Abstract
Background
“Primary papillary epithelial tumor of the sella (PPETS)” is a recently described rare tumor entity of the central nervous system (CNS) with stereotypic location in the sella. Comprehensive molecular investigations and epigenetic profiles of PPETS have not been performed to date.
Methods
We report a comprehensive clinical, histopathologic, and molecular assessment of 5 PPETS cases in comparison with a cohort composed of 7 choroid plexus papilloma (CPP), 7 central neurocytoma (CN), 15 posterior pituitary tumor (PPT) including 4 pituicytoma, 6 granular cell tumors of the sellar region (GCT), and 5 spindle cell oncocytoma.
Results
All PPETS had good outcomes. Immunohistochemically, PPETS tumors showed positive staining with TTF1, EMA, AE1/AE3, MAP2, and Vimentin, but were negatively stained with Syn, GFAP, CgA, and S100, and sporadically stained with Ki-67. In unsupervised hierarchical clustering and t-distributed stochastic neighbor embedding analyses of DNA-methylation data, PPETS and PPT tumors formed a distinct cluster irrespective of their histologic types. However, PPETS tumors did not cluster together with CPP and CN samples. Similar findings were obtained when our samples were projected into the reference cohort of the brain tumor classifier. Substantial fractions of the PPETS and PPT tumors shared broadly similar chromosomal copy number alterations. No mutations were detected using targeted next-generation sequencing.
Conclusions
Though more cases are needed to further elucidate the molecular pathogenesis of these tumors, our findings indicate that PPETS and PPT tumors may constitute a single neurooncological entity.
Keywords: DNA methylation profile, molecular classification, primary papillary epithelial tumor, sella, TTF1
Key Points.
Primary papillary epithelial tumor of the sella and posterior pituitary tumor share similar epigenetic and genetic features.
Primary papillary epithelial tumor of the sella and posterior pituitary tumor may constitute a single neurooncological entity.
Importance of the Study.
Primary papillary epithelial tumor of the sella (PPETS) is a type of rare tumor with obscure molecular genetic background. Comprehensive molecular characterizations of PPETS have not been reported so far. DNA methylation–based molecular classification of human central nervous system tumors has proven to be a reliable tool because individual tumor types may maintain an epigenetic “memory” of their unique cell of origin. Besides comprehensive immunohistochemical assessment and targeted next-generation sequencing, we here performed DNA methylation profiling of 3 PPETS in comparison with 7 choroid plexus papilloma, 7 central neurocytoma, and 15 PPT tumors using Infinium EPIC DNA methylation array. Our findings demonstrate that PPETS and PPT tumors show highly similar epigenetic profile. Further, PPETS and PPT tumors share similar chromosomal losses. Thus, PPETS and PPT tumors may constitute a single neurooncological entity.
“Primary papillary epithelial tumor of the sella (PPETS)” is a recently proposed tumor entity with stereotypic location in the sella.1 These tumors show histological features reminiscent of choroid plexus papilloma (CPP) and have previously been regarded as sellar ectopic CPP.2–5 Due to its rarity, PPETS is currently not described in the World Health Organization (WHO) classification of pituitary tumors.6,7 Their underlying driving molecular alterations are still unknown. It was hypothesized that PPETS, like posterior pituitary tumors (PPT), are also derived from pituicytes because of the intense and ubiquitous expression of TTF-1 in PPETS.1 The PPT, mainly including pituicytoma (PC), granular cell tumors of the sellar region (GCT), and spindle cell oncocytoma (SCO), are rare and account for 1% of primary sellar tumors.8 It is widely believed that all these three types of tumors originate from pituicytes.9,10 They are considered as one spectrum of a single nosological entity in the 2021 WHO classification of tumors of the central nervous system (CNS) and are assigned as WHO grade I neoplasms.7 This is substantiated by the nuclear expression of TTF-1, a robust marker expressed in PC, GCT, and SCO of the sellar area and in normal pituicytes.11–13 TTF-1 is a 38 kDa nuclear DNA-binding protein (as also known as Nkx2.1), which is also expressed in subependymal giant cell astrocytoma (SEGA), chordoid gliomas of the third ventricle, central neurocytoma (CN) in CNS.14–16 Interestingly, lung adenocarcinoma (LUAD) and papillary thyroid carcinoma (PTC) have also been shown to express TTF-1,17 brain metastasis of these tumors may confound the diagnosis. Though these CNS tumor types are currently diagnosed as separate clinical entities, assigning these tumors into distinct biological entities may greatly improve their diagnosis and treatment.
DNA methylation is an important early epigenetic modification that plays a significant role in the regulation of gene expression. By far, DNA methylation–based molecular classification of human CNS tumors has proven to be a reliable tool because individual tumor types may maintain an epigenetic “memory” of their unique cell of origin.18 Recent studies have eloquently demonstrated that a subset of the original DNA methylation signatures are retained during tumor progression.19 While epigenomic analyses have immensely contributed to our understanding of molecular mechanisms underlying many CNS tumors in the last years,18 current understanding on the molecular background of PPETS is limited.
