Abstract
Breast cancer is one of the most common cancers. Oridonin, a traditional Chinese medicine, is believed to inhibit tumor growth, but its particular effects on breast cancer remain unknown. In this study, we examined oridonin's effects on 4T1, MCF-7, and MDAMB-231 cellular activity using CCK8. Scratch assays were used to detect oridonin's effects on cellular migration. Oridonin's effects on the breast cancer cell cycle were studied using flow cytometry, and expression of cell cycle related proteins p53, CDK2, and p21 was detected using Western blot assays. Metabolomics assays were used to detect changes in small molecule metabolites and metabolic pathways in breast cancer cells after treatment with oridonin. Oridonin's effects on breast cancer growth were also studied using xenograft mice. Metabolomics assays were used to detect changes in metabolites and metabolic pathways in xenograft mouse plasma in a control group, model group, and drug administration group. Experimental results showed that oridonin could significantly inhibit breast cancer growth both in vivo and in vitro. Scratch experiments showed that oridonin could inhibit breast cancer cell migration. Oridonin was also able to arrest cells in S phase by affecting several cell cycle-related proteins, including p53, CDK2, and p21. Metabolomic analysis of 4T1 cells identified a total of 33 differential metabolites, including multiple amino acids (such as l-Glutamic acid, l-Asparagine, l-Histidine, l-Valine, and l-Isoleucine). KEGG pathway enrichment analysis showed significant changes in aminoacyl-tRNA biosynthesis, and in multiple amino acid metabolic pathways. Plasma metabolomic analyses of xenograft mice revealed 28 differentially-expressed metabolites between the different animal model groups, including multiple amino acids. KEGG pathway analysis showed significant alterations in multiple amino acid metabolic pathways in oridonin-treated mice. Additionally, changes in the expression of PI3K, AKT and mTOR proteins, as well as in branched amino acids, suggest that oridonin affects the PI3K/AKT/mTOR signaling pathway by inhibiting the biosynthesis of valine, leucine and isoleucine. Taken together, our results suggest that oridonin has strong anti-tumor activity in vitro and in vivo, and has potential as an adjuvant to breast cancer treatment regimens.
Keywords: Oridonin, Breast cancer, Metabolomics, Amino acid metabolism, PI3K/AKT/mTOR signaling pathway
1. Introduction
Breast cancer is one of the most common malignancies amongst women, has a high prevalence rate in low- and middle-income countries [1], and is highly heterogeneous in nature, thus requiring different treatment standards. Breast cancer has different stages, classifications, and types, but triple-negative breast cancer is the most dangerous, and tends to have poor prognoses and be difficult to treat. These patients have cancer which is negative for progesterone receptors (PR), estrogen receptors (ER), and ERBB2 receptors [2]. Current treatments for breast cancer include surgery, radiotherapy, and biological therapy. Systemic endocrine or immunotherapy is usually given at the preoperative stage if a tumor expresses ER, PR, or ERBB2 receptors [3]. Preoperative chemotherapy can be used when the tumor does not express any of these receptors. Although radiotherapy and chemotherapy can significantly improve breast cancer survival rates, they can also have serious adverse effects, including severe liver damage, renal damage, and gastrointestinal tract damage. In severe cases, intestinal homeostasis may also be affected, potentially leading to intestinal inflammation and tumors [4]. Therefore, looking for safe, efficient, and less toxic natural products when developing new therapeutic agents would help vastly improve quality of life.
Oridonin is a biologically active natural substance that is isolated from Isodon rubescens (Hemls.) Hara. It mainly consists of kaureane type tetracyclic diterpenoid natural organic compounds, and has been shown to have high anti-tumor activity. As a traditional Chinese medicine, Isodon rubescens (Hemls.) Hara has been used to treat esophageal cancer in China for more than 50 years [5]. It has been shown to have strong therapeutic effects on more than 20 different kinds of cancer, including esophageal cancer, liver cancer, oral cancer, and lung cancer [[6], [7], [8], [9]]. Oridonin has been reported to inhibit breast cancer growth by promoting apoptosis, inducing autophagy, and inhibiting angiogenesis [10,11]. However, the underlying anti-tumor mechanisms are still not fully understood.
