Summary
Human pluripotent stem cells (hPSCs) hold great promise for applications in regenerative medicine and disease modeling. Here, we present a protocol for establishing edited hPSC cell lines utilizing visualized orthogonal selective reporters. We describe steps for constructing plasmids, carrying out cell culture and electroporation, as well as performing drug-fluorescent dual enrichment, clone screening, and cell line characterization. This protocol facilitates the achievement of single-base homozygous mutations and reporter knockins, offering a reliable approach for precision genome editing.
Subject areas: Flow Cytometry, Molecular Biology, CRISPR, Stem Cells, Cell Differentiation
Graphical abstract

Highlights
-
•
Steps to perform CRISPR editing in hPSC from plasmid design to final characterization
-
•
Construct ready-to-use plasmid cloning backbones for genome editing
-
•
Guidance on drug-fluorescent dual selection to facilitate enrichment and clone picking
-
•
Characterization on the established cell lines
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Human pluripotent stem cells (hPSCs) hold great promise for applications in regenerative medicine and disease modeling. Here, we present a protocol for establishing edited hPSC cell lines utilizing visualized orthogonal selective reporters. We describe steps for constructing plasmids, carrying out cell culture and electroporation, as well as performing drug-fluorescent dual enrichment, clone screening, and cell line characterization. This protocol facilitates the achievement of single-base homozygous mutations and reporter knockins, offering a reliable approach for precision genome editing.
Before you begin
Genome editing via CRISPR-Cas9 system is now a routinely-applied tool.1 Yet, it is still challenging to monitor the whole enrichment process and pick a high-purity clone. To make it more user-friendly, this protocol provides a visualized genome editing approach utilizing orthogonal selective reporters to facilitate positive cell enrichment and clone picking. It is compatible with introducing heterozygous/homozygous mutations, tag insertions and reporter knock-ins. Here we demonstrate three examples to show the versatility of the approach: introducing homozygous mutation on B2M splice donor (SD), establishing a SOX17-NLS-tdTomato reporter line, and an INS-GFP-CD19 reporter line for pancreatic β cell induction. We will focus on the establishment of the B2M SD mutation cell line as it is more complicated.
B2M encodes β-2-immunoglobulin that forms heterodimer with human leukocyte antigen (HLA) class I protein (HLA-I). Since B2M knock-out led to HLA-I depletion, this strategy is for delaying graft rejection after allogeneic cell transplantation, which is meaningful for cell therapy.2 While a prior study using modRNA-based base editing to achieve this objective, the approach caused an unintended base edit occurring 4 base pairs away.3 Thus, we employ our editing approach to knock-out B2M via introducing a homozygous mutation on the splice donor (SD) located at the front of the first intron and establish a pure, scarless-edited hPSC line. SOX17 and insulin are the key markers during induction of pancreatic β cells.4,5 Thus, we utilized this approach to generate the reporter lines for direct monitoring of β cell differentiation and further purification of insulin-positive cells through fluorescence or magnetic-activated cell sorting (FACS or MACS).
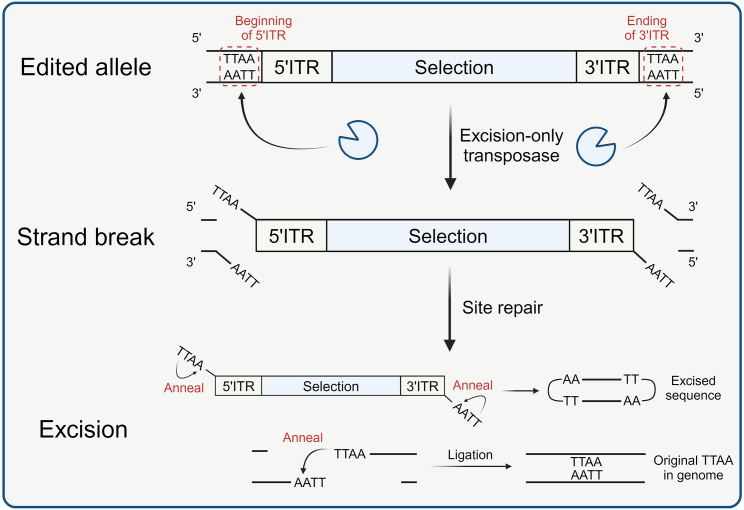
The conventional protocols for knock-in in genome editing typically employ a loxP-flanked (floxed) drug-resistant gene expression cassette to enrich targeted cells. For instance, a protocol utilizes the floxed PGK-Puro cassette to generate a TBXT-H2B-EGFP hPSC reporter line.6 However, the Cre-loxP excision system is incompatible with double knock-in procedures, as the presence of two floxed sites on different chromosomes may lead to translocation rather than deletion.7 Consequently, we have devised a piggyBac inverted terminal repeat (ITR)-flanked selection cassette for biallelic targeting, as it can be accurately excised after enrichment. Additionally, the use of a floxed cassette is not suitable for achieving scarless editing when introducing mutations. The Cre recombinase excision leaves a loxP sequence behind, potentially disrupting the coding sequence or regulatory elements. In contrast, the excision of the ITR-flanked sequence by piggyBac transposase leaves only a 4-bp TTAA scar.8 Identifying the TTAA sites in the genome and excluding the 4 bp sequence from the knock-in sequence allows for replenishing the sequence after ITR-flanked selection cassette excision, making the entire genome editing process scarless (Scheme 1). As a result, our ITR-flanked backbones offer a versatile solution for introducing mutations and routine reporter knock-ins.
Scheme 1.
Schematic of ITR-mediated excision via excision-only piggyBac transposase
The process involves the 4-bp "TTAA" motif located at the beginning of the 5′ITR and the end of the 3′ITR. Upon recognition of the ITR sequence, the transposase induces strand breaks, resulting in the generation of "TTAA" or "AATT" overhangs at the 5′ terminus of the edited allele and the ITR-flanked sequence. Subsequently, these overhangs anneal, facilitating the ligation of the targeted allele and leaving behind the 4-bp “TTAA” originally located in the genome. Moreover, the ITR-flanked sequence forms a hairpin structure and will not integrate back into the genome by the integration-deficient transposase.
The protocol below describes the detailed steps using human embryonic stem cell (hESC) line H1.9 This approach also achieved several successful editing on human induced pluripotent stem cell (hiPSC). Also, pluripotency and differentiation potential are monitored in the genetic modified cell lines.
Donor plasmid and sgRNA design
Timing: 1 day
This part of the protocol explains how to design the plasmid donor as well as get the primer pairs and sgRNA sequence via online tools. We utilize two established backbones for efficient cloning of donor plasmids (Figure 1A). Generally, the homology arm (HA) sequences can be inserted via digestion of the backbones with indicated restriction enzymes and ligation by Gibson Assembly.10
Note: Several restriction sites are located upstream or downstream of plasmid features. For cloning of HA sequences, digest the backbones with KpnI/NruI for left arm cloning, and HpaI/EcoRI or HpaI alone for right arm cloning. For scarless mutation introducing, the backbones can be digested with KpnI/EcoRV for the ligation of left arm and piggyBac 5′ inverted terminal repeat (5′ITR) seamlessly. For the exchange of antibiotic-resistant genes and fluorescent proteins, HindIII, NheI, XbaI and PmeI can be utilized.