To decipher the molecular background of PPETS, we here performed immunohistochemical staining with a broad scope of markers, DNA methylation analysis, and targeted next-generation sequencing in the PPETS tumors and the comparing cohort. DNA methylation profiles were analyzed using own data and the reference cohort of the brain tumor classifier.18 Our findings suggest that PPETS and PPT may belong to the same neurooncological entity.
Materials and Methods
Patient Cohort
Five PPETS were included in this study, of which one was collected from Sanbo Brain Hospital, Capital Medical University, one from The First Affiliated Hospital of Fujian Medical University, one from Chinese PLA General Hospital, one from Beijing Neurosurgical Institute, Capital Medical University, and one from Rui Jin Hospital, Shanghai Jiao Tong University School of Medicine. Comprehensive clinical, MRI (T1 weighted image/T1WI, T2 weighted image/T2WI, and enhancement following gadolinium injection), and histopathologic data were reviewed as available. The cases for comparing group included non-tumor brain tissues (NTB, n = 7), CPP (n = 7), CN (n = 7), and PPT (4 PC, 6 GCT, and 5 SCO). We included CPP for comparison because both PPETS and CPP had similar papillary architecture. PPT tumors were included because these tumors are also located in the sellar region, they originate from the posterior pituitary and express TTF1. CN tumors were included because PPETS and CN had similar TTF1 expressions. The NTB tissues were collected from patients with traumatic brain injury who underwent microsurgery. All cases in the comparing group were collected from Sanbo Brain Hospital, Capital Medical University. To exclude metastasis possibilities, 8 cases of lung adenocarcinoma were collected from Sanbo Hospital, and 8 cases of PTC from Beijing Neurosurgical Institute. Two experienced neuropathologists independently re-evaluated the hematoxylin-eosin staining slides of the PPETS and the comparing cases. Present study was conducted in accordance with the Declaration of Helsinki and under the guidelines of the institutional ethical board of the Sanbo Brain Hospital and the local ethical committees of respective hospitals. Written informed consent was obtained from all patients and/or their legal representatives.
Histology and Immunohistochemical Staining
All lesions located in the sellar region (including the PPETS tumors and the control cases) were diagnosed based on typical imaging findings by two experienced neuropathologists at the department of neuropathology in Sanbo Brain Hospital, Capital Medical University. Immunohistochemistry (IHC) were performed according to previous report.20 Sections of 4-μm thick were prepared from archived formalin-fixed and paraffin-embedded (FFPE) materials. IHC was performed on a Ventana BenchMark Ultra Immunostainer according to standard operational procedure. Primary antibodies used in this study were directed against the following markers: (i) neural markers: microtubule-associated protein 2 (MAP2) (Abcam, 1:1500 dilution), chromogranin A (CgA) (Dako, 1:300 dilution), synaptophysin (Syn) (Bio Genex, 1:100 dilution), S100 (Dako, 1:800 dilution), GFAP (Dako, 1:500 dilution); (ii) epithelial markers: epithelial membrane antigen (EMA) (Dako, 1:50 dilution), AE1⁄AE3 (Zymed, 1:200 dilution), CK7 (Dako; 1:700 dilution); (iii) pituitary neuroendocrine tumor-related markers: GH (Dako, 1:400 dilution), PRL (Dako, 1:300 dilution), ACTH (Dako, 1:500 dilution), βTSH (Dako, 1:500 dilution), βFSH (Dako, 1:600 dilution), βLH (Dako, 1:550 dilution), PIT1 (Thermo Fisher, 1:150 dilution), TPIT (Atlas Antibodies, 1:300 dilution), SF1 (Abcam, 1:150 dilution); (iv) TTF1: clone SPT24 (Leica, 1:500 dilution), clone 8G7G3/1 (Abcam, 1:200 dilution); (v) TTF1 positive tumor-related markers: napsin A (Dako, 1:250 dilution), PAX8 (Proteintech, 1:200 dilution), transthyretin (TTR) (Novocastra, 1:120 dilution); (vi) other markers: vimentin (Vim) (Dako, 1:500 dilution), Ki-67 (Dako, 1:100 dilution), phospho-p44/42 MAPK (p-Erk1/2) (Cell Signaling Technology, 1:400). One positive and one negative control slides were included in each IHC run. Two observers performed the IHC analysis according to the manufacturer’s criteria.
DNA Methylation Data Preparation
Using a protocol described by Suwala and colleagues21 and considering the availability of specimen, genomic DNA was extracted from FFPE samples of 3 PPETS, 7 CPP, 7 CN, 15 PPT (4 PC, 6 GCT, and 5 SCO), and 7 NTB. Illumina Infinium HumanMethylation 850 (850k) array was used to obtain genome-wide DNA methylation profiles, according to the manufacturer’s instructions (Illumina). The raw signal intensities were loaded, filtered, preprocessing, and normalized using the minfi R package (v1.30)22 in R version 4.1.2. The beta and M values were calculated. Samples with the mean probe detection P value >0.01 were removed. Probes with the following features were excluded from analysis: (i) probes with a detection P value >0.01, (ii) probes on X and Y chromosomes, (iii) probes of the CpG sites that include single-nucleotide polymorphisms (SNP), and (iv) probes mapping to multiple genome locations. Finally, all 39 samples and 612,060 probes were kept for analysis. The same procedures were performed in LUAD and PTC cases for excluding metastasis possibilities with methylation profiles.