Cancer development and progression are both closely related to the metabolic reprogramming of cells [12]. Small molecular metabolites have received a great deal of research attention recently because they uniquely reflect biological change processes such as cell differentiation and T cell survival [13]. Several research groups have harnessed metabolomics technology to explore the mechanism of action of anti-cancer drugs using small molecular metabolites. Building off this work, we aimed to use metabolomics technology to explore the anti-breast cancer mechanisms of oridonin.
Thus, here, we first determined oridonin's inhibitory effects on breast cancer in vitro and in vivo and next explored its anti-tumor mechanisms in breast cancer using metabolomics analysis.
2. Materials and methods
2.1. Chemicals and materials
Oridonin (purity 98%) was purchased from Shanghai Aladdin Biochemical Technology Co., (Shanghai, China) (Lot Number:L2103140). CCK8 kits were obtained from BOSTER Biological Technology Co., Ltd (Wuhan, China). Cell cycle kits, RIPA lysates, BCA protein quantitative detection kits and protein antibodies were all purchased from Servicebio (Wuhan, China). Primary antibodies for β-actin, p53, p-p53, CDK2, and mTOR were all purchased from Servicebio, AKT was from RuiYingBio (Suzhou, China), PI3K was from BIOSS (Beijing, China), and p21 was from proteintech (Wuhan, China). All secondary antibodies were procured from Servicebio (Wuhan, China). The UPLC-MS/MS reagents, including methanol, formic acid, acetonitrile, and ammonium bicarbonate, were purchased from ThermoFisher Scientific (Fair Lawn, NJ, USA). The internal standard materials with mixed isotope labels were purchased from the Dalian Institute of Chemical Physics, Chinese Academy of Sciences (Dalian, China).
2.2. Animal handling and experimental design
27 female BALB/c mice (mean body weight: 24 ± 3 g) were purchased from Hubei Bainte Biotechnology Co., Ltd. (Hubei, China; Rodent license SCXK2021-0027). The mice were kept under standard environmental conditions, including a 12 h/12 h light/dark cycle, temperature 22 ± 2 °C, and humidity 50 ± 10%. The animal study followed National Institutes of Health (NIH) guidelines and was approved by the Fifth Affiliated Hospital of Zhengzhou University Medical Ethics Committee; Ethics Review Number: KY2021036. The experiments were also carried out in accordance with established animal welfare guidelines. Throughout the experiment, we tried our best to minimize pain and suffering to the animals. All the mice were randomly divided into three groups: 1) a total of seven mice served as a normal control group, with daily 200 μl intraperitoneal saline injections; 2) 10 xenograft mice served as a xenograft control group, with daily 200 μl intraperitoneal saline injections; and 3) 10 xenograft mice served as an experimental group, and were treated with daily intraperitoneal 15 mg/kg oridonin injections. All subsequent interventions were given every 12 h for two weeks following 4T1 cell implantation, and then blood samples were taken from the eyeballs of each mouse. The tumors were isolated, weighed, and measured after all mice were sacrificed.
2.3. Cell culture
The 4T1 cells,MCF-7 cells, and MDAMB-231 cells were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China) and cultured in a RPMI-1640 medium (Sigma-Aldrich, USA) at 37 °C in a humidified incubator supplied with 5% CO2.
2.4. Cell proliferation assay
The 4T1 cells,MCF-7 cells, and MDAMB-231 cells were seeded in 96-well plates with a density of 8 × 103 cells per well. After 12 h of incubation, the cells were treated with a series of oridonin concentrations (0.5,1,2,3,4,6,8,10,16 μg/ml) for 24 h, 48 h or 72 h. After oridonin treatment, CCK8 solution (10 μL) was added to each well, and plates were incubated in the cell incubator for 1 h. After homogenization, the culture plate was examined at 450 nm for absorbance detection, and IC50 values were calculated by using GraphPad Prism 9 software.
