Abstract
Objectives:
Evaluate association between obesity and angiogenic-related gene expression in endometrial cancer (EC). Evaluate interaction between diet and metformin on angiogenic-related gene expression.
Methods:
We evaluated the association between 168 human angiogenic-related genes and body mass index (BMI) in the TCGA Uterine Corpus Endometrial Carcinoma cohort (endometrioid endometrial cancer (EEC) cohort n=290, and copy number high cohort n=60), an independent validation cohort from Gynecologic Cancer Center of Excellence (GYN-COE) (n=62), and corresponding 185 homologous mouse genes in an LKB1fl/flp53fl/fl mouse model of EC (n=20). Mice received 60% of calories from fat in a high-fat diet (HFD), mimicking diet-induced obesity, versus 10% of calories from fat in a low-fat diet (LFD). After tumor growth, HFD (n=5) and LFD (n=5) mice were treated with metformin (200 mg/kg/day) or control. Whole transcriptome analysis of mouse tumors was performed using RNA-Seq.
Results:
At a false-discovery rate of 10%, twenty-one angiogenic-related genes were differentially expressed with respect to BMI when adjusting for grade in the TCGA EEC cohort. Evaluation of these genes in the mouse model control group revealed association between increased Edil3 expression in HFD versus LFD mice (2.5-fold change (FC); unadjusted p=0.03). An interaction was observed for expression of Edil3 between diet and metformin treatment (unadjusted p=0.009). Association between BMI and increased expression of EDIL3 was validated in one of four EDIL3 probesets in the GYN-COE cohort (p=0.0011, adjusted p=0.0342).
Conclusions:
Obesity may promote tumor progression via differential modulation of angiogenic pathways in EEC. Our exploratory findings demonstrated that EDIL3 may be a candidate gene of interest.
INTRODUCTION
Endometrial cancer (EC), an obesity driven malignancy1, is the most common gynecologic cancer. Alarmingly, the incidence of endometrial cancer has continued to rise with 61,880 estimated new cases in 2019 and 12,160 expected deaths.2 The increased incidence and mortality of EC may be due to the obesity epidemic. Compared to women of normal weight, those with body mass indices (BMI) over 30 are three times more likely to be diagnosed with EC1 and severely obese EC patients (BMI ≥ 40 kg/m2) have a 6.25-fold increased risk of death.3 The mechanism underlying this is unclear; however, adipose tissue may promote altered regulation of angiogenic-related genes, which leads to an “angiogenic switch” and stimulates tumor growth, metastasis, and progression.4
Increased tumor angiogenesis is associated with worse survival in EC patients5, and overexpression of the potent pro-angiogenic factor, vascular endothelial growth factor (VEGF), is a risk factor for disease recurrence.6 We and others have identified an association between obesity, high fat diet (HFD), and angiogenesis in ovarian (publication pending) and breast cancers.7 We hypothesize that obesity and high fat diet are significantly associated with angiogenic-related gene expression in endometrial cancer. In this study, we sought to evaluate the association between BMI and differential expression of angiogenic-related genes in specimens from The Cancer Genome Atlas (TCGA) Uterine Corpus Endometrial Carcinoma (UCEC)8 cohort and in an innovative genetically-engineered EC mouse model (LKB1fl/flp53fl/fl) subjected to diet-induced obesity.9 We also sought to evaluate if metformin, an mTOR inhibitor with known anti-proliferative tumor activity, mitigates angiogenic-related gene expression.
MATERIALS AND METHODS
TCGA Analysis:
Using the Uterine Corpus Endometrial Cancer cohort in the TCGA database, our study included all patients with endometrioid histology (grades 1, 2 and 3), available BMI, and available RNA-Seq (Illumina GA RNASeqV2) data (N=290).8 TCGA mRNA data were retrieved from the Cancer Genomic Data Server (CGDS) through the Computational Biology Center Portal (cBio, http://www.cbioportal.org/). The cdgsr extension package cran.rproject.org/web/packages/cgdsr/ was used to execute the retrieval. The RSEM counts were used10. For the regression models, the log2 transformed normalized expressions were used. Analyses were restricted to a set of 168 angiogenic-related candidate human genes. This list was generated a priori based on literature review, known genes in the pathways of interest, and potential therapeutic targets (Supplemental Table 1).
The primary TCGA analysis (n=290) was based on the association between gene expression and BMI with grade as a covariate. A smoothly clipped absolute deviation penalty quantile regression was employed.11,12 For the effect size, we used the estimated coefficient for log(BMI) and the corresponding p-value. We calculated the FDR-adjusted p-values (q-value) for multiple testing correction13. Based on these analyses, we selected 21 genes with q-value<0.1 from the primary analysis for further mechanistic validation in the mouse model (Supplemental Table 2).
