Emerging sars-cov-2 variants of concern (Vocs): an impending global crisis. Biomedicines. 2021;9:1–25. doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.D.T., de Araujo W.R., de Lima L.F., Ferreira A.L., de la Fuente-Nunez C. Low-cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter. 2021;4:2403–2416. doi: 10.1016/j.matt.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran M.A., Zhang C., Morin T.J., Chang L., Barik S., Yuan Z., Lee W., Kim G., Malik A., Zhang Z., Guo J., Wang H., Shen B., Wu L., Vahala K., Bowers J.E., Park H., Komljenovic T. Extending the spectrum of fully integrated photonics to submicrometre wavelengths. Nature. 2022;610:54–60. doi: 10.1038/s41586-022-05119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünlü S., Üsküdar-Güçlü A., Başustaoǧlu A. Predominant mutations of SARS-CoV-2: their geographical distribution and ootential consequences. Mediterr. J. Infect. Microbes Antimicrob. 2021;10 doi: 10.4274/mjima.galenos.2021.2021.15. [DOI] [Google Scholar]
- Vaisocherová H., Brynda E., Homola J. Functionalizable low-fouling coatings for label-free biosensing in complex biological media: advances and applications. Anal. Bioanal. Chem. 2015;407:3927–3953. doi: 10.1007/s00216-015-8606-5. [DOI] [PubMed] [Google Scholar]
- van Belkum A., Bachmann T.T., Lüdke G., Lisby J.G., Kahlmeter G., Mohess A., Becker K., Hays J.P., Woodford N., Mitsakakis K., Moran-Gilad J., Vila J., Peter H., Rex J.H., Dunne W.M. Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2019;17:51–62. doi: 10.1038/s41579-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitoria M., Granich R., Gilks C.F., Gunneberg C., Hosseini M., Were W., Raviglione M., De Cock K.M. The global fight against HIV/AIDS, tuberculosis, and malaria: current status and future perspectives. Am. J. Clin. Pathol. 2009;131:844–848. doi: 10.1309/AJCP5XHDB1PNAEYT. [DOI] [PubMed] [Google Scholar]
- Wang J., Sanchez M.M., Yin Y., Herzer R., Ma L., Schmidt O.G. Silicon-based integrated label-free optofluidic biosensors: latest advances and roadmap. Adv. Mater. Technol. 2020;5:1901138. doi: 10.1002/admt.201901138. [DOI] [Google Scholar]
- Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K., Deng R., Li W. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal. Chem. 2021;93:3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Li Q., Liang Z., Li T., Liu S., Cui Q., Nie J., Wu Q., Qu X., Huang W. The significant immune escape of pseudotyped SARS-CoV-2 variant omicron, emerg. Microbes Infect. 2022;11:1–5. doi: 10.1080/22221751.2021.2017757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G., McLellan J.S., Wilson I.A., Bowden T.A., Ward A.B., Crispin M. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Zhang C., Du X., Zhang Z. Research progress of biosensors for detection of SARS-CoV-2 variants based on ACE2. Talanta. 2023;251 doi: 10.1016/j.talanta.2022.123813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases, 2019. 2020. pp. 1–7. [Google Scholar]
- Wu M., Wu S., Wang G., Liu W., Chu L.T., Jiang T., Kwong H.K., Chow H.L., Li I.W.S., Chen T.H. Microfluidic particle dam for direct visualization of SARS-CoV-2 antibody levels in COVID-19 vaccinees. Sci. Adv. 2022;8:1–12. doi: 10.1126/sciadv.abn6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.M., Lee S.Y. Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 2016;34:7–25. doi: 10.1016/j.tibtech.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Yoo S.M., Kim D.K., Lee S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta. 2015;132:112–117. doi: 10.1016/j.talanta.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J., Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1–3. doi: 10.1056/nejmc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., Meyer B., Rudich Y., Li C., Marton I., Dym O., Elad N., Lewis M.G., Andersen H., Gagne M., Seder R.A., Douek D.C., Schreiber G. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- Zanchetta G., Lanfranco R., Giavazzi F., Bellini T., Buscaglia M. Emerging applications of label-free optical biosensors. Nanophotonics. 2017;6:627–645. doi: 10.1515/nanoph-2016-0158. [DOI] [Google Scholar]
- Zeng L., Liu Y., Nguyenla X.H., Abbott T.R., Han M., Zhu Y., Chemparathy A., Lin X., Chen X., Wang H., Rane D.A., Spatz J.M., Jain S., Rustagi A., Pinsky B., Zepeda A.E., Kadina A.P., Walker J.A., Holden K., Temperton N., Cochran J.R., Barron A.E., Connolly M.D., Blish C.A., Lewis D.B., Stanley S.A., La Russa M.F., Qi L.S. Broad-spectrum CRISPR-mediated inhibition of SARS-CoV-2 variants and endemic coronaviruses in vitro. Nat. Commun. 2022;13:1–16. doi: 10.1038/s41467-022-30546-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Malekjahani A., Udugama B.N., Kadhiresan P., Chen H., Osborne M., Franz M., Kucera M., Plenderleith S., Yip L., Bader G.D., Tran V., Gubbay J.B., McGeer A., Mubareka S., Chan W.C.W. Surveilling and tracking COVID-19 patients using a portable quantum dot smartphone device. Nano Lett. 2021;21:5209–5216. doi: 10.1021/acs.nanolett.1c01280. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhao W., Chen X., Zhang X., Zhu J., Li S., Wu C., Tian Z., Sui G. Fully automated CRISPR-LAMP platform for SARS-CoV-2 Delta and omicron variants. Anal. Chem. 2022;94:15472–15480. doi: 10.1021/acs.analchem.2c03607. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability is available on GISAID: (https://www.gisaid.org/) and (https://ourworldindata.org/).