2.5. Cell migration analysis
4T1 cells were seeded in a 6-well plate marked with a marker pen. After the cells were covered with a monolayer, a 10 μl pipette tip was used to make cell scratches, and the medium was aspirated, gently washed 3 times with PBS, and replaced with a serum-free medium (control group and oridonin treat group). Next, cells were cultured in a 37 °C, 5% CO2 incubator and photographed regularly (0 h, 12 h and 24 h).
2.6. Cell cycle analysis
The 4T1 cells inoculated in the 6-well plate were divided into a control group and an oridonin-treated group (at both 1 μg/ml and 2 μg/ml concentrations). After the cells adhered, the old medium was discarded and replaced with new blank medium or medium containing oridonin, and the cells were placed in a 37 °C, 5% CO2 incubator for 24 h. Cells were collected and washed with PBS and placed in 70% ethanol overnight at 4 °C. RNase digestion and PI staining were performed in the dark, and flow cytometry was performed after incubation for 30 min. Modifit software was used to analyze the results.
2.7. Western blot analysis
Proteins were extracted and quantified with a BCA kit. Proteins were denatured in a boiling water bath and stored for later use. Protein separation was performed using 10% SDS-PAGE gel electrophoresis, and the target protein was transferred from the gel to a PVDF membrane. The transferred membrane was placed in an incubation tank with TBST and rinsed once quickly. Next, it was saturated with skim milk, placed on a destaining shaker, and blocked for 30 min at room temperature. After TBST washes, the membrane was incubated with special primary antibodies overnight at 4 °C. After another TBST wash, the membrane was incubated with secondary antibodies, and then developed and fixed in the dark room.
2.8. Metabolomics experiment
2.8.1. Preparation of animal serum samples
100 μl serum samples were isolated from all mice after two weeks of treatment with oridonin, and added to 400 μL methanol/water (6:1, vol/vol), then vortexed and placed in the centrifuge at 12,000 rpm for 15 min in 4 °C. Next, 300 μl of the supernatant was taken and dried using a vacuum concentrator. In the sample reconstitution stage, each sample was reconstituted with 100 μL of a methanol solution, and then the supernatants were separated and passed through a 0.22 μm filter membrane for analysis.
2.8.2. Preparation of cell samples
The cells were digested using trypsin in their logarithmic growth phase, and then ∼5 × 105 cells were platted into a 7 cm plate. 24 h later, cells were treated with oridonin. Six replicates were performed while the control group was treated with new culture medium. After 24 h of treatment, the cells were collected for metabolomic analysis as previously reported [14]. In brief, the cells were collected using a cell scraper, and intracellular metabolites were extracted with methanol-water (4: 1, vol/vol) and chloroform.
2.8.3. Compound identification and metabolic pathway analysis
To analyze the metabolomics data obtained from LC-MS/MS, we first imported the original file (.raw) into OSI-SMMS software. Qualitative and quantitative metabolites were obtained by spectral processing and database retrieval. Next, to reveal between-group differences, we used simca-13.0 software to conduct multivariate statistical analysis of the metabolites. Differential compounds fulfilling VIP>1, P < 0.05, were screened out and confirmed through functional enrichment analysis in the KEGG and HMDB databases, and major metabolic pathways were then sorted out using the Metaboanalyst website (https://www.metaboanalyst.ca).
2.9. Statistical analysis
All data, which were obtained from three independent experiments, were expressed as mean ± standard deviations (SD). One-way analyses of variance (ANOVAs) were used to assess the differences between the two groups. Two-way ANOVA tests and Tukey's post-hoc analyses were used compare multiple groups. SPSS 22.0 or GraphPad Prism 9 software was used for statistical analysis, with P values < 0.05 were considered significant.