Mouse model:
The LKB1fl/flp53fl/fl genetically engineered mouse model is a unique endometrial cancer mouse model that specifically and somatically deletes the tumor suppressor genes, Lkb1 and Trp53 in adult endometrial epithelial cells. Knockout of Lkb1 and Trp53 are conditional and only activated via injection of an adenoviral vector expressing Cre (AdCre) into the uterine horn of adult female mice. All mice were handled according to protocols approved by UNC-CH Institutional Animal Care and Use Committee (IACUC). To mimic diet-induced obesity (DIO), half of the mice were subjected to a high fat diet (HFD, 60% calories derived from fat), while the other half were subjected to a low fat diet (LFD, 10% calories derived from fat, Research Diets, New Brunswick, USA) at 3 weeks age. The mice were injected with recombinant adenovirus Ad5-CMV-Cre (AdCre, Transfer Vector Core, University of Iowa) into the left ovarian bursa cavity at 6–8 weeks age. Eight weeks after injection, HFD-fed and LFD-fed mice (n=20 per group) initiated treatment with either metformin (200 mg/kg, drinking water) or vehicle for 4 weeks. All mice were euthanized after 4 weeks of metformin and placebo treatment. Tumor tissues from HFD treated, LFD treated and controls (5 cases per group) were collected and extracted RNA. Whole-transcriptome analysis of the tumors was performed using RNA-Seq.
Mouse data analysis:
The University of North Carolina, Chapel Hill (UNC) institutional high throughput sequencing facility performed the RNA sequencing. The raw sequencing read data were aligned against the mouse mm 10 reference genome using the STAR14 aligner. Prior to this step, the quality of the sequencing reads was assessed using FastQC15 and adapter sequences were trimmed using Trimmomatic16. The aligned reads were mapped to genomic features using HTSeq17. The reference sequence and GTF file were obtained from the UCSC mm 10 bundle available from the iGenomes collection. Differential expression analysis was conducted using R18 and its extension package DESeq219. Estimates of the fold-change (FC) were obtained by transforming the corresponding log2 fold change estimates from the regression model (FC=2^[log2 FC estimate]).
The analysis for the mouse model was based on modeling the raw counts within a framework of a negative binomial model of diet with respect to: 1) diet, low versus high fat diet within the control group, and 2) interaction effect between diet and metformin treatment. We considered a gene to “validate” if the corresponding two-sided p-value was less than 0.05/K and the estimated effect size was in the same direction as that estimated for the gene in the TCGA data where K denotes the number of genes to be validated. The primary analysis was conducted on a set of K=23 mouse genes mapped by homology from the 21 human genes identified in the TCGA data. An exploratory analysis was performed for the 162 human angiogenic-related genes (six human genes did not find homologs in mouse genes), which mapped to 185 genes in the mice (Supplemental Table 1).
Exploratory analysis in the copy number high cohort of TCGA
The molecular biology of uterine cancers induced in the LKB1fl/flp53fl/fl genetically engineered mouse model most closely resembles the copy number high (CNH) cancers reported in TCGA,8 given that LKB1 loss is most commonly seen in grade 3 vs. grade 1/2 ECs and is accompanied by p53 loss about 20% of the time20. Therefore, an exploratory TCGA analysis was performed limited to the copy number high (CNH) cohort (N=60), based on the association between gene expression and BMI. The exploratory analysis evaluated the top 22 differentially expressed, HFD vs LFD genes (unadjusted p <0.05) identified in mice within the control group. The mouse genes were mapped to the corresponding human genes based on homology.21 An additional exploratory analysis of the CNH data was performed evaluating the 168 pre-selected human angiogenic-related genes using methodology described for the UCEC cohort.
Validation cohort analysis
Using an independent cohort from GYN-COE (n=62), the top twenty-one angiogenic-related genes associated with BMI (q-value<0.1) from the TCGA analysis were evaluated using Affymetrix Human Genome U133 Plus 2.0 Array data. The association between RMA-normalized expression of each gene’s probe set and log (BMI) was analyzed by adjusting for grade using the same model for the TCGA cohort.
All statistical analyses were carried out using the R statistical environment along with extension packages from the tidyverse ecosystem22 and the knitr extension package23 were used to facilitate adherence to the principles of reproducible analysis.
RESULTS
Association between obesity and angiogenic-related genes in endometrioid endometrial cancer
The TCGA patient cohort contained 290 patients. Notably, 72.4% of patients had stage 1 disease; however, there was a relatively even distribution of grade among the patients (29.3%, 32.8%, and 37.9% respectively for grade 1, 2 and 3 disease). Obesity (BMI of 30 or greater) was noted in 64.1% of patients. The majority of patients (78.3%) were post-menopausal and 83.8% were tumor free at the completion of primary treatment. Most patients were white (81.7%) and 7.9% were black. (Table 1)
Table 1.