Unsupervised Clustering Analysis of DNA Methylation Data
Unsupervised clustering in our cohort was performed using the 5000 most variably methylated probes as described by Johann and colleagues.23 Unsupervised hierarchical clustering analysis were performed and visualized using R package pheatmap (1.0.12), and the Pearson correlation coefficient was the distance measure. The t-distributed stochastic neighbor embedding (t-SNE) analysis was performed using Rtsne package (v.0.16) with default parameter.
The CNS tumor data set (GSE90496)18 was used as an external data set to assess the similarities with various neuropathological tumors. The 450K beta value matrix of reference set was downloaded. The probes shared by 450K and 850K were kept. The batch effect was reduced by combat algorithm (sva package, v3.14). The 32,000 most variably methylated probes were selected to performed t-SNE analysis (Rtsne package, v.0.16). The t-SNE parameters were set as previously described.18 Similarly, the data matrix of a recent large data set for posterior pituitary tumors (GSE185041) was also downloaded and processed. And the parameters of t-SNE analysis based on our data and GSE185041 together were set as previously described.24
Identification of Differentially Methylated Positions
The differentially methylated positions (DMPs) were predicted using the R package ChAMP (v.2.24).25 Only probes with Benjamini-Hochberg adjusted P <0.01 and |deltaBeta| >0.2 would be identified as DMPs. Additionally, the CpG probes mapped at promoter regions (including TSS1500, TSS200, 5’UTR, and 1st exon) were kept in gene ontology (GO) analysis. The GO analysis was performed using the DAVID website (https://david.ncifcrf.gov/).26
Copy Number Variation Analysis
Genome-wide DNA methylation array data was also used to assess copy number variation (CNVs). The NTB samples were used as control. The raw intensities (idat files) of each single sample were first analyzed separately with the “conumee” R package (v.1.18.0)27 by default parameters, the segment files were generated. To obtain cumulated CNVs per group, the R package GenVisR (v.1.16.1)28 was used to process segment files, which can generate plots displaying the proportion of copy number losses/gains at a group level. The amplitude threshold of ±0.15 was set to reduce the impact of noise.
Targeted Next-Generation DNA Sequencing
Targeted DNA NGS analysis was performed for 3 PPETS, 3 PC, 2 GCT, and 2 SCO. FFPE samples were used for DNA extraction using the QIAamp DNA FFPE Tissue Kit (Qiagen). DNA Concentration and purity were qualified using Nanodrop 2000 (Thermo Fisher Scientific) and quantified with Qubit 2.0 using the dsDNA HS Assay Kit (Life Technologies) according to the manufacturer’s protocol. Hybridization capture-based next-generation sequencing was performed using an assay that targets 494 cancer-related gene panel (Supplementary Table 1). The enriched libraries were sequenced on the HiSeq4000 NGS platform (Illumina) with 2 × 150 bp pair-end reads. Trimmomatic48 was used for FASTQ file quality control (QC).29 Sequencing data were aligned to the reference hg19 with the Burrows-Wheeler Aligner (BWA-mem, v0.7.12).30 SNVs and Indels were detected using SCALPEL (http://scalpel.sourceforge.net) and Genome Analysis Toolkit (GATK), respectively. The SNPs and indels were annotated by ANNOVAR31 against the following databases: dbSNP, 1000Genome, ExAC, COSMIC, ClinVAR, and SIFT.
Result
Clinical Features
Our cohort of 5 PPETS patients included one male and four females with a median age of 45 years (range: 29–66 years) (Table 1). All patients showed a benign clinical course, no recurrence was observed during the follow-up period (range: 28–123 months). Clinical information is summarized in Table 1. Three of the 5 patients (Case 1, Case 2, and Case 5) had one or more abnormal hormone levels, prolactin appeared to be the most vulnerable hormone. Two patients had vision loss. No other neurological defects were found in this cohort of PPETS patients. They had no other systemic symptoms and no history of other cancers. All patients underwent transsphenoidal surgery with gross total removal of the lesion. The cases for comparing group included 7 CPP, 7 CN, and 15 PPT (4 PC, 6 GCT, and 5 SCO). The clinical features of this series are summarized in Supplementary Table 2. The cases for comparing group had no recurrence during the entire follow-up period.
Table 1.