3. Results
3.1. Oridonin significantly inhibits the growth of breast cancer cells
To explore oridonin's effects on the proliferation of breast cancer cells, three breast cancer cell lines were selected, and cell viability was determined using CCK8 assays. Results showed that the viability of 4T1, MCF-7 and MDAMB-231 cells decreased in a time - and dose-dependent manner (Fig. 1(A-C)). GraphPad Prism software showed that the IC50 values of oridonin on 4T1 cells for 24 h, 48 h and 72 h were 3.53, 1.66 and 0.95 μg/ml, respectively, and the IC50 values of oridonin on MCF-7 cells were 8.38, 3.48 and 2.50 μg/ml, respectively, and the IC50 values of oridonin on MDAMB-231 cells, were 4.55, 1.14, and 0.35 μg/ml, respectively.
Fig. 1.
Oridonin's effects on the proliferation of 4T1,MCF-7 and MDAMB-231 cells. A. 4T1 cell survival rates. B. MCF-7 cell survival rates. C. MDAMB-231 cell survival rates.
3.2. Oridonin can inhibit 4T1 cell migration
Oridonin's effects on 4T1 cellular migration were analyzed using scratch experiments. The results showed that the cells in the control group had strong migration abilities, while the migration abilities of 4T1 cells were significantly inhibited after oridonin treatment (Fig. 2A and B).
Fig. 2.
A. 4T1 cell migration assay results after oridonin treatment. B. Wound area of cells at different concentrations and times. (ns, no significant difference, *P < 0.05, ***P < 0.001, ****P < 0.0001).
3.3. Oridonin affects the 4T1 cell cycle by interfering with the expression of cell cycle-related proteins
We examined the cell cycle to better understand why 4T1 cells proliferation was inhibited by oridonin. Cells were divided into a control group and an oridonin interference group (with treatment concentrations of both 1 μg/ml and 2 μg/ml). After oridonin treatment, cells were blocked in S phase (Fig. 3A, B). To further examine the etiology of the cell cycle arrest, we examined p53, p-p53, p21 and CDK2 expression levels with western blots. Results showed that, compared with the control group, the expression of the cell cycle-related proteins p53, p-p53 and p21 increased with increases in drug concentration, while the expression of CDK2 proteins gradually decreased in a dose-dependent manner (Fig. 3C). These findings suggest that oridonin may arrest 4T1 cells in S phase by regulating the expression of cyclins, thereby effectively inhibiting cell proliferation (Fig. 3D).
Fig. 3.
Effects of oridonin on the cell cycle and cycle-related proteins of 4T1 cells. A and B. Graphical representation of the cell cycle distribution of 4T1 cells after treatment with oridonin. C. Western blot analysis of the expression of the S phase arrest-related proteins p53, p-p53, p21 and CDK2. Relative expression levels of p53, p-p53, p21 and CDK2 proteins (ns, no significant difference,**P < 0.01,***P < 0.001****P < 0.0001). D. Schematic diagram of oridonin-induced cell cycle arrest in S phase.
3.4. Oridonin can effectively inhibit the progression of breast cancer tumors in mice
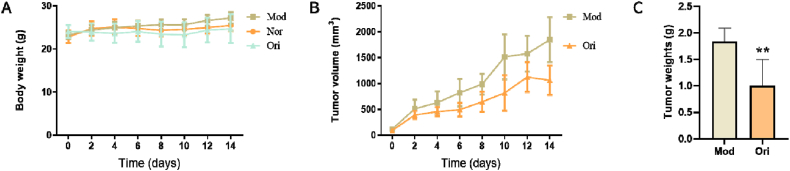
To determine the therapeutic effects of oridonin in vivo, xenograft models were created by injecting ∼6 × 105 breast cancer (4T1) cells subcutaneously into the right axilla of mice. After successfully establishing the model, we recorded the body weight and tumor volume of the mice every two days. Treatment was started after 12 h of cellular implantation, and mice in the oridonin group had significantly reduced overall body weight (Fig. 4A). Similarly, the overall tumor volume and weight in the oridonin group was significantly decreased compared to that in the model group (Fig. 4B,C).
Fig. 4.