-
1.Genome region annotation for HA and sgRNA design.Note: Acquire the genomic sequence encompassing exons, introns and UTRs of the target gene from reputable sources such as NCBI or Ensembl. Add features via SnapGene. This will facilitate defining the sgRNA targeting site.
-
a.Identify the location where the desired mutation is to be introduced.
-
b.To maintain allele integrity, exclude a 4-bp TTAA site from the homology arm to compensate for redundant TTAA sequences post-transposase excision.Note: Typically, TTAA sites can be found near the target site, and the nearest one is selected as the center of the homology arm and the sgRNA target site (Figure 1B).Note: Our backbones are compatible with 3′ knockin. Since the TTAA scar will normally not affect the expression of the edited allele, we suggest to take stop codon as the center of the homology arm as most knockin strategies do.
-
a.
-
2.Design the HA for PCR amplification.Note: Instead of de novo synthesis of the HA sequence, we amplify the HA from the extracted genomic DNA to accelerate plasmid construction (Figure 1C). Thus, it is essential to define a primer pair for efficient amplification.
-
a.Use Primer-BLAST to design the primer pairs. Copy the 1000-bp sequence located upstream and downstream of the target site into the “PCR Template” region.
-
b.Set the screening ranges of the primers (Forward primer: from 1 to 400, Reverse primer: from 1600 to 2000).
-
c.Set the PCR product size (Min: 1200, Max: 2000).
-
d.Set the database in “Primer Pair Specificity Checking Parameters” as Refseq representative genomes.
-
e.Pick one outputs for further PCR amplification.Note: We recommend setting the length of HA between 600 and 1000 bp, striking a balance between homology-directed repair (HDR) efficiency and the complexity of plasmid construction.Note: In a few cases, the designed primer pair does not work well and strong unspecific amplifications will be observed. More outputs from Primer-BLAST can be tested.
-
a.
-
3.Design the sgRNA using CRISPOR.Note: Given that the distance between the Cas9 cutting site and the HA center plays a crucial role in HDR efficiency, we recommend screening for sgRNAs within 40 base pairs upstream and downstream of the target site.11
-
a.Copy the 40-bp sequence located upstream and downstream of the target site as the input.
-
b.Select the genome as Homo sapiens GRCh37/hg19 or GRCh38/hg38.
-
c.Select the Protospacer Adjacent Motif (PAM) as 20bp-NGG - SpCas9.
-
d.Picking a sgRNA owning both specificity and efficiency.Note: The position of sgRNA determines whether the PAM mutation should be included in HA, as illustrated in Figure 1D. Mutate the second or the third “G” of the “NGG” PAM sequence can prevent the retargeting of the CRISPR system on the edited allele, which may cause potential insertions and deletions (indels). But if the sgRNA targeting sequence or PAM sequence are disrupted after HDR, introduce PAM mutation is optional.Note: For some genes, the sequence around the target site is A-T rich, which is hard to find PAM sequence. Thus, the screening range can be extended to 50-bp or 60-bp.
-
a.
Figure 1.
Design and cloning strategy of the donor and sgRNA plasmid
(A) Plasmid structure of the backbones and the constructed donors, and the process of cloning.
(B) A snapshot in SnapGene software to show the TTAA site and the splice donor site in the B2M locus, and the design of homology arm (HA) and sgRNA.
(C) Strategy to amplify HA and introduce point mutation in the PCR product.
(D) Two situations to determine whether the PAM mutation should be included in HA.
hPSC culture
Timing: 2 weeks
This part of the protocol describes the routine maintenance and passaging of hPSC cell lines.
-
4.Passage the hPSC in feeder-free conditions when reaches 70%–80% confluency by dissociate the cells into small aggregates using non-enzymatic dissociation buffer.12 After thawing the cryopreserved cells, passage at least twice before electroporation.
-
a.Thaw one bottle of hESC-qualified Matrigel on ice. Add 1 mL Matrigel into 500 mL DMEM/F12 to obtain a diluted Matrigel solution (1:500). Add 1 mL of the diluted solution into each well of a 6-well plate and place the plate at room temperature or 37°C for 30 min to coat the plate.Note: We find that the concentration of the Matrigel will not affect the coating when diluting at 1:500.
-
b.Warm-up mTeSR1 medium supplemented with 5 μM Y-27632, PBS and hPSC dissociation buffer at room temperature for at least 30 min before using.
-
c.Swirl the plate to collect the dead cells and aspirate the medium. Wash the cells with 1 mL PBS once and add 1 mL hPSC dissociation buffer. Place the plate in the 37°C incubator for 6–7 min.Optional: Gentle Cell Dissociation Reagent (GCDR) or Versene Solution can be good alternatives.Note: The incubation time may vary between different hPSC lines from 6 min to 10 min.
-
d.Aspirate the hPSC dissociation buffer and add 1 mL mTeSR1 medium supplemented with 5 μM Y-27632 to collect the cells.
-
e.Aspirate the coating solution and split the cells as 1:6. Adjust the volume of the cell suspension and pipette 2 mL suspension per well.
-
f.The next day, change the medium to 2 mL mTeSR1 per well. Renew the medium every day. Cells are maintained at 37°C in humidified atmosphere of 5% CO2.
 CRITICAL: Use healthy and proliferative hPSCs for further electroporation. Cells should be routinely checked to avoid mycoplasma contamination.
CRITICAL: Use healthy and proliferative hPSCs for further electroporation. Cells should be routinely checked to avoid mycoplasma contamination.
-
a.
FACS sorter setup
Timing: 30 min
This part of the protocol describes the preset of flow cytometer and gating strategy for sorting correctly knock-in cells or selection cassette-excised cells.
-
5.Set up the flow cytometer for single cell sorting. The steps below show how to set up BD FACSymphony S6 flow cytometer.
-
a.Perform routinely instrument maintenance and quality control according to the instruction of the manufacturer (https://www.bdbiosciences.com/content/dam/bdb/marketing-documents/products-pdf-folder/instruments/research-cell-sorters/UG-Symphony-S6.pdf).Optional: If the flow cytometer is not maintained in sterile condition, flush the fluidics line with 75% alcohol for 5 min and water with the maximum flow rate for 10 min instead.
-
b.Cool down the sample chamber of the sorting system to 4°C.
-
c.Put a 15-mL centrifuge tube containing 1 mL mTeSR1 medium supplemented with 5 μM Y-27632 inside the sample chamber to receive the sorted cell.
-
d.Load the sample and gate the desired population.
-
i.Gate on the hPSC and single cell population first (Figure 2A).
-
ii.For sorting cells with only EGFP or tdTomato, set the 488 nm laser with 515/30 filter for GFP and 561 nm laser with 610/20 filter for tdTomato. Gate on the cells with the strongest fluorescent. These cells will normally form a clear population (Figure 2B, left and middle panel).
-
iii.For sorting cells with dual fluorescent, set the laser and filter as described above. Gate on the double-positive population (Figure 2B, right panel).