Clinical Information of 5 PPETS Cases
| Patient | Age (Years) | Sex | Location | Clinical Findings | GTR | Adjuvant Therapy |
Recurrence | Follow-up (ms) | IHC | Infinium 850k | NGS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | 29 | F | Sellar region | Blurred binocular vision; Bitemporal hemianopsia; results of total body MRI, thyroid ultrasound and PET scan were negative; slight decrease of TSH |
Y | N | N | 28 | Y | N | Y |
| Case 2 | 52 | F | Sellar region | Sudden headache; nausea and vomiting; slight decrease of thyrotropin, adrenocorticotrophin and luteinizing hormone; slightly higher prolactin | Y | N | N | 30 | Y | Y | Y |
| Case 3 | 49 | F | Sellar region | Blurred binocular vision; normal pituitary hormones levels | Y | N | N | 120 | Y | Y | Y |
| Case 4 | 66 | M | Sellar region | Headache; normal pituitary hormones levels | Y | N | N | 108 | Y | Y | N |
| Case 5 | 31 | F | Sellar region | Amenorrhea and intermittent galactorrhoea for 2 years; headache; slightly higher prolactin | Y | N | N | 123 | Y | N | N |
F, female; M, male; GTR, gross total resection; IHC, immunohistochemistry; NGS, next-generation sequencing; WHO, World Health Organization.
Imaging Features
MRI revealed a well-circumscribed mass in the sellar region. Three PPETS cases (cases 1, 2, and 5) showed isointensity in most regions but focal hyperintensity on T1WI and heterogeneous hyperintensity on T2WI (e.g., Case 1, Figure 1a–c; Case 5, Figure 1d–f). Cases 3 showed isointensity on T1WI and heterogeneously hyperintensity on T2WI (Figure 1g–i). Case 4 showed heterogeneous hyperintensity on T1WI and slight hyperintensity on T2W1. And all cases showed heterogeneously enhanced contrast following gadolinium injection.
Figure 1.
MRI findings of representative PPETS cases. Case 1: a–c; Case 5: d–e; Case 3: g–i. Pre-operative MRI images for Case 1 (sagittal, a) and Case 5 (coronal, d) showed a well-circumscribed, T1-isointense, and slightly hyperintense mass within the sella. T2-weighted images for Case 1 (axial, b) and Case 5 (coronal, e) showed a heterogeneous hyperintensity mass. After gadolinium contrast injection, heterogeneous enhancement for Case 1 (sagittal, c) and Case 5 (coronal, f) was noted. For Case 3, pre-operative T1-weighted image showed an isointensity mass (sagittal, g), T2-weighted image showed a hyperintensity mass (axial, h). After gadolinium contrast injection, heterogeneous enhancement was noted over coronal view (sagittal, i).
Histological and Immunohistochemical Features
All 5 PPETS showed similar papillary architecture, contained cylindrical, cuboidal cells with a single layer or multiple layers of tightly packed lining fibro-vascular core (Figure 2a–e). The tumor cells were round to ovoid with eosinophilic cytoplasm. The cells had a slightly hyperchromatic nucleus with dense chromatin and inconspicuous nucleolus, but without ground glass nuclei (Figure 2f). Neither nuclear groves nor inclusions were found. Specimens from all patients contained interstitial edema and inflammatory cell infiltration (lymphocytic infiltrates admixed with plasma cells) (Figure 2a and b). In Case 5, the core of several papillae contained foamy histiocytes (Figure 2f). There were cholesterol clefts (Figure 2g), multiple giant cells, and foamy macrophages in cases 2, 3, and 5. Mitoses were not identified. Focal calcification was observed in Case 5 (Figure 2h). Neurohypophysis and bone invasion were not found. Degeneration and necrosis were found in cases 2, 3, and 5. These microscopic features are similar to previous reports.1–5
Figure 2.
Histologic features of PPETS samples examined. All 5 PPETS were composed of one or more layers of columnar epithelial cells (Cases 1–5, a–e, ×200) and the core of papillae contained lymphocytes or plasma cells (a and b, ×200). The neoplastic cells showed eosinophilic cytoplasm and round to ovoid nucleus, and the core of papillae contained foamy macrophages (arrows in panel f, ×400). Focal hemorrhage and necrosis with regions containing cholesterol crystals were present (g, Case 5, ×200). Focal calcified detritus was found in Case 5 (h, ×400).
The tumor cells were positively stained with TTF1, EMA, AE1/AE3, CK7, MAP2 and vimentin (Figure 3a–f), but were negative with neural marker Syn, thyroid carcinoma marker PAX8, and lung adenocarcinoma marker Napsin A (Figure 3g–i), as well as with TTR (a marker of CPP tumor), CgA, GFAP, TG, and S100. Cell proliferation, as detected with Ki-67 staining, was below 3% in all 5 cases (Figure 3j). No reactivities for pituitary hormones (GH, PRL, ACTH, βTSH, βLH, βFSH) and transcription factors of pituitary adenoma (PIT1, SF1, TPIT) were detected in tumor cells. Based on these histologic features and IHC results, we diagnosed these tumors as PPETS.1 In addition, unlike the previous reported positivity of p-Erk1/2, a reporter of RAS/MAPK pathway activation, in SCO and PC,32,33 the staining of p-Erk1/2 was negative in 4 PPETS cases. There was no staining result for Case 2 due to lack of material. The detailed immunohistochemical staining results of the 5 PPETS are summarized in Supplementary Table 3.