Tumor growth curves in xenograft mice (Nor-normal group, Mod-model group, Ori-oridonin group). A. Changes in body weights of mice. Over time, mouse body weight gradually increased, but the average body weight of the oridonin group was lower than that of the model group (Mod vs. Ori group. P = 0.0087). B. Mouse tumor volume also gradually increased in both groups, but the overall growth rate was significantly lower in the oridonin group (Mod vs. Ori. P = 0.0065). C, Tumor quality after two weeks of was significantly lower in the oridonin group than in the untreated mice (Mod vs. Ori group. **P < 0.01).
3.5. Effects of oridonin on the metabolic profiles of 4T1 cells and xenograft mice
To analyze the effects of oridonin on cellular metabolism, 4T1 cells, and the xenograft mice were treated with oridonin, and subsequent metabonomic analyses were performed. The 4T1 cells were randomly divided into a control group and an oridonin (2 μg/ml) treatment group, and the xenograft mice were divided into a normal group, a model group, and an oridonin treatment group. UPLC-MS/MS was used to detect and analyze endogenous metabolites collected from 4T1 cells. Similarly, serum from xenograft mice was collected and subjected to metabolomic analysis. All samples were dispersed in an OPLS-DA fractional plot. Cellular populations in different groups showed good separation trends in both positive and negative ion mode (Figs. S1A and B). Permutation testing verified that the model made sense and that there was no overfitting (Figs. S2A and B).
The metabolomic analysis showed that the levels of 33 endogenous metabolites in 4T1 cells were significantly changed after oridonin treatment (Fig. 5A). We also performed functional enrichment of the differentially-expressed metabolites using metaboanalyst, and identified the pathways affected by oridonin (Fig. 5C). Specifically, aminoacyl-tRNA biosynthesis was the top affected pathway, and seven out of the 48 differentially expressed metabolites were found to regulate this pathway. Those seven metabolites included l-Asparagine, l-Histidine, l-Valine, l-Isoleucine, L-Tryptophan, l -Tyrosine, and l-Glutamic acid. In addition, two of the six compounds involved in the d-Glutamine and d-glutamate metabolism pathways were identified.
Fig. 5.
Results of metabolomic analyses in cells and plasma of mice treated with oridonin. A. Cluster plot of differentially expressed metabolites in 4T1 cells. Yellow represents an elevated level of metabolites, and blue represents decreased levels of metabolites. (Con-control group, Ori- oridonin group) B. Cluster plot of differentially expressed metabolites in mice serum. Yellow represents increased metabolites, and blue represents decreased metabolites (Nor-normal group, Mod-model group, Ori-oridonin group). C. Enrichment of metabolites in various metabolic pathways in 4T1 cells. The darker the color, the smaller the P-value, and the larger the circle, the greater the number of differentially expressed metabolites. D. Overview of the enriched metabolome in mice serum.
Consistent with the metabolomics data derived from our 4T1 cell lines, analysis in xenograft mice identified 28 differentially expressed metabolites that were affected after oridonin treatment (Fig. 5B). Pathway enrichment analysis of these metabolites showed that a total of 14 metabolites were associated with arginine biosynthesis, three of which (l-Glutamic acid, ornithine, and N-Acetylglutamic acid) were detected. In addition, two of the six compounds were involved in the d-Glutamine and d-glutamate metabolism pathways, and the biosynthesis of unsaturated fatty acids was regulated by 3/36 metabolites (Fig. 5D).