-
iv.For sorting cells excised with piggyBac excision-only transposase, set the laser and filter as described above. Gate on the double-negative population and sort only a small part to avoid potential conflicted positive cells (Figure 2C).Note: Since the cells are collected as a bulk, staining the cell with DAPI to discriminate dead and alive cells is optional. Unstained control samples are dispensable due to the obvious clustering of positive cells.
-
i.
-
e.Proceed with single cell sorting using a flow rate under 4. Collect at least 0.2 million cells per sample.
-
a.
Figure 2.
Cell sorting strategy of single-fluorescent or dual-fluorescent cells
Representative flow cytometry dot plots showing (A) The total hPSC population (left) and single cell population (right); (B) Gating strategies for sorting EGFP positive cells or tdTomato positive cells or double positive cells.
(C) Gating strategies for sorting excision-only piggyBac transposase excised cell population. Double-negative cells are gating and only a small fraction of them is sorted to avoid conflicted cells.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-human β2-microglobulin antibody (dilution 1:100) | BioLegend | Cat#316311 |
| Mouse anti-Oct3/4 primary antibody (dilution 1:200) | Santa Cruz Biotechnology | Cat#sc-5279 |
| Rabbit anti-NANOG primary antibody (dilution 1:800) | Cell Signaling Technology | Cat#3580 |
| Donkey anti-mouse IgG (H + L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (dilution 1:500) | Thermo Fisher Scientific | Cat#A21206 |
| Donkey anti-rabbit IgG (H + L) highly cross-adsorbed secondary antibody, Alexa Fluor 568 (dilution 1:500) | Thermo Fisher Scientific | Cat#A10042 |
| Chemicals, peptides, and recombinant proteins | ||
| PrimeSTAR Max DNA polymerase | Takara | Cat#R045A |
| 2 × rapid Taq master mix | Vazyme | Cat#P222 |
| BbsI-HF | New England Biolabs | Cat#R3539 |
| EcoRI-HF | New England Biolabs | Cat#R3101 |
| KpnI-HF | New England Biolabs | Cat#R3142 |
| EcoRV-HF | New England Biolabs | Cat#R3195 |
| HindIII-HF | New England Biolabs | Cat#R3104 |
| XbaI | New England Biolabs | Cat#R0145 |
| HpaI | New England Biolabs | Cat#R0105 |
| T4 DNA ligase | New England Biolabs | Cat#M0202 |
| EDTA | Thermo Fisher Scientific | Cat15575020 |
| Bovine serum albumin | Proliant | Cat#68700 |
| Matrigel hESC-qualified matrix, LDEV-free | Corning | Cat#354277 |
| hPSC dissociation buffer | Nuwacell | Cat#RP01007 |
| mTeSR1 | STEMCELL | Cat#85850 |
| TrypLE express enzyme (1X), no phenol red | Thermo Fisher Scientific | Cat#12604021 |
| Penicillin-streptomycin | Procell | Cat#PB180120 |
| DMEM/F12 | Thermo Fisher Scientific | Cat#C11330500BT |
| Y-27632 | Topscience | Cat#T1870 |
| Puromycin | InvivoGen | Cat#ant-pr |
| Hygromycin B Gold | InvivoGen | Cat#ant-hg |
| Critical commercial assays | ||
| FastPure blood/cell/tissue/bacteria DNA isolation mini kit | Vazyme | Cat#DC112 |
| Mix & Go E. coli transformation kit & buffer set | Zymo Research | Cat#T3002 |
| FastPure Gel DNA extraction mini kit | Vazyme | Cat#DC301 |
| ClonExpress ultra one step cloning kit | Vazyme | Cat#C115 |
| Experimental models: Cell lines | ||
| Human embryonic stem cell line H1 | WiCell | WA01 |
| Oligonucleotides | ||
| B2M-HA-F | This paper | CACTAAAATTGCCGAGCCCTT |
| B2M-HA-R | This paper | GGACATGCGAACTTAGCGGG |
| B2M-HAL-F | This paper | CTTTCGTCGGCGCGGGTACC CACTAAAATTGCCGAGCCCT |
| B2M-HAL-R | This paper | GACTATCTTTCTAGGGTTAAACGCGTGCCCAGC |
| ITR-F | This paper | TTAACCCTAGAAAGATAGTCTGCGTAAAATTG |
| ITR-R | This paper | ATGGCTGTCCCTGATATCTATAACAAGAAAA TATATATATAATAAGTTAT |
| B2M-HAR-F1 | This paper | GATTATCTTTCTAGGGTTAATATAAGTGGAG GCGTCGCG |
| B2M-HAR-R1 | This paper | GCGCTGGATAGCCTCCAGG |
| B2M-HAR-F2 | This paper | GAGGCTATCCAGCGCGAGTCTCTCCTACCC TCCCG |
| B2M-HAR-R2 | This paper | CTTTTGCTCACATGGAATTCGGACATGCGA ACTTAGCGG |
| Hygro-tdT-F | This paper | TTCAGGTGTCGTGAAAGCTTGCCACCATG AAAAAGCCTGA |
| Hygro-tdT-R | This paper | TTAAACGGGCCCTCTAGATTACTTGTACAG CTCGTCCATG |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-Puro (PX459) V2.0 | Ran et al.1 | Addgene #62988 |
| SOX17-3′gRNA | This paper | Addgene #210465 |
| SOX17-NLS-tdTomato-EPG | This paper | Addgene #210466 |
| INS-3’gRNA | This paper | Addgene #210468 |
| INS-GFP-CD19-EPT | This paper | Addgene #210469 |
| B2M-SDMutation-EPG | This paper | Addgene #215545 |
| B2M-SDMutation-EhT | This paper | Addgene #215546 |
| B2M-SDMutation-gRNA | This paper | Addgene #215547 |
| P2A-NLS-tdtomato-EPG backbone | This paper | Addgene #215548 |
| P2A-mCherry-EhT backbone | This paper | Addgene #215549 |
| Software and algorithms | ||
| CRISPOR | TEFOR infrastructure | http://crispor.tefor.net/crispor.py |
| Primer-BLAST | NCBI | https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi |
| FlowJo software | FlowJo (Becton Dickinson) | https://www.flowjo.com/ |
| Other | ||
| Tissue culture plate, 6-well | Jet | Cat#TCP010006 |
| Tissue culture plate, 12-well | Jet | Cat#TCP010012 |
| Tissue culture plate, 24-well | Jet | Cat#TCP010024 |
| 15 mL centrifuge tube | LabSelect | Cat#CT-002-15 |
| 50 mL centrifuge tube | Jet | Cat#CFT011500 |
| Cryogenic tube | Corning | Cat#430659 |
| 5 mL serological pipette | Jet | Cat#GSP010005 |
| 10 mL serological pipette | Jet | Cat#GSP010010 |
| Lonza Nucleofector 2b | Lonza | N/A |
| TGem Pro spectrophotometer | Tiangen | N/A |
| FACSymphony S6 cell sorter | BD Biosciences | N/A |
Materials and equipment
Cell freezing medium
| Reagent | Final concentration | Amount |
|---|---|---|
| mTeSR | N/A | 9 mL |
| Y27632 (5 mM) | 5 μM | 10 μL |
| DMSO | 10% | 1 mL |
| Total | N/A | 10 mL |
Prepare fresh, sterile.