Disease status and characteristics of women in TCGA database and the GYN-COE database with endometrioid endometrial cancer with available BMI and RNA sequencing data
| TCGA N = 290 |
GYN-COE N=67 |
||
|---|---|---|---|
| Stage | 1 | 210 (72.4%) | 33 (49.3) |
| 2 | 19 (6.55%) | 0 (0.0) | |
| 3 | 49 (16.9%) | 22 (32.8) | |
| 4 | 12 (4.1%) | 12 (17.9) | |
| Grade | 1 | 85 (29.3%) | 12 (17.9) |
| 2 | 95 (32.8%) | 15 (22.4) | |
| 3 | 110 (37.9%) | 35 (52.2) | |
| Unknown | 5 (7.4) | ||
| BMI | 0 to < 25 | 48 (16.6%) | 7 (10.4) |
| 25 to < 30 | 56 (19.3%) | 20 (29.9) | |
| ≥30 | 186 (64.1%) | 40 (59.7) | |
| Menopause status | Indeterminatea | 14 (4.8%) | 0 (0.0) |
| Perib | 13 (4.5%) | 5 (7.5) | |
| Postc | 227 (78.3%) | 60 (89.5) | |
| Pred | 24 (8.3%) | 0 (0.0) | |
| Unknown | 12 (4.1%) | 2 (3.0) | |
| Tumor status | Tumor Free | 243 (83.8%) | 53 (79.1) |
| With tumor | 37 (12.8%) | 14 (20.9) | |
| Unknown | 10 (3.4%) | 0 (0.0) | |
| Race | White | 237 (81.7%) | 58 (86.6) |
| Black or African American | 23 (7.9%) | 6 (8.9) | |
| Asian | 16 (5.5%) | 2 (3.0) | |
| Unknown | 6 (2.1%) | 1 (1.5) | |
| Native Hawaiian or other Pacific Islander | 5 (1.7%) | 0 (0.0) | |
| American Indian or Alaska Native | 3 (1.0%) | 0 (0.0) | |
Indeterminate – Neither pre or post-menopausal
Peri – 6 to 12 months since last menstrual period (LMP)
Post – prior bilateral oophorectomy OR >12 months since LMP with no prior hysterectomy
Pre – <6 months since LMP AND no prior bilateral oophorectomy AND not on estrogen replacement
In the TCGA cohort, twenty-one genes, from the panel of 168 angiogenic-related genes, were found to be associated with BMI (Table 2) in the adjusted evaluation. All twenty-one of these genes were upregulated.
Table 2.
Association between BMI and differential expression of candidate angiogenic genes in the TCGA endometrioid endometrial cancer cohort
| Unadjusted evaluation | Adjusted evaluation | |||||
|---|---|---|---|---|---|---|
| Gene | Tau statistic | p-valuea | q-valueb | Estc | p-valued | q-valuee |
| JAG1 | 0.15 | <0.001 | 0.014 | 0.81 | <0.001 | 0.015 |
| NRG1 | 0.11 | 0.006 | 0.304 | 1.34 | 0.004 | 0.054 |
| ID1 | 0.1 | 0.011 | 0.304 | - | - | - |
| MMP14 | 0.1 | 0.014 | 0.304 | 0.99 | <0.001 | 0.006 |
| TGFA | 0.1 | 0.014 | 0.304 | 0.98 | 0.011 | 0.089 |
| HEY1 | 0.09 | 0.016 | 0.304 | 1.34 | 0.002 | 0.042 |
| CXCL12 | 0.09 | 0.018 | 0.304 | 1.31 | 0.002 | 0.042 |
| IL5RA f | 0.09 | 0.021 | 0.313 | - | - | - |
| HPRT1 | −0.09 | 0.031 | 0.324 | - | - | - |
| HSP90AA1 | −0.08 | 0.031 | 0.324 | - | - | - |
| CXCL1 | 0.08 | 0.035 | 0.324 | 2.36 | 0.007 | 0.068 |
| PLAUg | 0.08 | 0.042 | 0.324 | - | - | - |
| MAPK3 | 0.08 | 0.047 | 0.324 | - | - | - |
| CHGA h | 0.08 | 0.048 | 0.324 | - | - | - |
| COL15A1 | 0.08 | 0.048 | 0.324 | 1.27 | 0.002 | 0.039 |
| AKTIP | −0.08 | 0.050 | 0.324 | - | - | - |
| IL1B | 0.08 | 0.052 | 0.324 | 0.88 | <0.001 | 0.027 |
| ENPP2 | 0.07 | 0.063 | 0.324 | - | - | - |
| B2M i | 0.07 | 0.065 | 0.324 | - | - | - |
| LRDD | 0.07 | 0.065 | 0.324 | - | - | - |
| TGFB2 | - | - | - | 1.38 | <0.001 | 0.027 |
| TIMP1 | - | - | - | 1.16 | 0.001 | 0.027 |
| FLT4 | - | - | - | 0.78 | 0.002 | 0.042 |
| PDGFRB | - | - | - | 1.00 | 0.003 | 0.045 |
| PRL | - | - | - | 0.83 | 0.004 | 0.054 |
| CTGF | - | - | - | 0.96 | 0.005 | 0.054 |
| KDR | - | - | - | 0.67 | 0.006 | 0.068 |
| PROX1 | - | - | - | 1.41 | 0.007 | 0.068 |
| IL12A | - | - | - | 1.37 | 0.009 | 0.077 |
| VEGFC | - | - | - | 0.64 | 0.009 | 0.077 |
| EDIL3 j | - | - | - | 1.47 | 0.012 | 0.091 |
| PDCD1 | - | - | - | 1.39 | 0.013 | 0.092 |
Unadjusted p-value: unadjusted Kendall Tau p-value for association between BMI and gene expression.