3.6. Oridonin can inhibit valine, leucine, and isoleucine biosynthesis, and regulate the mTOR signaling pathway
Our metabolomic analyses showed that the aminoacyl-tRNA biosynthesis pathway was strongly inhibited by oridonin, and that a variety of amino acids were disturbed following oridonin treatment. We noticed that the content of several intracellular branched-chain amino acids (BCAAs) including valine, leucine and isoleucine, was decreased after oridonin treatment (Fig. 6A). BCAAs are essential amino acids for the human body, and tumor tissues acquire BCAAs from the surrounding tissues or blood circulation [15]. Thus, elevated plasma levels of BCAAs have been associated with the development of cancer [16]. Branched-chain amino acid transaminase 1 (BCAT1), which catalyzes the catabolism of essential BCAAs such as leucine, isoleucine, and valine, plays an important role in amino acid metabolism. It can convert BCAAs into their corresponding branched-chain α-keto acids and then into α-ketoglutarate by transferring amino acids to produce glutamic acid, which can also further promote the growth of breast cancer cells [17]. These branched-chain keto acids can be further oxidized to form acetyl-CoA and/or succinyl-CoA to fuel the TCA cycle or to contribute substrates to fatty acid synthesis [18]. After oridonin treatment, we found that intracellular BCAT1 expression was significantly decreased (Fig. 6B), which may have reduced intracellular nutrient and energy production. Because amino acids, including BCAAs, can also serve as upstream regulators of mTOR activity [19], we also examined the proteins involved in the mTOR signaling pathway. Western blot analysis also showed that mTOR, PI3K and AKT expression was significantly reduced after oridonin treatment (Fig. 6C).
Fig. 6.
Effects of oridonin on branched-chain amino acid metabolism and the mTOR pathway. A. Changes in l-Isoleucine, l-Valine, and Leucine in 4T1 cells after oridonin treatment. B. Western blot analysis of the expression of BCAT1, and relative expression levels of BCAT1 (**P < 0.01 versus the control group). C. Western blot analysis of mTOR, PI3K and AKT expression, and the relative expression levels of mTOR, PI3K and AKT proteins (**P < 0.01,****P < 0.0001).
4. Discussion
As one of the deadliest cancers in women, breast cancer can affect mental health and appearance, and can be life-threatening in severe cases. Natural products have been receiving increasing attention as cancer therapeutics because they have fewer toxic effects [20]. Oridonin, one of the natural tetracyclic diterpenoid compounds, has already been demonstrated to have strong anti-cancer effects [21]. In this study, we found that the traditional Chinese medicine compound oridonin could inhibit the growth of a variety of breast cancer cells. We also conducted further studies to understand the mechanism by which oridonin inhibited breast cancer growth. As cells divide and multiply, they need to go through the cell cycle. We found that oridonin could block the cell cycle of 4T1 cells in the S phase. P53, a multifunctional protein, is activated when there is DNA damage in cells. DNA damage can lead to aberrant phosphorylation of a variety of proteins, including p53, which then can accumulate in cells. As p53 and p-p53 levels rise, p21 production is triggered, which can then inhibit CDK2 activity. CDK2 regulates the transition of cells from G1 phase to S phase. Our data suggest that oridonin can arrest cells in S phase by inhibiting the expression of the CDK2 protein, which is closely related to the up-regulation of p21. Our results also confirm that oridonin increases p21 expression. In addition, oridonin can activate p53 and p-p53, suggesting that it arrests cells in S phase via a p53-mediated p21-CDK2 axis (Fig. 3D).
Analyzing small molecular changes, especially at the metabolic level, may help reveal better therapeutic targets for breast cancer. Based on these findings, we used metabolomics technology to study the inhibitory effects and mechanism of action of oridonin on breast cancer cells and tumors in xenograft mice at the level of endogenous small molecule metabolites. We also explored the mechanisms underlying oridonin's anticancer effects via metabolomics and found that oridonin treatment affected a variety of cellular amino acids, including l-Asparagine, l-Histidine, l-Valine, and l-Isoleucine. Pathway enrichment analysis of the differentially expressed metabolites showed that oridonin had a significant effect on the aminoacyl-tRNA biosynthesis and amino acid metabolism which ultimately inhibited breast cancer. However, this process requires further study. We also found that the levels of some BCAAs, including l-Isoleucine, l-Valine, and Leucine, were decreased after oridonin treatment, and metabolic pathway analysis suggested that valine, leucine, and isoleucine biosynthesis were all greatly affected. BCAT1 is highly expressed in various types of breast cancer [22], and can convert BCAAs into corresponding branched-chain keto acids, which can be further oxidized to form acetyl-CoA and succinyl-CoA, thus fueling the TCA cycle or to contributing substrates for fatty acid synthesis. Therefore, we next tested the enzyme BCAT1, which is involved in amino acid metabolism, and found that oridonin also significantly reduced its cellular expression. These results suggested that oridonin could inhibit the growth of breast cancer by inhibiting the biosynthesis of valine, leucine, and isoleucine. Inhibition of this metabolic pathway and reduced expression of BCAT1 may indirectly lead to reduced fuel for the TCA cycle and fatty acid biosynthesis (Fig. 7). However, this process still needs further study.