Blocking buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 × PBS | N/A | 50 mL |
| Bovine serum albumin | 2% | 1 g |
| Total | N/A | 50 mL |
Store at 4°C for up to 3 months.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 × PBS | N/A | 50 mL |
| Bovine serum albumin | 0.5% | 0.25 g |
| EDTA, 0.5 M | 5 mM | 500 μL |
| Total | N/A | 50 mL |
Store at 4°C for up to 3 months.
Step-by-step method details
Donor plasmid and sgRNA plasmid cloning
Timing: 1–2 weeks
In this section, we describe steps for restriction cloning of sgRNA plasmid and Gibson Assembly of the donor plasmid.
-
1.Cloning of sgRNA plasmid.
-
a.For the designed sgRNA, order a forward oligo (100 μM) with a 5′ CACC overhang and a reverse oligo (100 μM) with a 5′ AAAC overhang.
-
b.Mix 1 μL of each oligo together with 7 μL ddH2O and 1 μL 10 × T4 Ligation Buffer. Vortex to mix well. Anneal in a thermocycler using the following program:
Steps Temperature Time Cycles Initial Denaturation 95°C 3 min 1 Denaturation 94°C (−1°C/cycle) 10 s 70 cycles Hold 4°C forever -
c.Digest pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene #62988) with restriction enzyme BbsI for sgRNA cloning.
Reagent Amount Plasmid backbone 3 μg 10 × rCutSmart Buffer 4 μL BbsI-HF 3 μL ddH2O Up to 40 μL Digest in a thermocycle at 37°C for 1 h. -
d.Purify the digested backbone with a commercial kit (e.g., FastPure Gel DNA Extraction Mini Kit).
-
e.Ligate the annealed sgRNA duplex with the backbone:
Reagent Amount Digested PX459 1 μL sgRNA duplex 1 μL 10 × T4 Ligation Buffer 1 μL T4 DNA Ligase 1 μL ddH2O Up to 10 μL React in a thermocycle at 21°C for 30 min. -
f.The product is ready for transformation.
-
a.
-
2.Cloning the left HA into backbone for donor plasmid construction.Note: As described previously, we utilize two backbones containing a reporter construct and ITR-flanked selection cassette to efficiently clone donor plasmid. Here, we describe the cloning process of the donor plasmids for introducing B2M SD mutation.
-
a.PCR amplify the HA sequence.Note: Utilize a high-fidelity polymerase for PCR amplification of the HA from the extracted genome. To account for potential variations in the genome, especially among different hPSC lines, we recommended to use the genome of the specific cell undergoing genome editing. Here, we use genome of H1 hESC as template.
Reagent Amount Genome of H1 hESC 100 ng PrimeSTAR Max Premix (2×) 12.5 μL B2M-HA-F (10 μM) 1 μL B2M-HA-R (10 μM) 1 μL ddH2O Up to 25 μL Use touch-down PCR program to enable specific amplification of HA:Steps Temperature Time Cycles Initial Denaturation 98°C 2 min 1 Denaturation 98°C 10 s 20 cycles Annealing 62°C (−0.5°C /cycle) 5 s Extension 72°C 1 s per 100 bp Denaturation 98°C 10 s 10 cycles Annealing 50°C 5 s Extension 72°C 1 s per 100 bp Final extension 72°C 5 min 1 Hold 4°C forever Note: Unless otherwise specified, use the PCR program above to efficiently amplify DNA fragments for plasmid cloning. -
b.After the PCR reaction, add 5 μL gel loading dye, thoroughly mix, and separate the PCR product on a 1.5% agarose gel.
-
c.Excise the correct band and recover the DNA using a commercial kit.
-
d.Amplify left HA with primer pair B2M-HAL-F/B2M-HAL-R and 5′ITR sequence with primer pair ITR-F/ITR-R using the backbone as the template.Note: Amplify the left and right HA by utilizing the purified product as a template, and extend the product with a 15–20 bp adaptor for Gibson assembly with the digested backbone (Figure 1C).
-
e.Gel-purity the PCR product with a commercial kit.
-
f.Digest the P2A-NLS-tdtomato-EPG backbone (Addgene #215548). Choose the compatible restriction enzymes as described in donor plasmid and sgRNA design section (Figure 1A).Note: The restriction sites may be included in the HA sequence. Adjust the order of left and right HA ligation to avoid such circumstance.
Reagent Amount P2A-NLS-tdtomato-EPG backbone 3 μg 10 × rCutSmart Buffer 4 μL KpnI-HF 1.5 μL EcoRV-HF 1.5 μL ddH2O Up to 40 μL Digest in a thermocycle at 37°C for 1 h. -
g.Purify the digested backbone with a commercial kit.
-
h.Perform Gibson Assembly to ligate the left HA and 5′ITR sequence with the digested backbone using a commercial kit (e.g., ClonExpress Ultra One Step Cloning Kit).
-
i.The product is ready for transformation.
-
a.
-
3.Transformation and sequence validation.
-
a.Before transformation, prepare competent cells using the Mix & Go E. coli Transformation Kit following the manufacturer’s protocol (https://files.zymoresearch.com/protocols/_t3001_t3002_mix_go_e._coli_transformation_kit_buffer_set.pdf).Note: Use commercial or home-made competent cells as an alternative.Thaw one aliquot on ice. Add 2 μL of the ligated product into 20 μL of competent cells. Mix well and incubate at room temperature for 2 min.
-
b.Plate the mixture onto a 6-cm LB-agar plate containing 100 μg/mL ampicillin. Invert the plate and incubate overnight at 37°C.
-
c.The next day, inspect the plates for colony growth and pick three colonies per plate into 1 mL LB medium with 100 μg/mL ampicillin.
-
d.Shake the culture at 37°C for 4–5 h and perform Sanger sequencing on the culture using the primer upstream of the inserted fragment.Note: ITR sequence in donor plasmids can be unstable. To avoid potential rearrangement or deletion of plasmid DNA when growing bacterial cultures up for maxipreps, use SURE2 competent cells which are deficient in the related genes and grow the cultures at 30°C instead of 37°C.
-
e.Align the sequencing reads in SnapGene and choose the correctly inserted clone for strain preservation and plasmid extraction.Note: Use an endotoxin-free plasmid extraction kit for preparation of plasmids for electroporation to minimize the damage to the cells.
-
a.
-
4.Ligation of the right HA and selection cassette exchange.
-
a.After confirm the correct insertion of left HA, extract the plasmid for further cloning.
-
b.PCR amplify right HA with primer pair B2M-HAR-F1/B2M-HAR-R1 and B2M-HAR-F2/B2M-HAR-R2 to introduce mutation via PCR extension.
-
c.Digest the left HA-cloned plasmid with restriction enzyme HpaI and EcoRI.
-
d.Purify the PCR products and digested plasmid with a commercial kit.