Figure 3.
Immunohistochemical features of the 5 PPETS samples examined. All 5 PPETS were positively stained with TTF1 (a), EMA (b), AE1/AE3 (c), CK7 (d), MAP2 (e), and vimentin (f). These tumors were negative for Syn (g), PAX8 (h), and Napsin A (i). Ki-67 labeling was detected in <3% of the cells (j).
For comparison, we also characterized the microscopic features and TTF1 expressions in CPP (n = 7, Supplementary Figure 1a and b), CN (n = 7, Supplementary Figure 1c and d), and PTT tumors including PC (n = 5, Supplementary Figure 1e and f), GCT (n = 6, Supplementary Figure 1g and h), and SCO (n = 5, Supplementary Figure 1i and j). Although CPP showed a similar morphology to that of PPETS, unlike the PPETS that showed stable TTF1 expression, no CPP expressed TTF1. In addition, the CPP showed diffuse expression of S100 protein which was negative in PPETS. By comparison, CN and all PPT tumors (PC, GCT, and SCO) showed stable TTF1 expression. Detailed histologic features and immunophenotypes of these comparing cases are listed in Supplementary Tables 2 and 4.
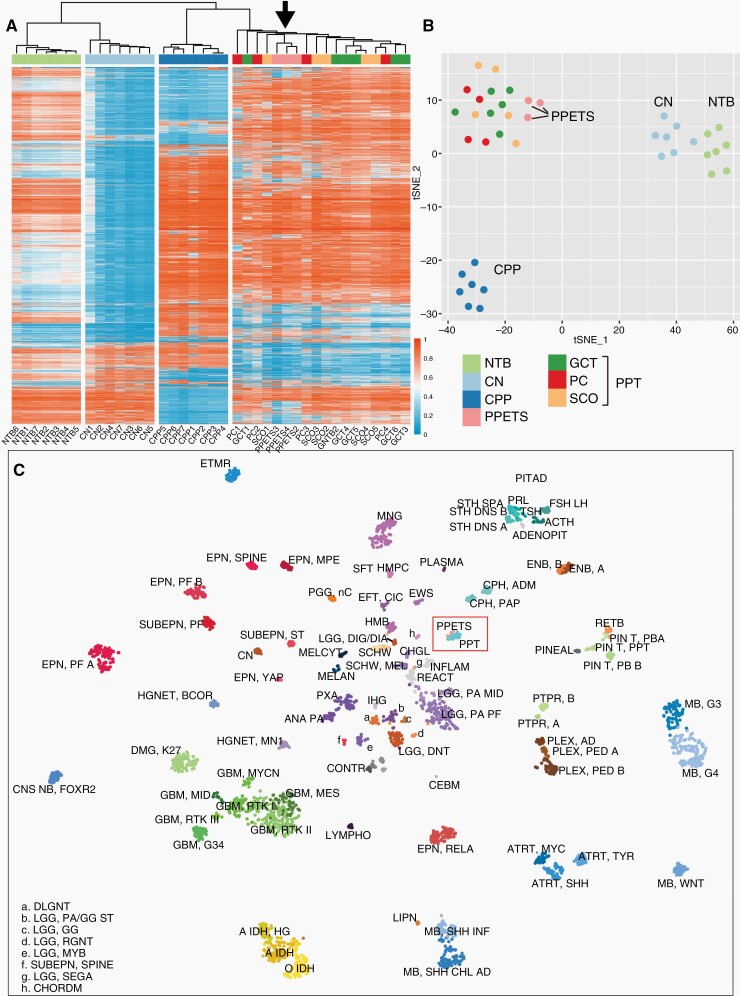
PPETS and PPT Tumors Form a Distinct Molecular Cluster
Based on the availabilities of the tumor materials, we performed DNA methylation analysis with 3 PPETS, 7 CN, 7 CPP, 15 PPT (4 PC, 6 GCT, and 5 SCO), and 7 NTB (Figure 4a and b). In both unsupervised hierarchical clustering and t-SNE analysis, DNA methylation profiles across the samples demonstrated that PPETS and PPT formed a distinct cluster irrespective of their histologic types (Figure 4a and b). However, DNA methylation profiles of PPETS were clearly distinct from that of CN and CPP tumors. Notably, CNs are derived from foramen of Monro with TTF1 expression, and CPPs are derived from choroid plexus epithelium, with morphological features similar to PPETS. Using t-SNE analysis, we further assessed the DNA methylation profile of these tumors in the context of the reference cohort of the brain tumor classifier,18 all PPTES tumors of our cohort and the 29 PPT tumors of the reference cohort formed a well-defined group, which is clearly distinct from all other CNS tumor classes (Figure 4c). The similar result was also found when our data was analyzed together with a recent large data set for posterior pituitary tumors24 (Supplementary Figure 2). To exclude the possibility of metastatic cancer, we also compared the methylation profile of PPETS with the profile of LUAD and PTC which also express TTF1. The result showed that methylation profiles of LUAD and PTC were distinct and completely different from PPETS (Supplementary Figure 3).