Fig. 7.
Mechanistic diagram of oridonin's effects on breast cancer cells.
Prior studies have shown that BCAA-driven metabolic reprogramming can generate intermediates and pathways that reconnect other pathways and change several metabolic factors, such as the mTOR signaling pathway, α-KG levels, and glutamate levels [[23], [24], [25], [26]], which can alter mitochondrial function and gene expression and promote cancer cell proliferation. The KEGG PATHWAY Database (https://www.kegg.jp) suggests that amino acids can regulate mTOR through the transformation of multiple regulatory factors. Therefore, we studied the expression of mTOR using western blots. As shown in the results, the expression of mTOR decreased after oridonin treatment. As one of the most common pathways in human cancer, the PI3K/AKT/mTOR signaling pathway plays an important role in cellular activity, and has been widely studied as an anticancer target. Thus, we next examined the levels of the important proteins in the classic PI3K/AKT/mTOR signaling pathway, and confirmed that oridonin could downregulate their expression, thereby inhibiting the modified signaling pathway (Fig. 7). These findings also indicate that oridonin may inhibit breast cancer growth by interfering with the PI3K/AKT/mTOR signaling pathway.
5. Conclusion
In summary, oridonin is a very promising antitumor drug with strong inhibitory effects on breast cancer in vitro and in vivo. Oridonin can inhibit cellular division by blocking the S-phase of the cell cycle. In addition, it can inhibit various amino acid metabolic pathways and affect the PI3K/AKT/mTOR signaling pathway. However, the connection between changes in metabolic pathways and changes in signaling pathways still needs further study. Although the effects of oridonin on macromolecular proteins remain unknown, the results of this study suggest that it may be a safe and reasonable addition to breast cancer treatment regimens.
Author contribution statement
Weijie Zhang: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Youcai Tang: Conceived and designed the experiments.
Xia Xu, Lei Shi, Wei Zhou: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yuan Xi, Xin Liu, Ya Li, Xinying Wang: Conceived and designed the experiments; Wrote the paper.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 31471330), National Key R&D Program of China (2020YFC2009006), the Henan Provincial Key R&D and Promotion Special Project (212102310033), the Zhengzhou University Discipline Key Special Project (XKZDQY202001), and the Joint construction project of Henan Provincial Health Commission (LHGJ20210494).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18046.
Contributor Information
Xia Xu, Email: xuxia@zzu.edu.cn.