-
e.Perform Gibson Assembly to ligate the two HAR fragments with the digested plasmid using a commercial kit.
-
f.Perform transformation and Sanger Sequencing as described above. The resulted plasmid is B2M-SDMutation-EPG (Addgene #215545).
-
g.Digested the resulted plasmid with restriction enzyme HindIII and XbaI for changing the selection cassette from Puro-EGFP to Hygro-tdTomato.
-
h.PCR amplify the Hygro-tdTomato sequence with primer pair Hygro-tdT-F/Hygro-tdT-R taking the P2A-mCherry-EhT backbone (Addgene #215549) as template.
-
i.Purify the PCR products and digested plasmid with a commercial kit.
-
j.Perform Gibson Assembly to ligate the Hygro-tdTomato sequence with the digested plasmid using a commercial kit.
-
k.Perform transformation and Sanger Sequencing as described above. The resulted plasmid is B2M-SDMutation-EhT (Addgene #215546).
-
a.
Delivery of donor and sgRNA plasmid via electroporation
Timing: 1 h
In this section, we describe steps for efficient delivery of plasmids into cultured hPSC.
-
5.
Prepare healthy and proliferative hPSCs for electroporation. Bad-grown hPSCs often survive poorly post-electroporation.
-
6.Three days after passage as 1:6, cells should reach 70%–80% confluency and are ready for experiment.
-
a.Coat the 6-well plate with Matrigel. Each well for one electroporation sample.
-
b.After coating, aspirate the coating solution. Add 2 mL mTeSR1 with 5 μM Y-27632 per well for receiving the electroporated cells.
-
c.Aspirate the medium, wash the cells once with PBS and add 1 mL hPSC Dissociation Buffer per well. Incubate at 37°C for 6–7 min.
-
d.Pipette to detach the cells and neutralize with 1 mL DMEM/F12.
-
e.Centrifuge the cells at 300 × g for 3 min at room temperature.
-
f.Aspirate the supernatant and resuspend the cell in 100 μL mTeSR1 per sample.
-
g.Pipette 5–7 times to mix 100 μL cell suspension (about 1 million cells) with 1 μg sgRNA plasmid and 2 μg donor plasmid.
-
h.Transfer all the mixture to an electroporation cuvette.
-
i.Electroporate the cells using the program “B-016” with Lonza Nucleofector 2b.
-
j.Use a long 200 μL tip to transfer the cells into one well of a 6-well plate.
-
k.Incubate the cells in a 37°C, 5% CO2 incubator.
-
a.
Note: To our test, program “B-016” retrieves the best survival rate. Other programs can be employed according to the instruction of the manufacturer (https://bioscience.lonza.com/lonza_bs/CH/en/document/29864).
Optional: Use Lonza Nucleofector 4D Nucleofector X Unit and P3 Primary Cell 4D-Nucleofector LV Kit as an alternative.
Drug-fluorescent dual selection
Timing: 2–3 weeks
In this section, we describe steps for enrichment of positive cells using the orthogonal selective reporters. The fluorescent will also facilitate further cloning picking to easily isolate the pure clones.
Note: 24 h post-electroporation, bright fluorescent should be observed under microscope, indicating successful introduction of the plasmids. Perform drug selection first to enrich correctly targeted cells. The day of electroporation is indicated as Day 0.
-
7.Perform the first-round drug selection with hygromycin:
-
a.Day 1: change the medium to mTeSR1 with 5 μM Y-27632.
-
b.Day 2: change the medium to mTeSR1.
-
c.Day 3: change the medium to mTeSR1 with 200 μg/mL hygromycin.
-
d.Day 4: change the medium to mTeSR1 with 200 μg/mL hygromycin.
-
e.Day 5: change the medium to mTeSR1.
-
a.
Note: Significant cell death will be observed at day 4, and cells with NLS-tdTomato signal will be enriched. For single-donor reporter knockin, one round drug selection with either 1 μg/mL puromycin or 200 μg/mL hygromycin is sufficient (select different antibiotics according to the resistant genes on the plasmid). Concentration of antibiotics may vary between different hPSC lines. If no cells survival after selection, lower the concentration to 0.5 μg/mL puromycin and 100 μg/mL hygromycin.
-
8.Once confluency reaches 80%, passage the cells at a 1:3 ratio for next round selection. Add selection antibiotics in only one well and the duplicates will help once no cell survives. The passage day is indicated as Day 0.
-
a.Day 1: change the medium to mTeSR1.
-
b.Day 2: change the medium to mTeSR1 with 1 μg/mL puromycin.
-
c.Day 3: change the medium to mTeSR1 with 1 μg/mL puromycin.
-
d.Day 4: change the medium to mTeSR1.
-
a.
Note: Puromycin has a much faster effect than hygromycin. Significant cell death will be observed at day 2. If the confluency at the end of day 2 is under 30%, stop selection by changing the medium without puromycin. Add 5 μM Y-27632 is also a feasible way.
-
9.
After the dual drug selection, significant enrichment of double-positive cells (EGFP+/tdTomato+) are expected (Figure 3A). Passage the cells at a 1:3 ratio to expand the cells.
Pause point: Cells can be cryopreserved after enrichment. We use mTeSR1 with 5 μM Y-27632 and 10% (v/v) DMSO as cell freezing media.
Note: Since our backbones utilize EF1α promoter, a relatively strong promoter to drive the expression of selection genes, cells will continuously appear to be fluorescent positive cells up to 7 days post-electroporation, although the cells may not undergo HDR events. Thus, sometimes one round drug selection may not be effective for enrichment. Repeat steps 7–8 to have another round selection is a good choice. As FACS sorting cannot achieve 100% efficiency, enhancing the purity of the cell mixture will improve sorting efficiency, resulting in a more homogeneous cell population post-sorting.
Optional: According to our test, cells with HDR events can resist up to 5 μg/mL puromycin and 500 μg/mL hygromycin. When the proportion of positive cells is more than 30%, this selection strategy can enrich the cells more efficiently. However, it may not be applicable to all hPSC lines since some cell lines undergo significant spontaneous differentiation under high concentration of antibiotics.
-
10.Perform FACS purification. When cells fully recover from drug selection, prepare 3 confluent wells of a 6-well plate for FACS sorting and one well for back-up.
-
a.Coat one well of a 12-well plate for plating the sorted cells.
-
b.Aspirate the medium, wash the cells with PBS once and add 1 mL TrypLE Express. Incubate at 37°C for 3–4 min.
-
c.Neutralize with 1 mL DMEM/F12 and pipette 3–5 times to dissociate the cells into single cell. Transfer the cells into a 15-mL tube.
-
d.Centrifuge the cells at 300 × g for 3 min at room temperature.
-
e.Aspirate the supernatant. Resuspend the cells in 1–2 mL PBS and filter the cell suspension through a 40-μm cell strainer.
-
f.Transfer the cell suspension into a sterile FACS tube. Place the tube on ice until FACS sorting.
-
g.Preset FACS sorter according to FACS sorter setup section.
-
h.Perform FACS sorting. Collect 0.2–0.4 million cells for one well of a 12-well plate.
-
i.After sorting, centrifuge the cells at 300 × g for 3 min at room temperature.