Figure 4.
The DNA methylation profiles of PPETS and PPT tumors together form a distinct “island.” Both unsupervised hierarchical clustering analysis (a) and t-SNE analysis (b) of our cohort show that PPETS (black arrow in panel a) and PPT tumors (including PC, GCT, and SCO) form a distinct methylome cluster. (c) PPETS and PPT tumors together form a distinct “island” in the context of the reference cohort of the brain tumor classifier.18 Plot of t-SNE analysis of the DNA methylation data of our cohort projected into the reference cohort of the brain tumor classifier is shown.
We further identified DMPs between the cluster of PPETS and PPT (n = 18) and the samples of NTB (n = 7) in our data. The 20.89% of DMPs was located in promoter region (Supplementary Figure 4). Gene ontology analysis was separately performed in the hypermethylated and hypomethylated DMPs. The hypermethylated DMPs, potentially involved in down-regulated gene expression, were enriched in axon guidance (GO:0007411), nervous system development (GO:0007399), and positive regulation of synapse assembly (GO:0051965). The hypomethylated DMPs, potentially involved in up-regulated gene expression, were enriched in inflammatory response (GO:0006954), signal transduction (GO:0007165), and cell–cell adhesion (GO:0098609) (Supplementary Table 5). These results indicated that inflammatory response may contribute to the maintenance of PPETS and PPT tumors.
Broadly Similar Chromosomal CNVs Between PPETS and PPT
Chromosomal CNVs were determined using DNA methylation array data. Notably, 2 of the 3 PPETS and 8 of the 15 PPT tumors shared losses of Chr16, Chr19, Chr20, and Chr22 (Figure 5a and b). Chromosomal losses were also more frequent than chromosomal gains in both PPETS and PPT tumors. Focal amplifications or deep deletions were not observed in any of the PPETS and PPT tumors. In contrast, different chromosomal CNV profiles were observed in the CN tumors (Figure 5c) and CPP tumors (Figure 5d). In CN tumors, no obvious chromosomal gains or losses were identified, whereas gain of chromosomes 5, 7, 8, 9 12, 15, 17, and 18, and loss of chromosome 10 predominated the CPP tumors. Detailed chromosomal alterations are listed in Supplementary Table 6.
Figure 5.
Summary chromosomal CNV profiles of PPETS, PPT, CN, and CPP tumors examined. Chromosomal CNV profiles were analyzed using DNA methylation data. PPETS and PPT tumors showed broadly similar chromosomal losses (a and b), whereas CN tumors contained no obvious chromosomal CNVs (c), CPP tumors however harbored a strikingly different set of chromosomal CNV profile (d).
Targeted Next-Generation DNA Sequencing Analysis
The 3 PPETS, 3 PC, 2 GCT, and 2 SCO tumors sequenced did not contain any relevant variants in the genes examined.
Discussion
Using DNA methylation profiling, our studies demonstrate that PPETS and PPT tumors form a distinct molecular cluster and share broadly similar chromosomal copy number alterations.
PPETS is extremely rare. We found only 1 case in over 26,000 tumors operated at Sanbo brain hospital and 4 cases out of over 4 million surgical specimens from the other 4 hospitals. MRI showed a sharply circumscribed mass in the sellar region. No primary tumors outside the sella were found in all 5 patients. The neuroimaging features included contrast enhancement, isointense, or isointense with slightly hyperintense on T1WI, and heterogeneous hyperintensity on T2WI. Recurrence was not observed in all 5 PPETS tumors, their clinical characteristics are similar to those in previous reports.1–5 Consistent with previous literatures,1–3 all 5 PPETS tumors examined exhibited a similar papillary architecture, which contained cylindrical, cuboidal cells with a single layer or multiple layers of tightly packed lining fibro-vascular core. These clinical findings suggest that PPETS tumor entity follows a benign course comparable to that of the WHO Grade I PPT tumors.
Immunohistochemically, lack of pituitary hormone and pituitary lineage-restricted transcription factors ruled out the possibility of a primary tumor of adenohypophyseal cells. Notably, TTR is a good marker for CPP34 and more than 90% of CPP are also positive for S100.35 The negativity of TTR and S100 distinguishes PPETS from the CPP tumors. Napsin A is a putative marker for lung adenocarcinoma.36 Similarly, PAX8 is a valuable marker for metastatic thyroid carcinoma and is expressed in more than 90% of thyroid carcinoma.37 The benign microscopic features and the negative staining for Napsin A and PAX8 distinguish PPETS tumors from the metastasis of lung adenocarcinoma and thyroid carcinoma.