Youcai Tang, Email: tangyoucai@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Khan S., et al. Stem cell therapy: a paradigm shift in breast cancer treatment. World J. Stem Cell. 2021;13(7):841–860. doi: 10.4252/wjsc.v13.i7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar P., Aggarwal R. An overview of triple-negative breast cancer. Archives of gynecology and obstetrics. Arch. Gynecol. Obstet. 2016;293(2):247–269. doi: 10.1007/s00404-015-3859-y. [DOI] [PubMed] [Google Scholar]
- 3.Maughan K.L., et al. Treatment of breast cancer. American family physician. Am. Fam. Physician. 2010;81(11):1339–1346. [PubMed] [Google Scholar]
- 4.Medema J.P., Vermeulen L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature. 2011;474(7351):318–326. doi: 10.1038/nature10212. [DOI] [PubMed] [Google Scholar]
- 5.Xiong H. Study on modern pharmacological and chemical components and clinical medication of Isodon rubescens. Inner Mongolia traditional. Chin. Med. 2014;33(22):2. [Google Scholar]
- 6.Xu L., et al. Oridonin inhibits the migration and epithelial-to-mesenchymal transition of small cell lung cancer cells by suppressing FAK-ERK1/2 signalling pathway. J. Cell Mol. Med. 2020;24(8):4480–4493. doi: 10.1111/jcmm.15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., et al. Oridonin inhibits oral cancer growth and PI3K/Akt signaling pathway. Biomed. Pharmacother. 2018;100:226–232. doi: 10.1016/j.biopha.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J.H., et al. Oridonin-induced mitochondria-dependent apoptosis in esophageal cancer cells by inhibiting PI3K/AKT/mTOR and Ras/Raf pathways. J. Cell. Biochem. 2019;120(3):3736–3746. doi: 10.1002/jcb.27654. [DOI] [PubMed] [Google Scholar]
- 9.Luo D., et al. Oridonin derivatives as potential anticancer drug candidates triggering apoptosis through mitochondrial pathway in the liver cancer cells. Eur. J. Med. Chem. 2019;178:365–379. doi: 10.1016/j.ejmech.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., et al. Oridonin phosphate-induced autophagy effectively enhances cell apoptosis of human breast cancer cells. Med. Oncol. 2015;32(1):365. doi: 10.1007/s12032-014-0365-1. [DOI] [PubMed] [Google Scholar]
- 11.Li J., et al. Oridonin synergistically enhances the anti-tumor efficacy of doxorubicin against aggressive breast cancer via pro-apoptotic and anti-angiogenic effects. Pharmacol. Res. 2019;146 doi: 10.1016/j.phrs.2019.104313. [DOI] [PubMed] [Google Scholar]
- 12.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guijas C., et al. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018;36(4):316–320. doi: 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., et al. Oridonin induces ferroptosis by inhibiting gamma-glutamyl cycle in TE1 cells. Phytother Res. 2021;35(1):494–503. doi: 10.1002/ptr.6829. [DOI] [PubMed] [Google Scholar]
- 15.Neinast M., et al. Branched chain amino acids. Annual review of physiology. Annu. Rev. Physiol. 2019;81:139–164. doi: 10.1146/annurev-physiol-020518-114455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayers J.R., et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014;20(10):1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Han J. Branched-chain amino acid transaminase 1 (BCAT1) promotes the growth of breast cancer cells through improving mTOR-mediated mitochondrial biogenesis and function. Biochem. Biophys. Res. Commun. 2017;486(2):224–231. doi: 10.1016/j.bbrc.2017.02.101. [DOI] [PubMed] [Google Scholar]
- 18.Mossmann D., et al. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer. 2018;18(12):744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 19.Jung M.K., et al. Role of branched-chain amino acid metabolism in tumor development and progression. Journal of cancer prevention, J Cancer Prev. 2021;26(4):237–243. doi: 10.15430/JCP.2021.26.4.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi C., et al. Natural products targeting cancer cell dependency. J. Antibiot. 2021;74(10):677–686. doi: 10.1038/s41429-021-00438-x. [DOI] [PubMed] [Google Scholar]
- 21.Liu X., et al. Oridonin and its derivatives for cancer treatment and overcoming therapeutic resistance. Genes Dis. 2020;8(4):448–462. doi: 10.1016/j.gendis.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ananieva E.A., Wilkinson A.C. Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care. 2018;21(1):64–70. doi: 10.1097/MCO.0000000000000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin S.B., et al. Leucine and branched-chain amino acid metabolism contribute to the growth of bone sarcomas by regulating AMPK and mTORC1 signaling. Biochem. J. 2020;477(9):1579–1599. doi: 10.1042/BCJ20190754. [DOI] [PubMed] [Google Scholar]
- 24.Ericksen R.E., et al. Loss of BCAA catabolism during carcinogenesis enhances mTORC1 activity and promotes tumor development and progression. Cell Metabol. 2019;29(5):1151–1165. doi: 10.1016/j.cmet.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raffel S., et al. BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature. 2017;551(7680):384–388. doi: 10.1038/nature24294. [DOI] [PubMed] [Google Scholar]
- 26.Tönjes M., et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat. Med. 2013;19(7):901–908. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.