-
j.Aspirate the supernatant. Resuspend the cells in 1 mL mTeSR1 with 5 μM Y-27632. Plate the cells into the coated well. Put the plate into the 37°C, 5% CO2 incubator.
-
a.
Note: After the attachment of cells, observe the cells under an inverted fluorescent microscope and almost all the cells should be double-positive cells (Figure 3B).
-
11.
After cells reach confluence, passage the cells into one well of a 6-well plate.
-
12.
The cells normally reach confluency again after 2–3 days. Passage the cells at 1:3 ratio.
Pause point: Cells can be cryopreserved after FACS sorting.
-
13.Continue to passage the cells until the cells fully recover from FACS sorting. During passaging, harvest one well of cells for genome extraction and PCR genotyping.
-
a.Extract the genome from the sorted cells and the wild-type (WT) parental cells with a commercial kit (e.g., FastPure Blood/Cell/Tissue/Bacteria DNA Isolation Mini Kit).
-
b.Design PCR primers to detect on-target knock-in events.Note: To detect the generated mutation while avoiding random integration, design a primer pair using Primer-BLAST with one primer located outside the HA to ensure amplification of the on-target knockin fragment (Figure 3C, primer pair P1). For primer P1-F, make it locate on the consensus sequence of the two donor plasmids (e.g., ITR or poly-A sequence) to ensure the primer pair is able to amplify the two knockin allele. Then design another primer pair for amplification of the WT allele (Figure 3C, primer pair P2). Keep the PCR product no more than 2000 bp since the amplification efficiency of Taq polymerase is limited.
-
c.Use both genome from sorted cells and WT cells as PCR template. Set the PCR reaction as follows:
Reagent Amount Genome template 100 ng 2 × Rapid Taq Master Mix (2×) 10 μL Genotyping Primer F (10 μM) 1 μL Genotyping Primer R (10 μM) 1 μL ddH2O Up to 20 μL Use touch-down PCR program for genotyping:Steps Temperature Time Cycles Initial Denaturation 95°C 3 min 1 Denaturation 95°C 15 s 20 cycles Annealing 62°C (−0.5°C /cycle) 15 s Extension 72°C 1 s per 100 bp Denaturation 95°C 15 s 10 cycles Annealing 50°C 15 s Extension 72°C 1 s per 100 bp Final extension 72°C 5 min 1 Hold 4°C forever -
d.Separate the PCR product on a 1.5% agarose gel.
-
e.Detect the correct band using a gel imaging system.Note: Compare the PCR results in sorted cells and WT cells to determine the specificity of the genotyping primer (Figure 4A).
-
f.If the primer pair specificity is good, for mutation detection, repeat the PCR reaction utilizing the high-fidelity DNA polymerase. Gel-purify the PCR product and proceed Sanger Sequencing.
 CRITICAL: Genotyping on the sorting mixture to check if the on-target knockin events happen. If the desired band is not observed, try two or three more primer pairs for genotyping. If the band is still not observed, it possibly means that the enriched cells are mostly random-integrated cells instead of on-target knockin cells and the knockin design needs to be optimized.Note: Random integration of donor plasmids may occur, which cannot be identified using the genotyping strategy mentioned above, particularly in cells harboring both on-target knockin alleles and randomly integrated sequences. For further detection, the copy number of the integrated constructs can be determined via quantitative PCR, droplet digital PCR or DNA-FISH. Additionally, cells with random integrations can be excluded by incorporating negative selection markers into the donor plasmids. For instance, a BFP-expression cassette for FACS sorting or a ΔTK-expression cassette for drug selection. Ensure that these cassettes are positioned outside of the HAs to prevent their insertion into the genome during HDR process. However, adding extra elements will increase the size of plasmid and may pose challenges during the cloning.
CRITICAL: Genotyping on the sorting mixture to check if the on-target knockin events happen. If the desired band is not observed, try two or three more primer pairs for genotyping. If the band is still not observed, it possibly means that the enriched cells are mostly random-integrated cells instead of on-target knockin cells and the knockin design needs to be optimized.Note: Random integration of donor plasmids may occur, which cannot be identified using the genotyping strategy mentioned above, particularly in cells harboring both on-target knockin alleles and randomly integrated sequences. For further detection, the copy number of the integrated constructs can be determined via quantitative PCR, droplet digital PCR or DNA-FISH. Additionally, cells with random integrations can be excluded by incorporating negative selection markers into the donor plasmids. For instance, a BFP-expression cassette for FACS sorting or a ΔTK-expression cassette for drug selection. Ensure that these cassettes are positioned outside of the HAs to prevent their insertion into the genome during HDR process. However, adding extra elements will increase the size of plasmid and may pose challenges during the cloning.
-
a.
Figure 3.
Drug-fluorescent dual selection and PCR genotyping
(A) Representative fluorescent image of cells with B2M splice donor mutation after drug selection.
(B) Representative fluorescent image of cells with B2M splice donor mutation after FACS sorting. Almost all cells are double positive.
(C) Primer pair designing strategy for identifying on-target knock-in and homozygosity.
Figure 4.
Validation and characterization of the edited cell lines
(A) Representative gel electrophoresis image of PCR genotyping with primer pairs designed as P1 and P2 in Figure 3C.
(B) Sanger Sequencing result on the B2M locus revealing the successful homozygous mutation introduction on splice donor site.
(C) Flow cytometry analysis on β2M expression between wild-type parental cells and the knock-out cells.
(D) Representative immunocytochemistry images of the pluripotency markers expression after genome editing.
(E) Representative fluorescent image of the SOX17-NLS-tdTomato reporter line established via this protocol when differentiation towards definitive endoderm.
(F) Representative fluorescent image of the INS-GFP-CD19 reporter line established via this protocol when differentiation towards pancreatic β cell.
(G) The schematic process of constructing the SOX17-NLS-tdTomato reporter line.
Single-cell cloning and PCR screening
Timing: 2 weeks
This section describes how to obtain pure edited hPSC clones via single cell cloning and PCR screening.
-
14.Use proliferative FACS sorted cell mixture for splitting and clone picking.
-
a.When cells reach 50%–70% confluency. Dissociate the cells into single cell according to the step 9.b-d.
-
b.Count the cells using a hemocytometer or an automated cell counter.
-
c.Split about 3000 cells into a Matrigel-coated 10-cm dish culturing with 8 mL mTeSR1 supplemented with 5 μM Y-27632.Note: Cell seeding density can be adjusted according to the viability of the edited cells. Lower seeding densities can be used to generate a more separated clone population. If few cells survive after seeding, 6000 cells per 10-cm dish can be a good adjustment.
-
d.On the next day, observe whether the cells attach on the plate with an inverted microscope.
-
e.Change the medium to 8 mL mTeSR1 3 days after splitting.
-
f.6 days after splitting, clones will emerge. Change the medium to 8 mL mTeSR1.
-
g.After another two days, the size of clones is suitable to pick. Coat 8 wells of a 24-well plate with Matrigel for clone picking.
-
h.After coating, aspirate the coating solution, add 500 μL mTeSR1 with 5 μM Y-27632 per well.