DNA methylation analyses in our own cohort and projection of our samples into the reference cohort of the brain tumor classifier18 concordantly show that PPETS and PPT tumors form a distinct cluster, indicating that PPETS and PPT are unlikely distinct entities. This is further supported by the broadly similar profiles in chromosomal CNVs, losses of Chr16, Chr19, Chr20, and Chr22 are common in a substantial fraction of PPETS and PPT tumors analyzed. Further, all PPETS and PTT tumors show TTF1 expression, indicating that PPETS and PTT tumors may share similar cells of origin. In contrast, PPETS tumors are epigenetically distinct from CPP, pituitary adenomas, and CN, though PPETS and CPP tumors share similar morphology, PPETS and pituitary adenomas locate in the same region, and CN tumors also all express TTF1.
In conclusion, PPETS and PPT (including PC, SCO, GCT) may constitute a single common neurooncological entity. In addition to the shared epigenetic features, these tumors share common features in chromosome CNVs and the expression of key transcriptional regulator TTF1.
Supplementary Material
Contributor Information
Jing Feng, Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Zejun Duan, Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Kun Yao, Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Qiuping Gui, The Department of Pathology, Chinese PLA General Hospital (301 Hospital), Beijing, China.
Xing Liu, Department of Pathology, Beijing Neurosurgical Institute, Capital Medical University, Beijing, China.
Xingfu Wang, Department of Pathology, The First Affiliated Hospital of Fujian Medical University, Fuzhou, China.
Zunguo Du, Department of Pathology, Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai, China.
Liwei Shao, The Department of Pathology, Chinese PLA General Hospital (301 Hospital), Beijing, China.
Benyan Zhang, Department of Pathology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Shanshan Cai, Department of Pathology, the Second Affiliated Hospital of Fujian Medical University, Fuzhou, China.
Mingwang Zhu, Department of Imaging, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Zejuan Hu, Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Lei Xiang, Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Xiaolong Fan, Beijing Key Laboratory of Gene Resource and Molecular Development, Laboratory of Neuroscience and Brain Development, School of Life Sciences Beijing Normal University, Beijing, China.
Xueling Qi, Department of Pathology, Sanbo Brain Hospital, Capital Medical University, Haidian District, Beijing, China.
Data Availability
The data sets supporting the current study have not been deposited in a public repository because of institutional ethics restrictions, but all the data supporting the findings of this study are available within the main article and supplementary files and from the corresponding author upon reasonable request. The requestors must complete a data request and when contacting the corresponding author (xszqxl169@mail.ccmu.edu.cn) details on data sharing criteria for requesting access will be sent.
Conflict of interest statement
The authors have no conflicting interests to disclose.
Authorship statement
X.Q., K.Y., and J.F. conceived the Study Conception and designed the experiments; X.L., X.W., Zu.D., L.S., B.Z., S.C., and Q.G. provided the Sample Resources. K.Y., J.F., Ze.D., Z.M., Z.H., and L.X. performed the experiments and collected the data. J.F., Ze.D., K.Y., and M.Z. participated in the analysis of the results. J.F., K.Y., and X.F. wrote and revised the paper.
References
- 1. Roncaroli F, Chatterjee D, Giannini C, et al. Primary papillary epithelial tumour of the sella: expanding the spectrum of TTF-1-positive sellar lesions. Neuropathol Appl Neurobiol. 2020;46(5):493–505. [DOI] [PubMed] [Google Scholar]
- 2. Bian LG, Sun QF, Wu HC, et al. Primary choroid plexus papilloma in the pituitary fossa: case report and literature review. Acta Neurochir. 2011;153(4):851–857. [DOI] [PubMed] [Google Scholar]
- 3. Kuo CH, Yen YS, Tu TH, et al. Primary choroid plexus papilloma over sellar region mimicking with craniopharyngioma: a case report and literature review. Cureus. 2018;10(6):e2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma YH, Ye K, Zhan RY, Wang LJ.. Primary choroid plexus papilloma of the sellar region. J Neurooncol. 2008;88(1):51–55. [DOI] [PubMed] [Google Scholar]
- 5. Tetsuro Sameshima RT, Sugimura T, Izumi N, et al. Choroid plexus papilloma originating in the sella turcica--case report. Neurologia Medico-chirurgica (Tokyo). 2010;50(2):3. [DOI] [PubMed] [Google Scholar]
- 6. Asa SL, Mete O, Perry A, Osamura RY.. Overview of the 2022 WHO classification of pituitary tumors. Endocr Pathol. 2022;33(1):6–26. [DOI] [PubMed] [Google Scholar]
- 7. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shibuya M. Welcoming the new WHO classification of pituitary tumors 2017: revolution in TTF-1-positive posterior pituitary tumors. Brain Tumor Pathol. 2018;35(2):62–70. [DOI] [PubMed] [Google Scholar]
- 9. Suess U, Pliska V.. Identification of the pituicytes as astroglial cells by indirect immunofluorescence-staining for the glial fibrillary acidic protein. Brain Res. 1981;221(1):27–33. [DOI] [PubMed] [Google Scholar]