Unadjusted q-value: q-value of Kendall Tau p-value that measures the proportion of false positives incurred when that particular test is called significant.
Est: Estimated effect size
Adjusted p-value: p-value of association between log(BMI) and gene expression adjusted for grade using penalty regression for median.
Adjusted q-value: q-value of adjusted p-value that measures the proportion of false positives incurred when that particular test is called significant
Gene validated in GYN-COE cohort (padj = 0.0284)
Gene validated in GYN-COE cohort (padj = 0.0284)
Gene validated in GYN-COE cohort (padj = 0.0250)
Gene validated in GYN-COE cohort (padj = 0.0284)
Probeset 207379 validated in GYN-COE cohort (padj = 0.0342)
Association between high-fat diet, metformin, and candidate angiogenic-related genes in LKB1fl/flp53fl/fl genetically engineered mice
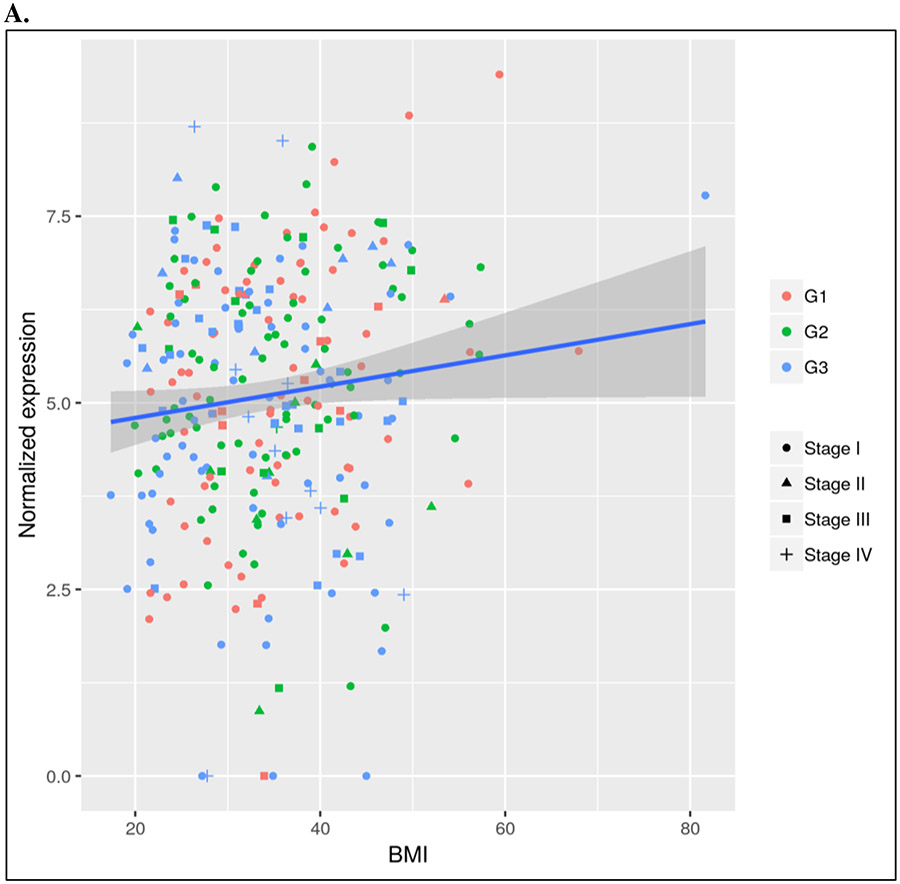
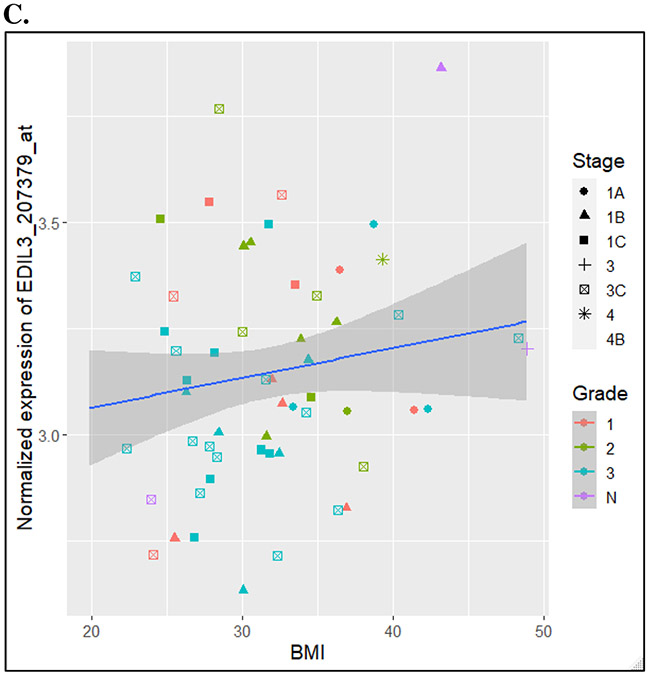
The twenty-one candidate angiogenic-related genes from the analysis adjusted for grade were mapped to twenty-three genes in mice by homology (Supplemental Table 2). Evaluation of these genes in our mouse model (n=10, with 5 HFD versus 5 LFD within the no treatment group) revealed strong signal of an association between increased Edil3 expression in HFD versus LFD mice (2.5FC; unadjusted p=0.03) (Figure 1). However, this did not meet our threshold of validation of p=0.05/21.