-
i.Aspirate the medium in the 10-cm dish. Wash the cells with 10 mL PBS once. And add 7 mL fresh mTeSR1. Move the plate to an inverted fluorescence microscope.
-
j.Observe the fluorescence of the clones to monitor the purity of clones. Scrape the ideal clone with a 200 μL tip and transfer the clone into one well of the prepared 24-well plate. Pipette three times to break the clone into small pieces.
-
k.Place the 24-well plate back to the 37°C, 5% CO2 incubator.Note: Since the positive clones are visualized via fluorescent, 8 clones are enough to screen out the desired editing.
-
a.
-
15.
On the next day, observe whether the picked clones survive. Change 500 μL fresh mTeSR1 medium every other day until 6 days after clone picking.
-
16.Passage the clones from 24-well plate to 12-well plate.
-
a.Aspirate the medium. Wash the cells with 500 μL PBS once. Add 500 μL hPSC Dissociation Buffer per well. Incubate at 37°C for 6 min.
-
b.Aspirate the buffer and add 1 mL mTeSR1 with 5 μM Y-27632 per well. Pipette to disperse the clones into small pieces.
-
c.Aspirate the coating solution in 12-well plate.
-
d.Directly transfer the cell suspension to the 12-well plate one-to-one.
-
a.
-
17.
On the next day, observe the attachment of the passaged clones. Change medium daily with 1 mL mTeSR1 until the confluency reach 70%–80%.
-
18.
Passage the clones from 12-well plate to 6-well plate while leaving half of the cells for genome extraction and genotyping using hPSC Dissociation Buffer.
-
19.
Perform PCR genotyping and Sanger sequencing according to step 12.
Pause point: All the clones can be cryopreserved until reliable PCR genotyping results are obtained.
Selection cassette excision
Timing: 2–3 weeks
After PCR and Sanger Sequencing validation, choose one clone for selection cassette excision and cryopreserve other clones.
-
20.
Prepare one well of a 6-well plate cells for electroporation.
-
21.
Perform electroporation according to step 5. We suggest using 3 μg excision-only piggyBac transposase expression plasmid.
-
22.
After electroporation, passage cells twice when confluency reaches 70%–80%.
-
23.
Perform FACS selection according to the description in step 12. Collect fluorescent-negative cells for further clone picking.
-
24.
Perform another round single cell cloning according to steps 13–17.
Characterization of the edited cells
Timing: Variable
After cell line establishment. Routinely check can be performed. For instance: mycoplasma test and karyotype analysis which are well described elsewhere. Here, we describe the approach for pluripotency marker staining and functional test of the scarless B2M SD mutation cell line.
-
25.Immunostaining on pluripotency marker OCT4 and NANOG.
-
a.Passage the edited cells into 48-well plate for immunostaining.
-
b.Cells after 1:6 passage 2 days are suitable for staining. Aspirate the medium and wash the cells with 200 μL PBS.
-
c.Fix the cells with 100 μL 4% paraformaldehyde at room temperature for 15 min.
-
d.Aspirate the paraformaldehyde. Wash the cells with 200 μL PBS once. Then permeabilize the cells with 100 μL 0.1% Triton X-100 at room temperature for 10 min.
-
e.Aspirate the Triton X-100. Wash the cells with 200 μL PBS once. Then add 100 μL blocking buffer at room temperature for 30 min.
-
f.Dilute OCT4 and NANOG primary antibodies in 100 μL blocking buffer (1:200 for OCT4 and 1:800 for NANOG).
-
g.Aspirate the blocking buffer and add the diluted antibodies. Stain at room temperature for 30 min.
-
h.Dilute secondary antibodies in 100 μL blocking buffer (1:500).
-
i.Aspirate the primary antibodies. Wash with 200 μL PBS three times. Add the diluted secondary antibodies. Stain at room temperature for 15 min. Avoid of light.
-
j.Aspirate the secondary antibodies. Wash with 200 μL PBS three times. Counterstain the cells with 1 μg/mL DAPI at room temperature for 5 min. Avoid of light.
-
k.Wash with 200 μL PBS and immediately observe under an inverted fluorescent microscope. Cells should display bright green and red fluorescent after excitation.
-
a.
Optional: Other pluripotent markers (SOX2, SSEA-4, TRA-1-60) can be good supplements. Usually staining for OCT4 and NANOG is enough.
-
26.Flow cytometry analysis on β2M expression.
-
a.Harvest both parental cells and edited cells for analysis. Usually one well of a 6-well plate (70%–80% confluency) is enough.
-
b.Dissociate the cells into single cell according to step 10 b-d.
-
c.Resuspend the pellets in 200 μL FACS buffer. Use 100 μL for β2M-APC antibody staining and the another 100 μL for IgG staining.
-
d.Add 1 μL antibody into the 100 μL cell suspension. Mix well and incubate at 4°C for 15 min. Avoid of light.
-
e.Wash cells once with PBS and resuspend the pellets in 500 μL FACS buffer.
-
f.Use a flow cytometer for analysis.
-
a.
Note: In our experiment, we observe clearly signal divergence between parental cells and B2M SD mutation cells, which means successful knockout (Figure 4B).
Expected outcomes
We have successfully generated a B2M SD homozygous mutation hPSC line, a SOX17-NLS-tdTomato reporter hPSC line and an INS-GFP-CD19 reporter hPSC line by applying the protocol described in this study. For single cell cloning, normally all the picked clones survived in 24-well plate. After PCR genotyping, a clear knock-in band should appear in the edited group with primer pair designed as primer P1, and no significant amplification appears in WT group. When applying homozygous knock-in, a WT band should appear in the WT group and no clear band appears in edited group due to the insertion of the selection cassette (Figure 4A). Since the orthogonal selective reporters are utilized and only fluorescent-positive clones are picked, normally all the clones are positive in junction PCR. For a few cases, the HDR efficiency is extremely low and random-integrated cells are massively enriched after orthogonal selection. We do meet this kind of circumstance but it can be detected by genotyping on the sorting mixture before single cell cloning. Overall, the positive rate is nearly 100% for cloning picking. When performing Sanger sequencing to validate the introduced mutation, the altered base should be observed (Figure 4C).
We also conducted pluripotency markers staining to ensure the maintenance of pluripotency after genome editing. The positive signals of OCT4 and NANOG is expected to be observed (Figure 4D). For characterization of our B2M SD homozygous mutation hPSC line, we perform flow cytometry analysis on β2M and observed significant peak shift after knock-out (Figure 4B). For the functional validation of the SOX17-NLS-tdTomato and INS-GFP-CD19 reporter lines, we differentiate the cells towards β cell lineage as previously described with slight modification.5 And we observe massive bright red fluorescence localized in the nuclei at stage 1 day 3 (Figure 4E) and bright green fluorescence at stage 6 day 6 (Figure 4F), indicating the successful generation of definitive endoderm and endocrine lineage. We present an editing scheme of SOX17-NLS-tdTomato cell line here to illustrate the generation of a promoter-less editing allele for monitoring gene expression using our method (Figure 4G). The left and right HA sequences are cloned into the plasmid backbone P2A-NLS-tdtomato-EPG as described previously. Notably, since the fluorescent protein expressed under the endogenous SOX17 promoter differs from that under the exogenous EF1α promoter, it is possible to differentiate the editing cell mixture to check if the reporter allele works well. After excision, 4-bp “TTAA” scar will be left at the 3′UTR of the targeted gene, minimizing the effect of the editing scar on gene expression level compared with floxed selection cassette.