- 10. Takei Y, Seyama S, Pearl GS, Tindall GT.. Ultrastructural study of the human neurohypophysis. II. Cellular elements of neural parenchyma, the pituicytes. Cell Tissue Res. 1980;205(2):273–287. [DOI] [PubMed] [Google Scholar]
- 11. Lee EB, Tihan T, Scheithauer BW, Zhang PJ, Gonatas NK.. Thyroid transcription factor 1 expression in sellar tumors: a histogenetic marker? J Neuropath Exp Neur. 2009;68(5):482–488. [DOI] [PubMed] [Google Scholar]
- 12. Zamecnik J, Chanova M, Kodet R.. Expression of thyroid transcription factor 1 in primary brain tumours. J Clin Pathol. 2004;57(10):1111–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lazzaro D, Price M, de Felice M, Di Lauro R.. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113(4):1093–1104. [DOI] [PubMed] [Google Scholar]
- 14. Kimura S, Hara Y, Pineau T, et al. The T/ebp null mouse thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Gene Dev. 1996;10(1):60–69. [DOI] [PubMed] [Google Scholar]
- 15. Lee BJ, Cho GJ, NorgrenRB, Jr, et al. TTF-1, a homeodomain gene required for diencephalic morphogenesis, is postnatally expressed in the neuroendocrine brain in a developmentally regulated and cell-specific fashion. Mol Cell Neurosci. 2001;17(1):107–126. [DOI] [PubMed] [Google Scholar]
- 16. Nakamura K, Kimura S, Yamazaki M, et al. Immunohistochemical analyses of thyroid-specific enhancer-binding protein in the fetal and adult rat hypothalami and pituitary glands. Brain Res Dev Brain Res. 2001;130(2):159–166. [DOI] [PubMed] [Google Scholar]
- 17. Ordonez NG. Value of thyroid transcription factor-1 immunostaining in tumor diagnosis: a review and update. Appl Immunohisto M M. 2012;20(5):429–444. [DOI] [PubMed] [Google Scholar]
- 18. Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim M, Costello J.. DNA methylation: an epigenetic mark of cellular memory. Exp Mol Med. 2017;49(4):e322–e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wefers AK, Stichel D, Schrimpf D, et al. Isomorphic diffuse glioma is a morphologically and molecularly distinct tumour entity with recurrent gene fusions of MYBL1 or MYB and a benign disease course. Acta Neuropathol. 2020;139(1):193–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suwala AK, Stichel D, Schrimpf D, et al. Primary mismatch repair deficient IDH-mutant astrocytoma (PMMRDIA) is a distinct type with a poor prognosis. Acta Neuropathol. 2021;141(1):85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johann PD, Bens S, Oyen F, et al. Sellar region atypical teratoid/rhabdoid tumors (ATRT) in adults display DNA methylation profiles of the ATRT-MYC subgroup. Am J Clin Pathol. 2018;42(4):506–511. [DOI] [PubMed] [Google Scholar]
- 24. Schmid S, Solomon DA, Perez E, et al. Genetic and epigenetic characterization of posterior pituitary tumors. Acta Neuropathol. 2021;142(6):1025–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian Y, Morris TJ, Webster AP, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33(24):3982–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang DW, Sherman BT, Lempicki RA.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 27. Hovestadt V, M Z. conumee: enhanced copy-number variation analysis using Illumina DNA methylation arrays. R package version 1.9.0; 2017.
- 28. Skidmore ZL, Wagner AH, Lesurf R, et al. GenVisR: genomic visualizations in R. Bioinformatics. 2016;32(19):3012–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164e164.–e164ee164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller MB, Bi WL, Ramkissoon LA, et al. MAPK activation and HRAS mutation identified in pituitary spindle cell oncocytoma. Oncotarget. 2016;7(24):37054–37063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Viaene AN, Lee EB, Rosenbaum JN, Nasrallah IM, Nasrallah MP.. Histologic, immunohistochemical, and molecular features of pituicytomas and atypical pituicytomas. Acta Neuropathol Commun. 2019;7(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herbert J, Cavallaro T, Dwork AJ.. A marker for primary choroid plexus neoplasms. Am J Pathol. 1990;136(6):1317–1325. [PMC free article] [PubMed] [Google Scholar]
- 35. Paulus W, Janisch W.. Clinicopathologic correlations in epithelial choroid plexus neoplasms: a study of 52 cases. Acta Neuropathol. 1990;80(6):635–641. [DOI] [PubMed] [Google Scholar]
- 36. Bishop JA, Sharma R, Illei PB.. Napsin A and thyroid transcription factor-1 expression in carcinomas of the lung, breast, pancreas, colon, kidney, thyroid, and malignant mesothelioma. Hum Pathol. 2010;41(1):20–25. [DOI] [PubMed] [Google Scholar]
- 37. Laury AR, Perets R, Piao H, et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am J Surg Pathol. 2011;35(6):816–826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the current study have not been deposited in a public repository because of institutional ethics restrictions, but all the data supporting the findings of this study are available within the main article and supplementary files and from the corresponding author upon reasonable request. The requestors must complete a data request and when contacting the corresponding author (xszqxl169@mail.ccmu.edu.cn) details on data sharing criteria for requesting access will be sent.