Figure 1.
Differential expression of Edil3 in TCGA endometrioid endometrial, in the LKB1fl/flp53fl/fl mice, and in the GYN-COE validation cohort
A. Differential expression of Edil3 with BMI in TCGA database
B. Differential expression of Edil3 in mouse model with high fat diet versus low fat diet in non-treatment group
C. Differential expression of Edil3 (probeset 207379) with BMI in GYN-COE validation cohort
An exploratory analysis in the mouse model evaluating the total list of 162 human genes mapping to 185 mouse genes by homology (Supplemental Table 1), generated a list of twenty two angiogenic-related genes (unadjusted p<0.05) that are differentially expressed in a high fat versus low fat diet within the control group of mice. Notably this list of genes included increased expression of several potent pro-angiogenic genes in the HFD mice, including leptin (Lep) (4.0 FC, unadjusted p=0.002) and vascular endothelial growth factor A (Vegfa) (2.8 FC, unadjusted p=0.003) (Table 3, Figure 2). In this list, none of the genes demonstrated q<0.1 when adjusted.
Table 3.
Association between diet and differential expression of angiogenic candidate genes in the LKB1fl/flp53fl/fl mice
| Gene | log2FoldChange | Fold Change | p-value | p-value (adjusted) |
|---|---|---|---|---|
| Lep | 2.01 | 4.03 | 0.002 | 0.209 |
| Vegfa | 1.50 | 2.83 | 0.003 | 0.209 |
| Pgf | 1.02 | 2.03 | 0.005 | 0.209 |
| Mmp2 | 1.35 | 2.55 | 0.005 | 0.209 |
| Tgfb1 | 0.94 | 1.92 | 0.007 | 0.222 |
| Cxcl9 | 1.52 | 2.87 | 0.010 | 0.278 |
| Bcl2l11 | −0.47 | −1.39 | 0.016 | 0.314 |
| Pdgfa | −0.47 | −1.39 | 0.017 | 0.314 |
| Notch4 | 0.76 | 1.69 | 0.020 | 0.314 |
| Hgf | 1.17 | 2.25 | 0.020 | 0.314 |
| Angpt1 | 1.31 | 2.48 | 0.023 | 0.314 |
| Edn1 | −0.98 | −1.97 | 0.026 | 0.314 |
| Serpinb5 | 1.41 | 2.66 | 0.027 | 0.314 |
| Cd44 | 0.81 | 1.75 | 0.029 | 0.314 |
| Edil3 | 1.30 | 2.46 | 0.032 | 0.314 |
| Lect1 | −1.35 | −2.55 | 0.032 | 0.314 |
| Tnmd | 1.35 | 2.55 | 0.032 | 0.314 |
| Angpt2 | 0.84 | 1.79 | 0.041 | 0.362 |
| Vash1 | 0.71 | 1.64 | 0.041 | 0.362 |
| Il1rl2 | 0.79 | 1.73 | 0.044 | 0.362 |
| Ang4 | 1.19 | 2.28 | 0.046 | 0.362 |
Figure 2.
Differential expression of top ten candidate angiogenic genes in mouse model. Specifically, gene expression is depicted in high fat diet mice versus low fat diet mice within the control group of no metformin treatment. n=5 mice per group. Each dot represents one mouse. Blue dots represent mice fed a low fat diet and red dots represent mice fed a high fat diet.
Further exploratory analysis of this list of 185 genes to determine an interaction effect between diet and metformin treatment on gene expression identified four proangiogenic genes: angiopoietin 4 (Ang4) (adjusted p=0.001), notch4 (Notch 4) (adjusted p=0.03), chromogranin A (Chga) (adjusted p=0.04), Vegfa (adjusted p=0.04) (Supplemental Figure 1). Notably, there was also an interaction effect between diet and metformin treatment on gene expression of Edil3 (unadjusted p=0.009), but this did not maintain significance when adjusted.