Limitations
The described approach is a user-friendly CRISPR-based system for biallelic targeting and typical reporter knock-in. The visualized selection can help with targeted cell enrichment. However, it is more time-consuming than ssODN-based approach or prime editing since it needs selection cassette excision and another round of clone picking. For a complete cell line establishment, it usually takes 2 months. Thus, this protocol emphasizes on the effective enrichment and less laborious works on clone picking. Thus, the choice between this protocol and other alternative ways depends on the time requirements.
Another limitation to consider is that for scarless mutation usage, a TTAA site must be found near the mutation site. However, for some instance, the TTAA site can only be found far away from the mutation site (>200 bp). We found that for this circumstance, our approach is still compatible but the mutation may not be successfully introduced due to the distance between the two sites. This means that more clones need to be screened via Sanger Sequencing. As a result, choose the nearest TTAA site can increase the overall successful rate.
In certain scenarios, mutations must be introduced into essential genes without disrupting the coding sequence. To achieve this, targeted sites can be strategically placed within introns rather than exons, thereby preserving the integrity of the coding sequence and transcribed mRNA. However, while intron mutations typically minimize disruption to the coding sequence, they could still potentially influence gene expression due to the actively transcribed cassette. Moreover, successful scarless editing depends on suitable TTAA sites, which may further limit compatibility of our protocol with constitutively expressed essential genes in hPSCs. Consequently, these considerations must be taken into account when implementing our protocol on essential genes, and selection-free methods such as ssODN-based approach or prime editing can be good alternatives in such cases.13,14
Troubleshooting
Problem 1
The Sanger Sequencing results reveals the backbone sequence without target sequence insertion when constructing donor plasmids (Steps 2 and 4).
Potential solution
Make sure that the correct band is excised and purified. Extend the digestion of the backbone from 1 h to 2 h or even longer to ensure that no circular DNA exist after purification. Separate the digested linearized DNA with gel electrophoresis is also a good way.
Problem 2
Low cell viability after electroporation (Step 6).
Potential solution
Make sure that the cultured hPSCs are healthy and proliferative. If the cells are extremely difficult to dissociated with non-enzymatic approaches, the hPSCs are possibly over-grown and stressed. Optimize the passage ratio can avoid over-confluency.
Problem 3
Cells are contaminated after FACS sorting (Step 10).
Potential solution
For FACS sorters that are not maintained in biosafety cabinet, cell sorting may cause contamination. Remember to clean the fluidics line and cryopreserve some vials before FACS sorting.
Problem 4
No knock-in events detected after PCR genotyping (Step 13).
Potential solution
The knock-in events rely on the efficient cutting of Cas9-sgRNA complex and the structures of donor template, as well as the targeted site. Try the following possible solutions.
-
•
Test other sgRNA candidates to identify an efficient sgRNA. The activity of sgRNAs can be evaluated by amplifying the target locus and performing TIDE analysis on Sanger traces, which will facilitate the rational design of sgRNA.15
-
•
Try other CRISPR editing systems like CRISPR-Cas12a.16
-
•
For reporter knock-in, if possible, make a 5′ knock-in instead of 3’.
-
•
Amplify another set of HAs to generate new donor templates.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kai Wang, kai.wang88@pku.edu.cn.
Technical contact
Technical questions about this protocol should be directed to the technical contact, Yun Zhao, zy157@pku.edu.cn.
Materials availability
All the plasmids generated in this study have been deposited to Addgene and are available upon request.
Data and code availability
This study did not generate any unique datasets or code.
Acknowledgments
This work was supported by the National Key R&D Program of China (2022YFA1104800); the Beijing Natural Science Foundation (JQ23029, L234024, and L234021); the Beijing Nova Program (20220484100 and 20230484448); the National Natural Science Foundation of China (82370514); the Beijing Municipal Science & Technology Commission (Z231100007223001); the open research fund of the State Key Laboratory of Cardiovascular Disease, Fuwai Hospital (2022KF-04); the Clinical Medicine Plus X-Young Scholars Project, Peking University (PKU2022LCXQ003); the Emerging Engineering Interdisciplinary-Young Scholars Project, Peking University, the Fundamental Research Funds for the Central Universities (PKU2023XGK011); the Scientific and Technological Innovation project of the China Academy of Chinese Medical Science (C12023C056YLL); the open research fund of the State Key Laboratory of Digital Medical Engineering, Southeast University (2023K-01); the open research fund of the Beijing Key Laboratory of Metabolic Disorder Related Cardiovascular Disease, PR China (DXWL2023-01); and the open research fund from the State Key Laboratory of Female Fertility Promotion, Center for Reproductive Medicine, Peking University Third Hospital (BYSYSZKF2023023).
Author contributions
Methodology, Y.Z. and Z.P.; experiments, Y.Z., Z.P., Z.H., M.S., and Y.H.; writing, Y.Z., X.P., X.L., X.W., and K.W.; funding acquisition, K.W.; supervision, X.W. and K.W.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Yun Zhao, Email: zy157@pku.edu.cn.
Xi Wang, Email: xi_wang@bjmu.edu.cn.
Kai Wang, Email: kai.wang88@pku.edu.cn.
References
- 1.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H., Wang B., Ono M., Kagita A., Fujii K., Sasakawa N., Ueda T., Gee P., Nishikawa M., Nomura M., et al. Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell. 2019;24:566–578.e7. doi: 10.1016/j.stem.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Haideri T., Howells A., Jiang Y., Yang J., Bao X., Lian X.L. Robust genome editing via modRNA-based Cas9 or base editor in human pluripotent stem cells. Cell Rep. Methods. 2022;2 doi: 10.1016/j.crmeth.2022.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of Functional Human Pancreatic β Cells In Vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O’Dwyer S., Quiskamp N., Mojibian M., Albrecht T., et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 6.Zhong A., Li M., Zhou T. Protocol for the Generation of Human Pluripotent Reporter Cell Lines Using CRISPR/Cas9. STAR Protoc. 2020;1 doi: 10.1016/j.xpro.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauer B., Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B., et al. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. 2013;110:E2279–E2287. doi: 10.1073/pnas.1305987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 10.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 11.Koch B., Nijmeijer B., Kueblbeck M., Cai Y., Walther N., Ellenberg J. Generation and validation of homozygous fluorescent knock-in cells using CRISPR–Cas9 genome editing. Nat. Protoc. 2018;13:1465–1487. doi: 10.1038/nprot.2018.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beers J., Gulbranson D.R., George N., Siniscalchi L.I., Jones J., Thomson J.A., Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 14.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma H., Jaenisch R. Multiplex genome editing of human pluripotent stem cells using Cpf1. bioRxiv. 2022 doi: 10.1101/2022.04.13.488123. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.