Association between obesity and angiogenic-related genes in copy number high endometrial cancer
Twenty-two candidate angiogenic-related genes from the mouse analysis were mapped to 21 human genes and analyzed for the association of gene expression and log(BMI) in an exploratory analysis of the CNH cohort (N=60). Only three genes demonstrated unadjusted p < 0.05 and concordant direction of effect: tenomodulin (TNMD) (1448 FC, unadjusted p=3.4 x 10−9), ANG (2.7 FC, unadjusted p=0.041), and EDIL3 (2.5 FC, unadjusted p=0.048). An exploratory analysis of the 168 genes in the CNH TCGA cohort demonstrated that 29 genes were differentially expressed in association with log(BMI) (unadjusted p<0.05). (Supplemental Table 3)
Association between obesity and EDIL3 expression in the GYN-COE validation cohort
An independent cohort of 62 patients was evaluated for validation (see table 1 for disease status and characteristics of this cohort). One of four EDIL3 probesets demonstrated an association between log(BMI) and increased gene expression (adjusted p = 0.0342). The aggregate of expression of the four EDIL3 probesets was not statistically significant. CHGA (adjusted p=0.0250) as well as PLAU (adjusted p=0.0284), IL5RA (adjusted p=0.0284), and B2M (adjusted p=0.0284) demonstrated significant differential gene expression in this validation cohort.
DISCUSSION
Improving our understanding of the tumor microenvironment, specifically of angiogenic pathways, may assist with the identification of novel targets in endometrial cancer, particularly in obese patients who are most often affected. Our data suggest that obesity may be associated with differential expression of angiogenic-related genes in tumor specimens in women with endometrial cancer and in the innovative endometrial cancer mouse model. Many of these angiogenic-related genes are known to promote a pro-angiogenic tumor microenvironment and have been successfully targeted by anti-angiogenic therapies. Some of our candidate genes also have a role in immunology.
The top hit in our analyses was EDIL3. The EDIL3 gene encodes an extracellular matrix protein secreted by endothelial cells and is associated with vascular remodeling during angiogenesis. This protein has been demonstrated to play a role in the enhancement of cell invasion and acceleration of lung metastases in breast cancer mouse models as well as human patients with metastatic breast cancer.24 EDIL3 expression is up-regulated in pancreatic ductal adenocarcinoma25, hepatocellular carcinoma26, and increased expression is correlated with decreased overall survival in these patients. Similar findings have been demonstrated in colorectal malignancies27 and high-grade bladder cancers.28 To our knowledge, this is the first reporting of the association between obesity, defined as BMI, and EDIL3 in endometrial cancer patients. The findings regarding association between BMI and Edil3 expression were validated in one probeset using the independent GYN-COE cohort; therefore, we plan to prioritize further evaluation of this gene in future studies in independent endometrial cancer cohorts and mechanistic studies of Edil3 function.
In addition, our data identified multiple candidate genes from the TCGA dataset including matrix metallopeptidase 14 (MMP14), jagged 1 (JAG1), transforming growth factor beta 2 (TGFB2), TIMP metallopeptidase inhibitor 1 (TIMP1), interleukin 1 beta (IL1B), collagen type XV alpha 1 chain (COL15A1), fms related tyrosine kinase 4 (FLT4), hes related family bHLH transcription factor with YRPW motif 1 (HEY1), C-X-C motif chemokine ligand 12 (CXCL12), and platelet derived growth factor receptor beta (PDGFRB) which all demonstrated increased expression with increasing BMI. We were unable to validate our findings in the mice given limited sample size and large number of genes in our analysis. However, this exploratory analysis revealed genes of interest warranting further exploration in endometrial cancer. For example, JAG1 is an essential ligand in the Notch signaling pathway, which is fundamental in cell-cell communication and cellular differentiation.29 It has recently been implicated in endometrial carcinogenesis and may portend a poorer prognosis compared to patients without high expression levels.30 In breast cancer mouse models, increased Jag1 levels were demonstrated in obese, compared to lean, mice (as were several other components of the Notch pathway).31 Notch signaling inhibitors, particularly with gamma-secretase inhibitors (GSIs) are currently being investigated in phase I and II clinical trials in a variety of solid malignancies.32 Additionally, CXCL12 and its receptor CXCR4 have been implicated in malignant transformation, invasion, and metastasis in multiple cancer types including pancreatic cancer33 and glioblastomas.34 In fact, NOX-A12 (olaptesed pegol), an inhibitor of CXCL12, is currently under development for treating multiple types of malignancies. These novel therapeutics may warrant further investigation in endometrial cancers as well.
Our exploratory analysis in the mouse model suggests that several potent pro-angiogenic genes are overexpressed in mice subjected to diet-induced obesity, including four-fold increased Lep and approximate three-fold increased Vegfa. Lep encodes the protein leptin, which is made by adipose cells and regulates energy balance and signals satiety. Increasingly, leptin’s pro-angiogenic effects in cancer have been suggested to play a role in breast cancer.35 Vegfa encodes a protein critical in angiogenesis, inducing proliferation and migration of vascular endothelial cells. It is upregulated in many solid malignancies and is associated with poor outcome in endometrioid endometrial cancers.36 While anti-angiogenic agents targeting VEGFA (bevacizumab) are known active biologic therapies in various cancers including endometrial cancer, further studies are needed to elucidate the timing and circumstance of their optimal use. Aghajanian et al. reported single agent activity of bevacizumab in recurrent endometrial cancer.37 Combination bevacizumab with chemotherapy was associated with increased overall survival in the GOG-86P study (NCT00977574)38, as well as increased progression free survival and overall response rate in the MITO Group END-2 trial.39 We are currently investigating the efficacy of bevacizumab in obese versus non-obese patients in women who participated in GOG-86P in a multi-institutional exploratory analysis.
Metformin is a long-standing diabetic agent that has recently been identified as an antitumor therapy. Preliminary data from GOG286B evaluating combination paclitaxel, carboplatin, and metformin did not demonstrate improved disease control with the addition of metformin (verbal communication, NRG Oncology meeting January 2018); however, final results are pending and this analysis was performed in an unselected patient population. Therefore, the benefit of metformin has yet to be determined in obese women with endometrial cancer. Given that metformin may decrease endometrial cancer cell growth via inhibition of mTOR signaling, and that the mTOR pathway plays a central role in regulating angiogenesis, we evaluated if an interaction effect exists between diet and metformin treatment with respect to angiogenic-related gene expression. In an exploratory analysis of our primary set of 23 mouse genes, treatment with metformin did demonstrate reduction of Edil3 expression in mice fed a high fat diet. Expression of several additional pro-angiogenic genes (Notch 4, Chga, Vegfa, Tnni1, Fgfr3, and Tgfb1) was reduced with metformin treatment in mice fed a high fat diet; however, the effects are not consistent. Metformin could play a role in mitigating the effect of obesity on increased angiogenic-related gene expression. Alternatively, this may be a spurious association rather than an off-target metformin effect. Further analysis using matrigel angiogenesis assays may help determine if metformin has an anti-angiogenic effect.
There were several limitations to our study, including limitations inherent to the TCGA database with only short-term clinical follow-up, often limited or missing clinical data, and tissue processing occurring in multiple institutions. The sample size of our mouse model was quite small with only five mice in each of the four groups. With such a small sample size of mice, our power was limited to definitively validate candidate genes or the interaction effect of high fat diet and metformin treatment on gene expression. Furthermore, several genes noted to be significant by definition had extreme outlying expression values, potentially distorting the outcome. Additionally, there was overlap between only one gene of interest in the human TCGA data and the mouse model. Finding overlap in cross-species studies can be difficult given differences in human exposures, environmental effects, and baseline physiology that are more uniform in mouse models.
Despite these limitations, our study adds to the growing literature surrounding the tumor microenvironment and obesity-driven modulation of angiogenic pathways. Specifically, our results provide additional evidence of an association between obesity, diet, and differential angiogenic-related gene expression suggesting that obesity and high fat diet may promote tumor progression through differential regulation of angiogenic pathways in endometrioid endometrial cancer. Our results, while exploratory, identified several candidate genes of interest in the human and mouse analyses and specifically uncovered a common gene, EDIL3, that was overexpressed in obese humans and mice subjected to diet-induced obesity. EDIL3 may play an important role in this microenvironment and serve as a novel target that should be further explored in future studies. While metformin’s role with these specific genes still needs further elucidation, our study suggests that metformin may influence the effect of diet on modulating angiogenic-related gene expression. However, further mechanistic studies are needed to determine if metformin can reduce diet-induced obesity driven overexpression of pro-angiogenic genes in endometrial cancer.
Supplementary Material
ACKNOWLEDGEMENTS
As Duke Cancer Institute members, we acknowledge support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: P30 CA014236), specifically the Duke Cancer Institute’s Bioinformatics Shared Resource. This work was also supported by the Charles B. Hammond Research Fund through the Duke University Department of Obstetrics and Gynecology.
Footnotes
CONFLICT OF INTEREST
The authors have no relevant conflicts of interest to report.
REFERENCES
- 1.Chang SC, Lacey JV Jr., Brinton LA, et al. Lifetime weight history and endometrial cancer risk by type of menopausal hormone use in the NIH-AARP diet and health study. Cancer Epidemiol Biomarkers Prev. 2007;16(4):723–730. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 3.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. The New England journal of medicine. 2003;348(17):1625–1638. [DOI] [PubMed] [Google Scholar]
- 4.Roque DR ML, Chen TH, Rashid N, Hayes DN, Bae-Jump V. Association between differential gene expression and body mass index among endometrial cancers from The Cancer Genome Atlas Project. Gynecol Oncol. 2016;16:30811–30813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giatromanolaki A, Sivridis E, Brekken R, et al. The angiogenic "vascular endothelial growth factor/flk-1(KDR) receptor" pathway in patients with endometrial carcinoma: prognostic and therapeutic implications. Cancer. 2001;92(10):2569–2577. [DOI] [PubMed] [Google Scholar]
- 6.Chen CA, Cheng WF, Lee CN, et al. Cytosol vascular endothelial growth factor in endometrial carcinoma: correlation with disease-free survival. Gynecologic oncology. 2001;80(2):207–212. [DOI] [PubMed] [Google Scholar]
- 7.Gu JW, Young E, Patterson SG, et al. Postmenopausal obesity promotes tumor angiogenesis and breast cancer progression in mice. Cancer biology & therapy. 2011;11(10):910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H, Kong W, Zhang L, et al. Reversal of obesity-driven aggressiveness of endometrial cancer by metformin. American journal of cancer research. 2019;9(10):2170–2193. [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koenker R. quantreg: Quantile Regression. R package version 5.35. https://CRAN.R-project.org/package=quantreg. 2018.
- 12.Fan J, Li Runze. Variable Selection via Nonconcave Penalized Likelihood and its Oracle Properties. Journal of the American Statistical Association. 2001;96(456):1348–1360. [Google Scholar]
- 13.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 16.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England). 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonanni B, Puntoni M, Cazzaniga M, et al. Dual effect of metformin on breast cancer proliferation in a randomized presurgical trial. J Clin Oncol. 2012;30(21):2593–2600. [DOI] [PubMed] [Google Scholar]
- 19.MI Love, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Co NN, Iglesias D, Celestino J, et al. Loss of LKB1 in high-grade endometrial carcinoma: LKB1 is a novel transcriptional target of p53. Cancer. 2014;120(22):3457–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffen D SP, Birney E, Huber W. Nature Protocols 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickham H. tidyverse: Easily Install and Load the 'Tidyverse'. R package version 1.2.1. https://CRAN.R-project.org/package=tidyverse. 2017.
- 23.Xie Y. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.20. 2018. [Google Scholar]
- 24.Lee JE, Moon PG, Cho YE, et al. Identification of EDIL3 on extracellular vesicles involved in breast cancer cell invasion. Journal of proteomics. 2016;131:17–28. [DOI] [PubMed] [Google Scholar]
- 25.Jiang SH, Wang Y, Yang JY, et al. Overexpressed EDIL3 predicts poor prognosis and promotes anchorage-independent tumor growth in human pancreatic cancer. Oncotarget. 2016;7(4):4226–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun JC, Liang XT, Pan K, et al. High expression level of EDIL3 in HCC predicts poor prognosis of HCC patients. World J Gastroenterol. 2010;16(36):4611–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou X, Qiao H, Jiang X, Dong X, Jiang H, Sun X. Downregulation of developmentally regulated endothelial cell locus-1 inhibits the growth of colon cancer. Journal of biomedical science. 2009;16:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beckham CJ, Olsen J, Yin PN, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. The Journal of urology. 2014;192(2):583–592. [DOI] [PubMed] [Google Scholar]
- 29.Braune EB, Lendahl U. Notch -- a goldilocks signaling pathway in disease and cancer therapy. Discov Med. 2016;21(115):189–196. [PubMed] [Google Scholar]
- 30.Mitsuhashi Y, Horiuchi A, Miyamoto T, Kashima H, Suzuki A, Shiozawa T. Prognostic significance of Notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology. 2012;60(5):826–837. [DOI] [PubMed] [Google Scholar]
- 31.Battle M, Gillespie C, Quarshie A, et al. Obesity induced a leptin-Notch signaling axis in breast cancer. International journal of cancer Journal international du cancer. 2014;134(7):1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan X, Wu H, Xu H, et al. Notch signaling: An emerging therapeutic target for cancer treatment. Cancer Letters. 2015;369(1):20–27. [DOI] [PubMed] [Google Scholar]
- 33.Sleightholm RL, Neilsen BK, Li J, et al. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacology & therapeutics. 2017;179:158–170. [DOI] [PubMed] [Google Scholar]
- 34.Wang S, Zhang S, Li J, et al. CXCL12-induced upregulation of FOXM1 expression promotes human glioblastoma cell invasion. Biochemical and biophysical research communications. 2014;447(1):1–6. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Perez RR, Lanier V, Newman G. Leptin’s Pro-Angiogenic Signature in Breast Cancer. Cancers. 2013;5(3):1140–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamat AA, Merritt WM, Coffey D, et al. Clinical and biological significance of vascular endothelial growth factor in endometrial cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(24):7487–7495. [DOI] [PubMed] [Google Scholar]
- 37.Aghajanian C, Sill MW, Darcy KM, et al. Phase II trial of bevacizumab in recurrent or persistent endometrial cancer: a Gynecologic Oncology Group study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(16):2259–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aghajanian C, Filiaci VL, Dizon DS, et al. A randomized phase II study of paclitaxel/carboplatin/bevacizumab, paclitaxel/carboplatin/temsirolimus and ixabepilone/carboplatin/bevacizumab as initial therapy for measurable stage III or IVA, stage IVB or recurrent endometrial cancer, GOG-86P. Journal of Clinical Oncology. 2015;33(15_suppl):5500–5500. [Google Scholar]
- 39.Lorusso D, Ferrandina G, Colombo N, et al. Randomized phase II trial of carboplatin-paclitaxel (CP) compared to carboplatin-paclitaxel-bevacizumab (CP-B) in advanced (stage III-IV) or recurrent endometrial cancer: The MITO END-2 trial. Journal of Clinical Oncology. 2015;33(15_suppl):5502–5502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.